The first adult-repopulating hematopoietic stem cells (HSCs) emerge in the mouse aorta-gonad-mesonephros (AGM) region at embryonic day 10.5 prior to their appearance in the yolk sac and fetal liver. Although several genes are implicated in the regulation of HSCs, there are gaps in our understanding of the processes taking place in the AGM at the time of HSC emergence. To identify genes involved in AGM HSC emergence, we performed differential display reverse transcriptase–polymerase chain reaction (DD RT-PCR). Differentially expressed genes included β-catenin and homologs of human TM9SF2 and TAB2. We characterized the expression pattern of Wnt/β-catenin signaling,mTM9SF2, and mTAB2 in the embryo and adult. Interestingly, the expression of mouse TAB2 (mTAB2) in the E11 dorsal aorta endothelium suggests a role for mTAB2 in HSC emergence and/or regulation. The identification of differentially expressed genes in the AGM region should yield further insights into the development of this tissue and into the emergence and regulation of HSCs.
Introduction
Hematopoietic stem cells (HSCs) form the basis of the adult hematopoietic hierarchy.1 Adult-repopulating HSCs first appear at embryonic day 10.5 (E10.5) within the mouse embryo proper in the aorta-gonad-mesonephros (AGM) region.2,3More detailed studies show that HSCs are localized to the dorsal aorta.4-6 Because de novo production of HSCs is thought to take place during a limited time within the embryo, the AGM region is of interest to study the genes involved in the generation and regulation of the earliest HSCs.
To date, research on HSC development has focused on known transcription factors and signaling molecules. Transcription factors AML-1 and GATA-2 were shown to be important regulators of HSCs in the embryo.7-9 Also, members of the Hedgehog and Wnt (signaling) families have been implicated in playing instructive and/or regulatory roles on HSCs.10-12 To further increase our understanding of AGM HSC development, we set out to identify differentially expressed genes by using differential display reverse transcriptase–polymerase chain reaction (DD RT-PCR). We found several differentially expressed genes in the AGM region at the time of HSC induction, including β-catenin and novel mouse homologs of human TM9SF2 and TAB2. Characterization of mTM9SF2 and mTAB2 suggests a role for these genes in hematopoiesis. Interestingly, expression of mTAB2 in the E11 aortic endothelium suggests a role for mouse TAB2 (mTAB2) in HSC emergence and/or regulation.
Study design
Embryo generation and cell lines
DD RT-PCR and RNA expression analysis
RNA was isolated with Ultraspec (Biotecx, Houston, TX) or Trizol (Invitrogen/Life Technologies, Carlsbad, CA). For cDNA synthesis, 1 to 5 μg DNAse-treated RNA was reverse transcribed with Superscript II RT (Invitrogen/Life Technologies).
DD RT-PCR was essentially performed as described15 with 8 arbitrary primers. RT-PCRs were performed with Amplitaq (Roche, Branchburg, NJ) and 40 ng RNA equivalent of cDNA at optimized conditions for each primer set. Primer sequences are available on request.
For Northern blots 10 to 25 μg RNA, and for cDNA dot blots the equivalent of 1 μg RNA, was blotted to Genescreen membrane (NEN Technologies, Boston, MA) and probed with32P-labeled nick-translated DNA fragments (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom).
Western blot and immunohistochemistry
For Western blotting and immunohistochemistry (6 to 8 μM cryosections) anti-TAB2 (sc-11851; Santa Cruz Biotechnology, CA) antibody was used according to the manufacturer's guidelines. Immunohistochemistry staining was visualized with ABC staining kit (Santa Cruz Biotechnology).
Results and discussion
Identification of differentially expressed genes in the AGM region
Three independent DD RT-PCR screenings were performed on early E10 (30 to 35 somite pairs) and E11 (40 to 50 somite pairs) AGM material to identify genes expressed prior to and after the induction of HSCs. For the first screening, cDNA populations from E10 and E11 AGM were compared. In the second and third screenings, E10 and E11 aorta and complete E10 AGM and subdissected E11 AGM cDNA populations were compared, respectively. In total, 39 differential bands were detected, and 62 cloned sequences were compared against National Center for Biotechnology Information (NCBI) nucleotide databases.
RT-PCR analyses (Figure 1) confirm that clones corresponding to fetal globin ε, enolase, TRAP150(not shown), β-catenin, mTM9SF2, andmTAB2 are differentially expressed between E10 and E11 in the AGM region. As expected, some clones are not differentially expressed, such as Dvl2, BUB1B, and Cu/Zn SOD.16 Hence, in 3 DD RT-PCR screenings, we identified a small number of differentially expressed genes. Most importantly, we were able to identify 2 novel genes not previously implicated in development or hematopoiesis. A complete overview of the DD RT-PCR clones is available on the Blood website; see the Supplemental Table link at the top of the online article.
Expression patterns of DD RT-PCR clones and Wnt signaling members.
(A) RT-PCR analysis was performed to verify the differential expression of several clones derived from the DD RT-PCR screening. RNA isolated from AGM and fetal liver at E10, E11, and E12 was used for cDNA synthesis and RT-PCR under standard conditions. For each primer set, optimal PCR conditions and annealing temperature were determined and PCR was performed for 30 to 40 cycles. Three of the genes,mTAB2, mTM9SF2, and β-catenin (Figure 1B) are up-regulated between E10 and E11 in the AGM. The other genes(BUB1B and Cu/Zn SOD), not showing a differential expression pattern, represent false-positive DD RT-PCR clones. (B) RT-PCR analysis was performed for several components of the Wnt signaling pathway. Several components of the Wnt signaling pathway can be detected in the AGM and fetal liver between E10 and E12. Besides the differential expression of β-catenin between E10 and E11 in the AGM, we could also detect up-regulation of Dvl3and Wnt5 in the AGM between E10 and E11. + and − indicate with and without reverse transcriptase, respectively; H, H2O.
Expression patterns of DD RT-PCR clones and Wnt signaling members.
(A) RT-PCR analysis was performed to verify the differential expression of several clones derived from the DD RT-PCR screening. RNA isolated from AGM and fetal liver at E10, E11, and E12 was used for cDNA synthesis and RT-PCR under standard conditions. For each primer set, optimal PCR conditions and annealing temperature were determined and PCR was performed for 30 to 40 cycles. Three of the genes,mTAB2, mTM9SF2, and β-catenin (Figure 1B) are up-regulated between E10 and E11 in the AGM. The other genes(BUB1B and Cu/Zn SOD), not showing a differential expression pattern, represent false-positive DD RT-PCR clones. (B) RT-PCR analysis was performed for several components of the Wnt signaling pathway. Several components of the Wnt signaling pathway can be detected in the AGM and fetal liver between E10 and E12. Besides the differential expression of β-catenin between E10 and E11 in the AGM, we could also detect up-regulation of Dvl3and Wnt5 in the AGM between E10 and E11. + and − indicate with and without reverse transcriptase, respectively; H, H2O.
Expression of Wnt signaling components in the AGM region
Wnt family members are known regulators of developmental processes and hematopoiesis.10,12,17 Several Wnts and Wnt receptors are expressed in the yolk sac and fetal liver, both known hematopoietic sites during development. Identification of β-cateninand Dvl2 in our DD RT-PCR prompted us to examine whether other Wnt signaling members are expressed in the AGM and fetal liver (Figure 1B). In addition to β-catenin, Dvl3, Wnt5, andDvl1 (not shown) are up-regulated between E10 and E11 in the AGM. Additionally, TCF1 and LEF1 expression could be detected in the AGM and fetal liver. The expression ofTCF1 and LEF1 corresponds to previous findings of Oosterwegel et al.18 The simultaneous expression of several Wnt signaling components and the up-regulation of the key component β-catenin in the E11 AGM and liver suggests that the Wnt signaling pathway is functional. Further studies are required to address the role of Wnt signaling in the AGM, in particular in HSCs.
Characterization of the conserved transmembrane encoding themTM9SF2 gene
Protein sequence alignment of TM9SF2 homologs from several species, including mouse, human, Drosophila, andCaenorhabditis elegans, reveals high-level homology, indicative of a conserved function for this protein (data not shown). Human TM9SF2 is localized in endosomes and is proposed to function as channel or small molecule transporter.19 Alignment of themTM9SF2 mRNA sequence with genomic sequences reveals 17 exons (Figure 2A). Northern and cDNA dot blot analyses show mTM9SF2 expression in several adult tissues and hematopoietic cells lines. The highest levels of expression are observed in kidney and liver tissue (Figure 2B) and myeloid and B-lymphoid cell lines (Figure 2C). The expression pattern ofhTM9SF219 is partially consistent with our data in the mouse. Thus, the expression of mTM9SF2 in the AGM region and in hematopoietic cell lines makes this conserved gene an interesting candidate for future studies concerning its function and role in development and regulation of the hematopoietic system.
Structure and expression pattern of the mouse
TM9SF2 and mTAB2 genes.(A) Schematic representation of the mouse TM9SF2 gene locus. The 2746-bp mRNA sequence of mTM9SF2 (BC003862) was aligned against the Celera mouse genome database to retrieve information about the genomic structure. The gene contains at least 17 exons spanning approximately 53 kb and is located at mouse chromosome 14. The human and mouse TM9SF2 genes have a similar structure, and the hTM9SF2 is located on chromosome 13q11-34. (B) Northern blot with RNA derived from several adult mouse tissues probed with mTM9SF2 probe. Specific expression in all tissues is observed, with the highest expression in liver and kidney. The probe hybridizes to an RNA of approximately 3 to 3.5 kb. (C) Complementary DNA dot blot analysis shows mTM9SF2 andmTAB2 expression in several hematopoietic cell lines. FormTM9SF2, the highest expression levels were observed in myeloid (32D.cl3, DA3) and B-lymphoid (DA8) and lowest expression in T-lymphoid (DA2) and erythroid (MEL) cell lines. For mTAB2the highest levels of expression were observed in the myeloid cell lines. (D) Schematic representation of the mTAB2 gene locus. The 3480 bp mRNA sequence (BC004813) was aligned against the Celera mouse genome database to retrieve information about the genomic structure. The gene contains at least 6 exons spanning approximately 20 kb and is located at mouse chromosome 10. The gene structure ofmTAB2 and hTAB2 is similar, and hTAB2is located on human chromosome 6q25. (E) Northern blot containing RNA from E10 and E11 AGM and E11 liver tissues probed with anmTAB2 probe. The probe hybridizes to an RNA of approximately 3.5 to 4 kb. Differential expression of mTAB2 in the AGM between E10 and E11 is observed. (F) Northern blot containing RNA from adult mouse tissues reveals high levels of mTAB2 expression in the kidney, thymus, spleen, and BM.
Structure and expression pattern of the mouse
TM9SF2 and mTAB2 genes.(A) Schematic representation of the mouse TM9SF2 gene locus. The 2746-bp mRNA sequence of mTM9SF2 (BC003862) was aligned against the Celera mouse genome database to retrieve information about the genomic structure. The gene contains at least 17 exons spanning approximately 53 kb and is located at mouse chromosome 14. The human and mouse TM9SF2 genes have a similar structure, and the hTM9SF2 is located on chromosome 13q11-34. (B) Northern blot with RNA derived from several adult mouse tissues probed with mTM9SF2 probe. Specific expression in all tissues is observed, with the highest expression in liver and kidney. The probe hybridizes to an RNA of approximately 3 to 3.5 kb. (C) Complementary DNA dot blot analysis shows mTM9SF2 andmTAB2 expression in several hematopoietic cell lines. FormTM9SF2, the highest expression levels were observed in myeloid (32D.cl3, DA3) and B-lymphoid (DA8) and lowest expression in T-lymphoid (DA2) and erythroid (MEL) cell lines. For mTAB2the highest levels of expression were observed in the myeloid cell lines. (D) Schematic representation of the mTAB2 gene locus. The 3480 bp mRNA sequence (BC004813) was aligned against the Celera mouse genome database to retrieve information about the genomic structure. The gene contains at least 6 exons spanning approximately 20 kb and is located at mouse chromosome 10. The gene structure ofmTAB2 and hTAB2 is similar, and hTAB2is located on human chromosome 6q25. (E) Northern blot containing RNA from E10 and E11 AGM and E11 liver tissues probed with anmTAB2 probe. The probe hybridizes to an RNA of approximately 3.5 to 4 kb. Differential expression of mTAB2 in the AGM between E10 and E11 is observed. (F) Northern blot containing RNA from adult mouse tissues reveals high levels of mTAB2 expression in the kidney, thymus, spleen, and BM.
Characterization of mTAB2: a component of the IL-1 signaling pathway
Human TAB2 is a binding partner of TAK1 and is involved in nuclear factor–κB (NFκB) and Jun N-terminal kinase (JNK) activation, in the interleukin-1 (IL-1) and receptor activator of NFκB (RANK) signaling pathways.20,21 Both IL-1 and RANK function within the hematopoietic system, and NFκB is an important hematopoietic transcription factor.22-24 Interestingly, studies in Xenopus reveal that TAK1 negatively regulates the Wnt pathway,25 making mTAB2 an interesting candidate for further study.
Alignment of the mTAB2 mRNA sequence with genomic sequences reveals 6 exons in the mTAB2 gene (Figure 2D). Northern blotting verifies the differential expression of mTAB2 in the AGM (Figure 2E) and demonstrates mTAB2 expression in several adult mouse tissues, including spleen, thymus, and bone marrow (BM) (Figure 2F). Additionally, mTAB2 is expressed at high levels in myeloid cell lines and at lower levels in lymphoid and erythroid cell lines (Figure 2C). Thus, mTAB2is expressed in several hematopoietic tissues and cell lines.
mTAB2 is expressed in the dorsal aorta endothelium at E11 and E12
To investigate the expression pattern of mTAB2 in the AGM, we performed immunohistochemistry. First, a commercial antibody against hTAB2 was tested for its ability to recognize mTAB2 by Western blotting. In human and mouse cell lines, a single TAB2 protein band was detected at the predicted size of 77 kDa (Figure3A). Immunohistochemistry on cryosections of E10-E12 embryos reveals an increasing level of mTAB2 protein in the endothelium of the E11 (not shown) and E12 dorsal aorta and some underlying mesenchymal cells (Figure 3B). No mTAB2 protein expression was found in the E10 dorsal aorta, consistent with undetectable mTAB2 mRNA in the E10 AGM (not shown).
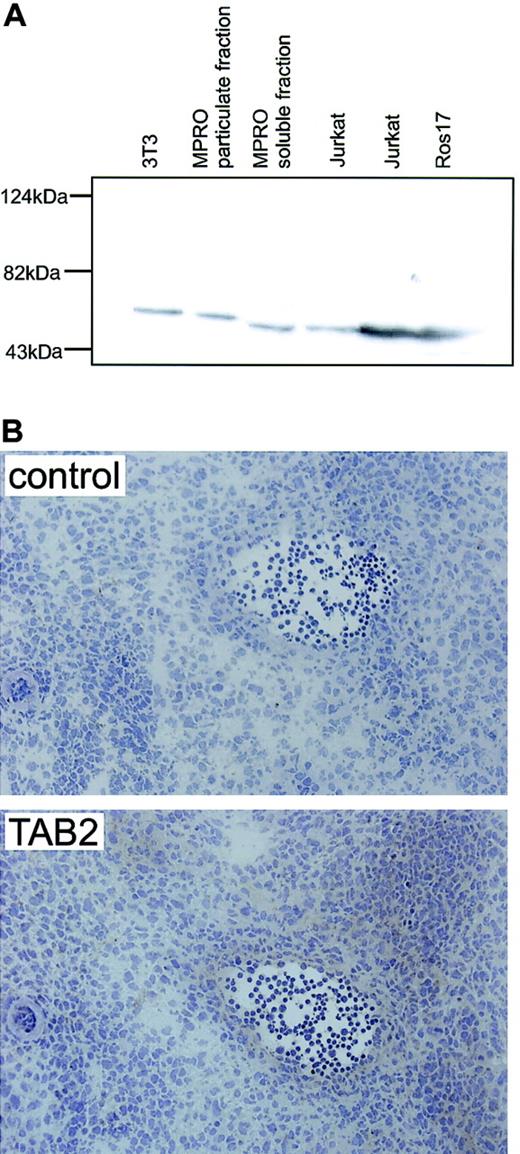
AGM immunostaining for mTAB2.
(A) Western blot analysis on mouse fibroblast (NIH-3T3) and myeloid (MPRO) cell lines and human B-lymphoid (ROS17) and T-lymphoid (Jurkat) cell lines shows that the antibody raised against human TAB2 recognizes a single protein band with the expected molecular mass of 77 kDa in both the human and mouse protein lysates. (B) Immunohistochemistry with the anti-TAB2 antibody was performed on 6 to 8 μM transverse cryosections of E12 embryos at the level of the AGM. Expression of mTAB2 was detected in the endothelium of the E11 (not shown) and E12 dorsal aorta and in some cells surrounding the dorsal aorta. Original magnification, × 200. TAB2 staining was visualized with peroxidase substrate DAB chromogen and sections were counterstained with hematoxylin.
AGM immunostaining for mTAB2.
(A) Western blot analysis on mouse fibroblast (NIH-3T3) and myeloid (MPRO) cell lines and human B-lymphoid (ROS17) and T-lymphoid (Jurkat) cell lines shows that the antibody raised against human TAB2 recognizes a single protein band with the expected molecular mass of 77 kDa in both the human and mouse protein lysates. (B) Immunohistochemistry with the anti-TAB2 antibody was performed on 6 to 8 μM transverse cryosections of E12 embryos at the level of the AGM. Expression of mTAB2 was detected in the endothelium of the E11 (not shown) and E12 dorsal aorta and in some cells surrounding the dorsal aorta. Original magnification, × 200. TAB2 staining was visualized with peroxidase substrate DAB chromogen and sections were counterstained with hematoxylin.
Taken together, the up-regulation of mTAB2 in the AGM coincides with the emergence of HSCs in this region, particularly in the dorsal aorta.4-6 Based on this spatial and temporal expression pattern of mTAB2, we suggest that mTAB2 is either directly or indirectly involved in the induction or regulation of HSCs in the AGM. Future studies should reveal whether mTAB2 is involved in IL-1 or RANK signaling and whether cross-talk between TAK1-TAB2 and Wnt signaling takes place in the AGM region.
Note added in proof. Recently, another group independently identified mouse TAB2 in neuronal cells.26
The authors thank Drs Marella de Bruijn and Marian Peeters for help with embryo dissections, Karin van der Horn for embryo sectioning, and Sjozef van Baal and Ton Verkerk for help with the Celera database/bioinformatics. Drs R. Delwel, S. Tsai, and T. Langerak generously provided hematopoietic cell lines, and Drs M. de Bruijn, M. Peeters, and D. Meijer provided critical comments on the manuscript.
Prepublished online as Blood First Edition Paper, November 14, 2002; DOI 10.1182/blood-2002-07-2260.
Supported by the Human Frontiers Scientific Program (RG00345/1999M) and the National Institutes of Health (DK54077).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Elaine Dzierzak, Erasmus University Medical Center, Department of Cell Biology and Genetics, PO Box 1738, 3000 DR Rotterdam, the Netherlands; e-mail:dzierzak@ch1.fgg.eur.nl.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal