A number of studies have implicated a role for the cell surface glycoprotein CD44 in several biologic events, such as lymphopoiesis, homing, lymphocyte activation, and apoptosis. We have earlier reported that signaling via CD44 on naive B cells in addition to B-cell receptor (BCR) and CD40 engagement generated a germinal center–like phenotype. To further characterize the global role of CD44 in B differentiation, we examined the expression profile of human B cells cultured in vitro in the presence or absence of CD44 ligation, together with anti-immunoglobulin (anti-Ig) and anti-CD40 antibodies. The data sets derived from DNA microarrays were analyzed using a novel statistical analysis scheme created to retrieve the most likely expression pattern of CD44 ligation. Our results show that genes such as interleukin-6 (IL-6), IL-1α, and β2-adrenergic receptor (β2-AR) were specifically up-regulated by CD44 ligation, suggesting a novel role for CD44 in immunoregulation and inflammation.
Introduction
B-cell differentiation is highly regulated by components in the surrounding microenvironment1 and is a central process in the humoral immune response. This often involves germinal center reactions eventually leading to population of the memory compartment as well as to generation of plasma cells.2 Important players in this process are, for example, adhesion molecules from the families of integrins, selectins, and immunoglobulins, as well as chemoattractants, such as the chemokines.3
The cell surface glycoprotein CD44 is a member of the hyaladherin or link protein superfamily (LPSF) that interacts with the polysaccharide hyaluronan (HA) in the extracellular matrix (ECM). It is widely distributed in the body and mediates cell-cell and cell-matrix interactions. In the hematopoietic system CD44 is expressed on all cell types and has been shown to play a role in lymphopoiesis and lymphocyte homing4 as well as in lymphocyte activation5,6 and apoptosis.6,7 CD44 has furthermore been associated with several different pathologic states where the linkage to cancer and autoimmune diseases is the most notable.4 8
All mechanisms supporting B-cell differentiation from a mature naive B cell to an immunoglobulin (Ig)–producing plasma cell are still not entirely understood, although many components have been identified.2 We and others have previously investigated the role of CD44 in the regulation of T-cell–dependent B-cell activation9 and subsequent germinal center formation,10 where ligation of CD44 was shown to contribute to the induction of a phenotype closely resembling a germinal center (GC) B cell.
B cells up-regulate CD44 upon activation,9,11,12 and HA-CD44 interactions induce activation of mature B cells in vivo.5 However, CD44 is down-regulated during the germinal center reaction.13-15 Interestingly, ectopic GC-like structures are found in several autoimmune and inflammatory diseases,16,17 indicating that a microenvironment promoting formation of GC-like follicular structures is generated in these pathologic states. Apart from T cells and follicular dendritic cells at these GC-like structures,18,19 an increase in expression levels of CD44 was evident.20 21
To further elucidate the functional effects of CD44 ligation on B cells, we assessed the transcriptional profiles from anti-CD44-stimulated naive B cells also costimulated via CD40 and the B-cell receptor (BCR). The transcriptional profile of approximately 6800 genes was evaluated by using a high-density DNA microarray technique. To facilitate the analysis of the complex patterns in the data set, we developed a novel statistical analysis scheme that accounts for inherent errors in the experimental handling and process often seen in these types of studies.22 23 In summary, the CD44-dependent regulated genes in our analysis were found to mainly pertain to proteins involved in inflammation and immunomodulation. Interestingly, the expression patterns regulated by CD44 fall into 2 groups: (1) genes augmented or repressed by CD44 in a temporal fashion and (2) genes directly regulated—that is, induced—by CD44 alone. Our results suggest a role for CD44 in the modulation of the mature B-cell response and may also have implications in inflammatory and autoimmune diseases, because genes such as the β2-adrenergic receptor for norepinephrin, platelet-derived endothelial cell growth factor, as well as interleukin-6 (IL-6) and IL-1α were induced by CD44.
Materials and methods
Antibodies
R-phycoerythrin (RPE)–conjugated anti-CD38 antibodies were obtained from Biosciences (BD; San Jose, CA). Anti-IgD-fluorescein isothiocyanate (FITC) and anti-CD3–phycoerythrin (PECy5) was obtained from Dako (Glostrup, Denmark), and anti-CD38-PECy5 antibody was purchased from PharMingen (San Diego, CA). Mouse antihuman IgM (AF6) and mouse antihuman CD44 (BU52) antibodies were kindly provided by I. MacLennan (University of Birmingham, United Kingdom). Mouse antihuman CD40 (S2C6) was a gift from S. Pauli (Stockholm University, Sweden).
Cells
Human tonsils were obtained from 4 different pediatric patients undergoing routine tonsillectomy at the University Hospitals of Lund or Malmö. Briefly, tonsils were minced and T cells were removed by rosetting with neuraminidase-treated sheep red blood cells. Mononuclear T-cell–depleted cells were isolated by density centrifugation using Ficoll-Isopaque (Amersham Pharmacia Biotech, Uppsala, Sweden). The interphase fraction, containing predominantly B cells, was washed in phosphate-buffered saline (PBS) containing fetal bovine serum (FBS) (10%) and incubated with precoated anti-CD38 Dynabeads, sheep antimouse IgG, or pan-mouse IgG (Dynal Biotech, Oslo, Norway) for 45 minutes on ice. Cells depleted for CD38 were stained with anti-CD38-PECy5, anti-CD3-PECy5, and anti-IgD-FITC antibodies. Positive selection using flow cytometric cell sorting of IgD+/CD38− B cells to a purity exceeding 98% was performed on a FACS Vantage SE cell sorter (BD).
Cell culture condition
IgD+/CD38− B cells (2.0 × 106) from the 4 different human donors were separately cultured at 37°C, 5% CO2, for a total of 6 hours, 24 hours, and 72 hours on 1.5 × 105CD32-transfected fibroblasts (162CG7) with 0.2 μg/mL anti-IgM (AF6) and 1.0 μg/mL anti-CD40 (S2C6) antibodies, with or without 1.0 μg/mL anti-CD44 (BU52) antibodies. The culture was performed in flat-bottomed 24-well plates (Costar, Corning, NY) using complete medium: RPMI 1640 supplemented with 2 mM l-glutamine, 1% nonessential amino acids, 50 μg/mL gentamicin (Gibco Life Technologies, Gaithersburg, MD), and 10% FBS (Hyclone Laboratories, Logan, UT).
Isolation of mRNA
Freshly isolated cells from each separate culture were lysed in Trizol (Life Technologies). The RNA was extracted from the cell lysate by adding 0.2 vol chloroform. The aqueous phase containing the RNA was separated and subsequently precipitated with isopropanol and washed in 75% ethanol. The RNA pellet was dissolved in diethylene pyrocarbonate (DEPC)-treated water and further purified with the RNeasy Mini Kit (QIAGEN, Hilden, Germany). The total RNA content was assessed spectrophotometrically (GeneQuant II, Amersham, Pharmacia Biotech) within a 260/280 nm OD (optic density) ratio of 1.9:2.1. After a second precipitation step in 2.5 vol ethanol and subsequent wash, the RNA was resuspended in DEPC-treated water. Five micrograms of total RNA was used for the cDNA and cRNA synthesis, as previously described24; cRNA is quality controlled by gel electrophoresis.
Hybridization and scanning of the DNA chips
A hybridization cocktail was prepared with the biotinylated and fragmented cRNA at 50 μg/mL, as described previously,24and hybridized onto HuGeneFL microarrays (Affymetrix, Santa Clara, CA). The probe array was then stained with a solution of 2 mg/mL acetylated bovine serum albumin (BSA) and 10 μg/mL streptavidin R-phycoerythrin (Molecular Probes, Eugene, OR). A secondary stain was performed with acetylated BSA, normal goat IgG (Sigma Chemical, St Louis, MO), and biotinylated goat antistreptavidin antibody (Vector Laboratories, Burlingame, CA) for amplification. A final staining step with streptavidin R-phycoerythrin was performed before the probe arrays were scanned in the gene array scanner and checked using the Micro Array Suite 4.0 (Affymetrix), as described previously.24 Several controls assessing the overall processing, the hybridization, and the quality of the material are included on the microarray. A total of 6 arrays were run for every donor, one for every time point for both the anti-CD44–stimulated and nonstimulated cells. In total, 24 arrays were evaluated.
Statistical analysis
A brief review of the mathematical details of the statistical data analysis is given here, and a more detailed description is available elsewhere (S.B., T.B., and M. Sigvardsson, unpublished data, 2002). The data consist of 6 different biologic varieties: anti-CD44-treated cells at 6, 24, and 72 hours after the onset of treatment and untreated cells sampled at the same time points. Each variety consists of 4 samples—that is, from the 4 different human donors. For each variety, data are discretized using the Affymetrix “present/absent” calls so that each gene is represented by a vector S ∈ {0,1}4. The distribution of observed states S in the variety is assumed to be generated by 3 underlying biologic states: ς0 for expression below the detection level of the chips, ς1 for expression above detection level, and ςT for genes with varying expression levels in the 4 samples. The latter state is assumed to give rise to random vectors S with equal probabilities of 0 or 1 at each position. For each measurement, a certain noise characteristic is assumed. This noise characteristic is modeled by misclassification probabilities {PP}.
To simplify the notation, we introduce the S dependent variables:
The distribution of observed states S is then modeled by
This distribution is fitted to the observed data by χ2 minimization of the unweighted errors. With the parameter estimates generated from the fit, we may now calculate the belief in terms of probability for an underlying state given the observed one. P(ς¦S) = [P(S¦ς)P(ς)]/P(S) whereς ∈ {ς0,ςT,ς1}. For the 36 = 729 possible expression profiles over the whole set of varieties υ ∈ {6h+, 24h+, 72h+, 6h−, 24h−, 72h−}, we calculate the probability of each one of them by Π P(ςυ ¦ Sυ).
For simplicity, the underlying states ς0,ςT, and ς1 are throughout the paper referred to by their index symbols 0, T, and 1. Further, the expression profile of a gene over the 6 varieties will be denoted, for example, 0T1000, where the position of 0, T, or 1 in this pattern refers to the expressional state in the variety 6h+, 24h+, 72h+, 6h−, 24h−, 72h−, respectively.
Flow cytometry and ELISA
Freshly isolated cells from each separate culture were assayed for IL-1α surface expression by flow cytometry using a FACScan (BD) and for IL-6 expression in the supernatant using enzyme-linked immunosorbent assay (ELISA). Anti-IL-1α-PE was purchased from PharMingen, while the human IL-6 ELISA kit was purchased from R&D Systems (Minneapolis, MN).
Results
To study the transcriptional changes associated with a CD44 stimuli on naive B cells, the entire cell population from ectomized pediatric tonsils was fractionated and a naive B-cell subset, defined by immunoglobulin (Ig) D cell surface expression and a lack of CD38, was collected. These cells were propagated in the presence of anti-CD40 and anti-IgM antibodies and immobilized on CD32-transfected fibroblasts.25 The phenotypic and functional changes then attributed to a CD44 stimulation were evaluated at 6, 24, and 72 hours over 4 separate samples using high-density DNA microarrays displaying probes for approximately 6800 genes and several hundred expressed sequence tags (ESTs).
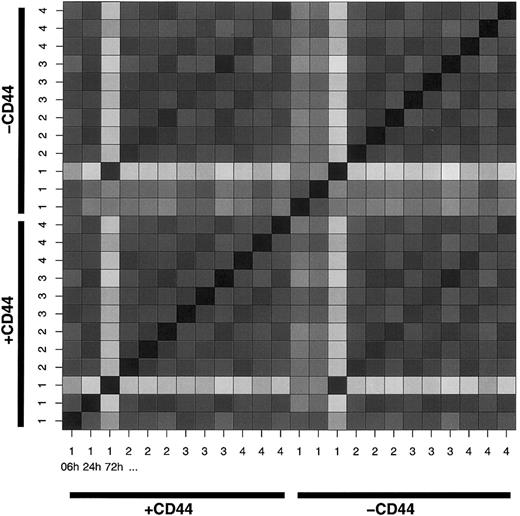
To investigate the sample coherence, we constructed a Hamming distance matrix of all samples (Figure 1). This validation clearly shows how samples labeled 1 in both treated and untreated cells at 72 hours deviate from the other samples. The deviation is also noted as raised misclassification probabilities for these samples (data not shown). Because the algorithm is designed to handle deviations even of this magnitude, the samples were retained.
Hamming distance matrix visualizing the number of genes differing between any 2 samples.
The color scale ranges from black, indicating no difference, to white, indicating a 2828-gene difference. The ordering is as follows (from the lower left corner up and right): CD44-stimulated (indicated as +CD44) sample no. 1 at 6, 24, and 72 hours; CD44-stimulated sample no. 2, at 6, 24, and 72 hours; sample no. 3 at 6, 24, and 72 hours; and sample no. 4 at 6, 24, and 72 hours. In a corresponding series is the control receiving no CD44 stimuli (indicated -CD44).
Hamming distance matrix visualizing the number of genes differing between any 2 samples.
The color scale ranges from black, indicating no difference, to white, indicating a 2828-gene difference. The ordering is as follows (from the lower left corner up and right): CD44-stimulated (indicated as +CD44) sample no. 1 at 6, 24, and 72 hours; CD44-stimulated sample no. 2, at 6, 24, and 72 hours; sample no. 3 at 6, 24, and 72 hours; and sample no. 4 at 6, 24, and 72 hours. In a corresponding series is the control receiving no CD44 stimuli (indicated -CD44).
To assess the difference in gene expression induced by CD44 ligation, the genes were clustered according to their most probable expression profile over the 6 varieties. These expression profiles can be viewed as 6-letter strings composed by 0, T, or 1 in the following order: 6h+, 24h+, 72h+, 6h−, 24h−, 72h−. For example, a gene with most probable expression profile 01T000 is below the detection level of the chip at 6 hours in the treated group, above the detection level at 24 hours in the treated group, and transient (varying) in the treated group at 72 hours. However, the expression level is below the detection level at all times in the untreated group.
To select genes directly regulated by CD44 (ie, induced or repressed by CD44) at all times, we extracted those having an expression pattern matching (1/T, 1/T, 1/T, 0, 0, 0) and (0, 0, 0, 1/T, 1/T, 1/T). To select temporally regulated genes (ie, genes having an earlier or delayed induction by CD44), we extracted those having an expression pattern matching (1/T, 1/T/0, 1/T/0, 0, 1/T/0, 1/T/0) and (0, 1/T/0, 1/T/0, 1/T, 1/T/0, 1/T/0). The selected clusters were further refined by applying a ranking of the genes having the highest appearance frequency within each specific pattern. In this way the most significant members were assessed. Tables1 and2 show genes directly regulated by CD44, and Tables 3 and4 show genes temporally regulated by CD44.
Genes directly regulated and induced by anti-CD44
| HGNC* . | Annotation . | Accession no. . | Pattern . |
|---|---|---|---|
| IL1A | Interleukin 1, alpha | M28983 | TTT000 |
| NA | Human clone 23908 mRNA sequence | U79290 | TTT000 |
| NA | Human (BSF-2/IL6) gene for B cell stimulatory factor-2 | Y00081 | TTT000 |
| PDE6A | Phosphodiesterase 6A, alpha subunit | M26061 | TT1000 |
| NA | H sapiens mRNA sequence (15q11-13) | X69636 | TT0000 |
| C18B11 | C18B11 homolog | U67934 | TT0000 |
| MLLT3 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 | L13744 | T1T000 |
| SOX5 | SRY (sex-determining region Y)-box 5 | S83308 | T10000 |
| ATP1A2 | ATPase, Na+K+ transporting, alpha-2 polypeptide | J05096 | T10000 |
| AQP1 | Aquaporin 1 | U41518 | T0T000 |
| STX1A | Syntaxin 1A | L37792 | T0T000 |
| PLG | Plasminogen | M34276 | T01000 |
| IF | I factor | Y00318 | T00000 |
| AFAP | Actin filament associated protein | D25248 | 1TT000 |
| SH3BP2 | SH3-domain binding protein 2 | AB000462 | 11T000 |
| GCNT2 | Glucosaminyl (N-acetyl) transferase 2, I-branching enzyme | L41607 | 0TT000 |
| PYGL | Phosphorylase, glycogen; liver | M14636 | 0TT000 |
| ADRB2 | Adrenergic, beta-2-, receptor, surface | M15169 | 0TT000 |
| PIGA | Phosphatidylinositol glycan, class A | S78467 | 0TT000 |
| ECGF1 | Endothelial cell growth factor 1 | S72487 | 0T1000 |
| NBL1 | Neuroblastoma, suppression of tumorigenicity 1 | D28124 | 0T1000 |
| MITF | Microphthalmia-associated transcription factor | Z29678 | 0T0000 |
| STK9 | Serine/threonine kinase 9 | X89059 | 00T000 |
| ALB | Albumin | U22961 | 00T000 |
| CAMK2D | Calcium/calmodulin-dependent protein kinase (CaM kinase) II delta | U50361 | 00T000 |
| MAPK14 | Mitogen-activated protein kinase 14 | L35253 | 00T000 |
| RGS3 | Regulator of G-protein signaling 3 | U27655 | 00T000 |
| SERPINC1 | Serine (or cysteine) proteinase inhibitor, clade C (antithrombin), member 1 | M21642 | 00T000 |
| HGNC* . | Annotation . | Accession no. . | Pattern . |
|---|---|---|---|
| IL1A | Interleukin 1, alpha | M28983 | TTT000 |
| NA | Human clone 23908 mRNA sequence | U79290 | TTT000 |
| NA | Human (BSF-2/IL6) gene for B cell stimulatory factor-2 | Y00081 | TTT000 |
| PDE6A | Phosphodiesterase 6A, alpha subunit | M26061 | TT1000 |
| NA | H sapiens mRNA sequence (15q11-13) | X69636 | TT0000 |
| C18B11 | C18B11 homolog | U67934 | TT0000 |
| MLLT3 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 | L13744 | T1T000 |
| SOX5 | SRY (sex-determining region Y)-box 5 | S83308 | T10000 |
| ATP1A2 | ATPase, Na+K+ transporting, alpha-2 polypeptide | J05096 | T10000 |
| AQP1 | Aquaporin 1 | U41518 | T0T000 |
| STX1A | Syntaxin 1A | L37792 | T0T000 |
| PLG | Plasminogen | M34276 | T01000 |
| IF | I factor | Y00318 | T00000 |
| AFAP | Actin filament associated protein | D25248 | 1TT000 |
| SH3BP2 | SH3-domain binding protein 2 | AB000462 | 11T000 |
| GCNT2 | Glucosaminyl (N-acetyl) transferase 2, I-branching enzyme | L41607 | 0TT000 |
| PYGL | Phosphorylase, glycogen; liver | M14636 | 0TT000 |
| ADRB2 | Adrenergic, beta-2-, receptor, surface | M15169 | 0TT000 |
| PIGA | Phosphatidylinositol glycan, class A | S78467 | 0TT000 |
| ECGF1 | Endothelial cell growth factor 1 | S72487 | 0T1000 |
| NBL1 | Neuroblastoma, suppression of tumorigenicity 1 | D28124 | 0T1000 |
| MITF | Microphthalmia-associated transcription factor | Z29678 | 0T0000 |
| STK9 | Serine/threonine kinase 9 | X89059 | 00T000 |
| ALB | Albumin | U22961 | 00T000 |
| CAMK2D | Calcium/calmodulin-dependent protein kinase (CaM kinase) II delta | U50361 | 00T000 |
| MAPK14 | Mitogen-activated protein kinase 14 | L35253 | 00T000 |
| RGS3 | Regulator of G-protein signaling 3 | U27655 | 00T000 |
| SERPINC1 | Serine (or cysteine) proteinase inhibitor, clade C (antithrombin), member 1 | M21642 | 00T000 |
Annotation based on HUGO Gene Nomenclature Committee (HGNC):http://www.gene.ucl.ac.uk/nomenclature/.
NA indicates not applicable.
Genes directly regulated and repressed by anti-CD44
| HGNC* . | Annotation . | Accession no. . | Pattern . |
|---|---|---|---|
| SSTR4 | Somatostatin receptor 4 | L14856 | 000TTT |
| NA | H sapiens CREB gene | X68994 | 000TTT |
| PALM | Paralemmin | D87460 | 000TT0 |
| NA | H sapiens mRNA for NK receptor | X97230 | 000TT0 |
| SLC14A1 | Solute carrier family 14 member 1 | U35735 | 000T0T |
| PRDM2 | PR domain containing 2 | U17838 | 000T0T |
| HLCS | Holocarboxylase synthetase | D87328 | 000T01 |
| APC | Adenomatosis polyposis coli | M73548 | 000T00 |
| MLLT10 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 | U13948 | 0001T0 |
| PLS1 | Plastin 1 | L20826 | 0000TT |
| LU | Lutheran blood group | X80026 | 0000TT |
| SERPINA1 | Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | K01396 | 0000T0 |
| LAMP2 | Lysosomal-associated membrane protein 2 | L09717 | 0000T0 |
| GUCY2F | Guanylate cyclase 2F, Retinal | L37378 | 00000T |
| EPHB3 | EphB3 | X75208 | 00000T |
| HGNC* . | Annotation . | Accession no. . | Pattern . |
|---|---|---|---|
| SSTR4 | Somatostatin receptor 4 | L14856 | 000TTT |
| NA | H sapiens CREB gene | X68994 | 000TTT |
| PALM | Paralemmin | D87460 | 000TT0 |
| NA | H sapiens mRNA for NK receptor | X97230 | 000TT0 |
| SLC14A1 | Solute carrier family 14 member 1 | U35735 | 000T0T |
| PRDM2 | PR domain containing 2 | U17838 | 000T0T |
| HLCS | Holocarboxylase synthetase | D87328 | 000T01 |
| APC | Adenomatosis polyposis coli | M73548 | 000T00 |
| MLLT10 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 | U13948 | 0001T0 |
| PLS1 | Plastin 1 | L20826 | 0000TT |
| LU | Lutheran blood group | X80026 | 0000TT |
| SERPINA1 | Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | K01396 | 0000T0 |
| LAMP2 | Lysosomal-associated membrane protein 2 | L09717 | 0000T0 |
| GUCY2F | Guanylate cyclase 2F, Retinal | L37378 | 00000T |
| EPHB3 | EphB3 | X75208 | 00000T |
Annotation based on HUGO Gene Nomenclature Committee (HGNC):http://www.gene.ucl.ac.uk/nomenclature/.
NA indicates not applicable.
Genes temporally regulated by induction by CD44
| HGNC3-150 . | Annotation . | Accession no. . | Pattern . |
|---|---|---|---|
| PRPF18 | PRP18 pre-mRNA processing factor 18 | U51990 | 1TT00T |
| NFKB2 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 | U20816 | 1T10TT |
| KDELR1 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 | X55885 | 1T10TT |
| PNMT | PhenylethanolamineN-methyltransferase | X52730 | 1T10T1 |
| DDX7 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 7 (RNA helicase, 52 kDa) | D26528 | 1T100T |
| MME | Membrane metallo-endopeptidase (neutral endopeptidase, enkephalinase, CALLA, CD10) | J03779 | 11T0TT |
| MPP3 | Membrane protein, palmitoylated 3 | U37707 | 11T0T1 |
| KIAA0121 | KIAA0121 gene product | D50911 | 1110T1 |
| TNFRSF6 | Tumor necrosis factor receptor superfamily, member 6 | X63717 | 11101T |
| NA | Human XIST | X56199 | 111011 |
| DIPA | Hepatitis delta antigen-interacting protein A | U63825 | 111011 |
| PRIM2 | Primase, polypeptide 2A | X74331 | 111011 |
| GPR31 | G protein-coupled receptor 31 | U65402 | 10T0T0 |
| HR44 | Hr44 antigen | X91103 | 10T0T0 |
| HGNC3-150 . | Annotation . | Accession no. . | Pattern . |
|---|---|---|---|
| PRPF18 | PRP18 pre-mRNA processing factor 18 | U51990 | 1TT00T |
| NFKB2 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 | U20816 | 1T10TT |
| KDELR1 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 | X55885 | 1T10TT |
| PNMT | PhenylethanolamineN-methyltransferase | X52730 | 1T10T1 |
| DDX7 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 7 (RNA helicase, 52 kDa) | D26528 | 1T100T |
| MME | Membrane metallo-endopeptidase (neutral endopeptidase, enkephalinase, CALLA, CD10) | J03779 | 11T0TT |
| MPP3 | Membrane protein, palmitoylated 3 | U37707 | 11T0T1 |
| KIAA0121 | KIAA0121 gene product | D50911 | 1110T1 |
| TNFRSF6 | Tumor necrosis factor receptor superfamily, member 6 | X63717 | 11101T |
| NA | Human XIST | X56199 | 111011 |
| DIPA | Hepatitis delta antigen-interacting protein A | U63825 | 111011 |
| PRIM2 | Primase, polypeptide 2A | X74331 | 111011 |
| GPR31 | G protein-coupled receptor 31 | U65402 | 10T0T0 |
| HR44 | Hr44 antigen | X91103 | 10T0T0 |
Annotation based on HUGO Gene Nomenclature Committee (HGNC):http://www.gene.ucl.ac.uk/nomenclature/.
NA indicates not applicable.
Genes temporally regulated by repression by CD44
| HGNC4-150 . | Annotation . | Accession no. . | Pattern . |
|---|---|---|---|
| KIAA0196 | KIAA0196 gene product | D83780 | 00T1TT |
| CRYGEP1 | Crystallin, gamma E pseudogene 1 | K03008 | 00T1TT |
| LYZ | Lysozyme | J03801 | 00T1TT |
| KIAA0141 | KIAA0141 gene product | D50931 | 00TT0T |
| TERF1 | Telomeric repeat binding factor | U40705 | 00TT1T |
| EFNA3 | Ephrin-A3 | U14187 | 00TTT0 |
| RNTRE | Related to the N terminus of tre | D13644 | 00TTT1 |
| NUCB2 | Nucleobindin 2 | X76732 | 00TTT1 |
| PAFAH2 | Platelet-activating factor acetylhydrolase 2 | D87845 | 00TTTT |
| GCG | Glucagon | J04040 | 00TTTT |
| ELL | ELL gene | U16282 | 00TTTT |
| MNAT1 | Menage a trois 1 (CAK assembly factor) | X87843 | 00TTTT |
| NR4A3 | Nuclear receptor subfamily 4, group A, member 3 | X89894 | 01T1TT |
| SLC18A1 | Solute carrier family 18, member 1 | U39905 | 0T0TTT |
| NA | Human clone 23907 mRNA sequence | U90907 | 0T111T |
| PLK | Pololike kinase | U01038 | 0T1T11 |
| CCNG2 | Cyclin G2 | U47414 | 0T1T11 |
| MX2 | Myxovirus (influenza virus) resistance 2 | M30818 | 0T1T1T |
| HTR1A | 5-hydroxytryptamine (serotonin) receptor 1A | M83181 | 0TT11T |
| FRDA | Friedreich ataxia | U43747 | 0TT11T |
| MEF2A | MADS box transcription enhancer factor 2, polypeptide A | X68505 | 0TT1T1 |
| SNCA | Synuclein, alpha | U46901 | 0TTT11 |
| APOC3 | Apolipoprotein C-III | X01388 | 0TTT1T |
| PRKACA | Protein kinase, cAMP-dependent, catalytic, alpha | M80335 | 0TTTT1 |
| AUH | AU RNA binding protein/enoyl-coenzyme A hydratase | X79888 | 0TTTT1 |
| HGNC4-150 . | Annotation . | Accession no. . | Pattern . |
|---|---|---|---|
| KIAA0196 | KIAA0196 gene product | D83780 | 00T1TT |
| CRYGEP1 | Crystallin, gamma E pseudogene 1 | K03008 | 00T1TT |
| LYZ | Lysozyme | J03801 | 00T1TT |
| KIAA0141 | KIAA0141 gene product | D50931 | 00TT0T |
| TERF1 | Telomeric repeat binding factor | U40705 | 00TT1T |
| EFNA3 | Ephrin-A3 | U14187 | 00TTT0 |
| RNTRE | Related to the N terminus of tre | D13644 | 00TTT1 |
| NUCB2 | Nucleobindin 2 | X76732 | 00TTT1 |
| PAFAH2 | Platelet-activating factor acetylhydrolase 2 | D87845 | 00TTTT |
| GCG | Glucagon | J04040 | 00TTTT |
| ELL | ELL gene | U16282 | 00TTTT |
| MNAT1 | Menage a trois 1 (CAK assembly factor) | X87843 | 00TTTT |
| NR4A3 | Nuclear receptor subfamily 4, group A, member 3 | X89894 | 01T1TT |
| SLC18A1 | Solute carrier family 18, member 1 | U39905 | 0T0TTT |
| NA | Human clone 23907 mRNA sequence | U90907 | 0T111T |
| PLK | Pololike kinase | U01038 | 0T1T11 |
| CCNG2 | Cyclin G2 | U47414 | 0T1T11 |
| MX2 | Myxovirus (influenza virus) resistance 2 | M30818 | 0T1T1T |
| HTR1A | 5-hydroxytryptamine (serotonin) receptor 1A | M83181 | 0TT11T |
| FRDA | Friedreich ataxia | U43747 | 0TT11T |
| MEF2A | MADS box transcription enhancer factor 2, polypeptide A | X68505 | 0TT1T1 |
| SNCA | Synuclein, alpha | U46901 | 0TTT11 |
| APOC3 | Apolipoprotein C-III | X01388 | 0TTT1T |
| PRKACA | Protein kinase, cAMP-dependent, catalytic, alpha | M80335 | 0TTTT1 |
| AUH | AU RNA binding protein/enoyl-coenzyme A hydratase | X79888 | 0TTTT1 |
Annotation based on HUGO Gene Nomenclature Committee (HGNC):http://www.gene.ucl.ac.uk/nomenclature/.
NA indicates not applicable.
The ligation of CD44 on a naive B-cell population was recently shown by us to partially induce a germinal center phenotype,10 as defined by an up-regulation of CD10, CD77, and CD95. The present analysis confirmed the induced presence of CD10 (MME) and CD95 (TNFRSF6) on the transcriptional level (Table 2). The neutral glycosphingolipid CD77 is not represented on the microarray. In the present setting the difference in CD10 and CD95 expression was attributed to a temporal induction, because the CD95 transcript was found in the 11101T cluster and the CD10 is found in the 11T0TT cluster. This indicated that CD44-dependent induction of these molecules occurred at the earlier time points.
The list of directly regulated genes represents several novel members. In the cluster designated 00T000, representing genes differentially expressed at 72 hours by CD44 ligation, is the SERPINC1 transcript for antithrombin III found. This cluster also contains the RGS3 transcript for the G-protein signaling regulator 3 protein, which is involved in the regulation of chemoattractant-mediated migration in lymphocytes.26,27 The cluster 0TT000, representing genes that are differentially up-regulated after 6 hours by the CD44 ligation, contains the transcript for the β2-adrenergic receptor (ADRB2). This receptor binds the sympathetic nervous system (SNS) mediator norepinephrine. This is of interest, because norepinephrine innervation of lymphoid organs plays a central regulatory role in the immune system.28
In the cluster 0T1000 we found the platelet-derived endothelial cell growth factor (ECGF1). This cytokine is known for its involvement in cancer and rheumatoid arthritis and has, for instance, been shown to induce inflammation and hyperplasia in synovial cells.29In the cluster described by TTT000, the immunomodulating interleukins IL-6 and IL-1α (IL1A) were found. IL-6 is secreted upon inflammation and has a wide array of functions both centrally and peripherally.30 IL-1α is another cytokine that has been implicated in functions both centrally and peripherally. In this context, it is a cytokine that has been considered to have overlapping functions with IL-1β,31 although IL-1α has been found not to compensate for IL-1β loss.32 However, several studies have reported a role for IL-1α in T helper cell regulation,33,34 suggesting a more confined functionality.
The up-regulation of IL-1α and IL-6 mRNA led us to investigate their presence at the protein level. Flow cytometry analysis was used to detect surface expression of IL-1α at the various time points after CD44 ligation. The population of IL-1α-expressing cells increased by a factor of 3.9 (SEM 0.44) at 24 hours. In total, 10% (SEM 0.16) of the total CD44-stimulated B-cell population was IL-1α+ at 24 hours (data not shown). The IL-6 production at the various time points after CD44 ligation was assessed by ELISA. The average net increase of CD44-induced IL-6, observed at 24 and 72 hours, was 1.6 times (SEM 0.26), reaching a total concentration of 300 pg/mL (SEM 30.0). These findings validate the CD44 involvement in the induction of these 2 immunomodulatory cytokines.
Discussion
The hyaluronan (HA) receptor CD44 has been implicated in the regulation of the immune system, and as a member of the hyaladherin family of proteins its role in adhesion and extracellular interactions has been extensively investigated.4 Furthermore, a crucial role of CD44 in inflammation has also lately been demonstrated.35 In the present study we assessed the transcriptional outcome of ligation of CD44 on mature naive tonsillar B cells. The dataset was analyzed employing a novel statistical analysis scheme created to retrieve the most likely expression pattern of an observed distribution. We were thus able to extract the patterns for genes classified as directly or temporally regulated by the anti-CD44 stimuli (Tables 1-2).
Our previous observation that CD44 ligation in naive B cells in fact induced a GC B-cell–like phenotype was corroborated in the present transcriptional analysis, in that CD10 (MME) and CD95 (TNFRSF6) were shown to be temporally regulated by CD44. The neutral endopeptidase CD10 has a strong association to activated germinal center B-cell populations,36 and the only other stage in B-cell ontogeny where CD10 is expressed is at the pro-B-cell stage in bone marrow. The functional role of CD10 is associated to inflammation, and CD10 provides protection to several neuropeptides and other mediators of inflammation in tissue. Some of the substrates include endothelin, bradykinin, substance P, calcitonin gene-related peptide, neuropeptide Y, and IL-1β.37 The CD95 expression is also highly associated with activation, especially in lymphocytes.38This receptor is a prerequisite for a physiological control of the activated immune system, enabling the possibility to clear malfunctioning cells by apoptosis. If this control is perturbed, the imbalance in the immune system could lead to autoimmunity. Interestingly, an increased susceptibility to CD95-mediated activation-induced cell death has been linked to CD44 functionality on thymocytes.6 7
Among the components that were found to be directly regulated by CD44 were the cytokines IL-6 and IL-1α. IL-6 is a pleiotropic molecule involved as a regulator of inflammation both centrally and peripherally.30 A role of IL-6 in the induction and progression of arthritis has also been demonstrated in IL-6–deficient mice.39 IL-6–deficient mice, furthermore, show impaired antibody production and reduced numbers of both B and T cells in lymph nodes as well as reduced germinal center formation.40 The induction of IL-6 is not unique to CD44, because IL-6 induction has been associated also with CD40-mediated signaling.41However, CD44 has been shown to augment IL-6 production in human rheumatoid synovial cells42 and polymorphonuclear cells,43 which is in agreement with our findings where the CD44 ligation generated an increase in IL-6 protein production. Interestingly, it has also recently been reported that IL-6 production can be specifically attributed to CD44 v6 at least in macrophages.44
Direct regulation by the CD44 stimuli was evident also for the IL-1α transcript (IL1A). This induction seems to be present relatively early in time and is sustained throughout the 72 hours as it follows the TTT000 pattern. Several studies have demonstrated a CD44-dependent induction of IL-1.45,46 However, in contrast to previous studies, the CD44 ligation on the BCR and CD40-activated B cells induced IL-1α. The immunomodulatory function of IL-1α is mostly associated with T helper cell proliferation. It induces T helper-2 (TH2) cell proliferation independently of IL-4 and initiates autocrine IL-1α production,34 which furthermore has been shown to increase the TH2 responsiveness to IL-4.47 Our confirmation that IL-1α was up-regulated at the protein level also demonstrated that a CD44 ligation augmented this surface expression, and a nearly 4-fold increase in the number of IL-1α–producing cells was evident. Taken together, these results suggest an attractive role of CD44-activated B cells in the regulation of T-cell proliferation.
CD44 has also been implicated in lymphocyte migration. In this study SERPINC1 and RSG3, both potent regulators of migration, were shown to be regulated by CD44. The serpin, antithrombin III (SERPINC1), is an important regulator of the coagulation cascade as well as being involved in inflammation.48 Antithrombin III has also been found to inhibit chemokine-mediated migration through the heparan sulfate proteoglycans syndecan-4 49 and by a similar interaction also to interfere with nuclear factor-κB (NF-κB) activation,50 which explains the anti-inflammatory effects of the enzyme. Another regulator of chemokine-mediated migration with a differential expression pattern is the G-protein signaling regulator RGS3. This molecule is a modulator of chemotaxis and cell adhesion mediated by G protein-coupled chemokine receptors26 and has been found to efficiently inhibit B-cell chemotaxis mediated by the CXCR4, CXCR5, and CCR7.27 Thus, the regulatory properties of SERPINC1 and RSG3 may be of critical importance, because a balance in the chemokine-mediated migration of lymphocytes has shown to be instructive in the formation of the secondary lymphoid organ structures.3
Another up-regulated immunoregulatory gene dependent on CD44 stimulation is the β2-adrenergic receptor (β2-AR). The adaptive response to inflammation involves components of the central nervous system (CNS). The systemic release of proinflammatory cytokines triggers the enrollment by CNS of the hypothalamic-pituitary-adrenal (HPA) axis and the sympathetic nervous system (SNS). These systems in concert modulate the immune response by a subsequent release of glucocorticoids from the adrenal glands and norepinephrine (NE) by postganglionic noradrenergic nerve endings, respectively. The peripheral discharge of glucocorticoids and NE will inhibit TH1 responses and promote a TH2 response thereby counteracting the inflammatory reaction.43,51,52 The sympathetic regulation of the immune system is mediated mainly by the β2-AR, which has been found on all lymphocytes except for TH2 cells. In B cells it has been reported that the β2-AR facilitates TH2-dependent IgG1 and IgE production quantitatively after antigen stimulation in mice.53,54 It is also proposed that the β2-AR plays a role in germinal center formation, because NE depletion attenuates GC formation.53 The β2-AR transcript ADRB2 is contained in a cluster characterizing genes induced after 24 hours (0TT000). This indicates that in line with proinflammatory cytokines55 the CD44 stimuli seem to be a regulator of the β2-AR expression of B cells. The β2-adrenergic receptor uses the cyclic adenosine monophosphate (cAMP) and the protein kinase A (PKA) pathway. This pathway has been reported to regulate TH1 and TH2 responses and TH2-dependent IgE and IgG production in B cells by a number of effectors.56 Thus, the induced presence of the β2-AR suggests a B cell involved in a TH2-dependent response.
Interestingly, a recent study demonstrated rapid down-regulation of CD44 on NK cells by NE treatment,57 and it is possible to speculate on a similar mechanism in the heavily innervated marginal zone. Such a mechanism presents a possible scenario where the CD44 participates in the decision making of the B-cell faith. For example, antigen-induced CD44 9,11,12 will as a costimultory molecule9 participate in a T cell-B cell cognate interaction with a sequential up-regulation of the β2-AR. This β2-AR up-regulation will mount the pivot point in this regulatory process at which the neuroendocrine system decides whether a germinal center formation should be supported or not. The β2-AR signaling will consequently induce cellular events facilitating GC formation53 54 characterized by a CD44 down-regulation.
In summary, our results suggest a novel role for CD44 in immunoregulation and inflammation. More specifically, possible mechanisms for CD44 involvement in the humoral response and the formation of germinal center reactions based on the up-regulation of transcript for CD10, CD95, IL-6, IL-1α, and β2-AR are proposed.
Prepublished online as Blood First Edition Paper, October 31, 2002; DOI 10.1182/blood-2002-06-1837.
Supported by Cancerfonden.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Carl A. K. Borrebaeck, Department of Immunotechnology, Lund University, PO Box 7031, SE-220 07 Lund, Sweden; e-mail: carl.borrebaeck@immun.lth.se.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal