Granulocyte colony-stimulating factor (G-CSF) is the major regulator of neutrophil production. Studies in cell lines have established that conserved tyrosines Tyr704, Tyr729, Tyr744, Tyr764 within the cytoplasmic domain of G-CSF receptor (G-CSF-R) contribute significantly to G-CSF–induced proliferation, differentiation, and cell survival. However, it is unclear whether these tyrosines are equally important under more physiologic conditions. Here, we investigated how individual G-CSF-R tyrosines affect G-CSF responses of primary myeloid progenitors. We generated G-CSF-R–deficient mice and transduced their bone marrow cells with tyrosine “null” mutant (m0), single tyrosine “add-back” mutants, or wild-type (WT) receptors. G-CSF–induced responses were determined in primary colony assays, serial replatings, and suspension cultures. We show that removal of all tyrosines had no major influence on primary colony growth. However, adding back Tyr764 strongly enhanced proliferative responses, which was reverted by inhibition of ERK activity. Tyr729, which we found to be associated with the suppressor of cytokine signaling, SOCS3, had a negative effect on colony formation. After repetitive replatings, the clonogenic capacities of cells expressing m0 gradually dropped compared with WT. The presence of Tyr729, but also Tyr704 and Tyr744, both involved in activation of signal transducer and activator of transcription 3 (STAT3), further reduced replating efficiencies. Conversely, Tyr764 greatly elevated the clonogenic abilities of myeloid progenitors, resulting in a more than 104-fold increase of colony-forming cells over m0 after the fifth replating. These findings suggest that tyrosines in the cytoplasmic domain of G-CSF-R, although dispensable for G-CSF–induced colony growth, recruit signaling mechanisms that regulate the maintenance and outgrowth of myeloid progenitor cells.
Introduction
Granulocyte colony-stimulating factor (G-CSF) supports the proliferation, survival, and differentiation of neutrophilic progenitor cells.1-4 G-CSF–deficient mice manifest a selective neutropenia, with blood neutrophil levels at 30% of those in wild-type (WT) mice. Blood neutrophil levels in mice lacking G-CSF receptors (gcsfr−/−) are also severely reduced, that is, approximately 15% of WT littermates.5 In addition, the numbers of myeloid progenitor cells in the bone marrow ofgcsfr−/− mice are significantly decreased.5 These observations have established that the G-CSF receptor (G-CSF-R) provides nonredundant signals for maintaining steady-state neutrophil levels.5 6
The G-CSF-R belongs to the cytokine receptor superfamily and possesses a single transmembrane region.1 Signaling molecules downstream of the G-CSF-R include Jak1, Jak2, and Tyk2; the signal transducer and activator of transcription (STAT) proteins, STAT1, STAT3, and STAT57-13; the Src kinases p55Lynand p56/59Hck14-16; components of the p21Ras/Raf/mitogen-activated protein kinase (MAPK) pathway17-19; and the SH2 domain-containing protein tyrosine phosphatases SHP-1 and SHP-2.19-21 The cytoplasmic domain of human G-CSF-R contains 4 conserved tyrosine residues, at positions 704, 729, 744, and 764 (equivalent to 703, 728, 743, and 763 in mouse G-CSF-R). Three of these tyrosines are located in the carboxy-terminal region implicated in the control of differentiation.22,23 On receptor activation, these tyrosines are phosphorylated and become docking sites for multiple SH2-containing signaling proteins, for example, STAT3 (Tyr704 and Tyr744), Shc (Tyr764), and Grb2 (Tyr764).21,24 25
We previously constructed a series of tyrosine (Tyr) to phenylalanine (Phe) substitution mutants of the G-CSF-R and expressed these in 32D cells to study their involvement in G-CSF signaling.25-27 These studies demonstrated that G-CSF-R substitution mutants lacking just 1 of the 4 tyrosines were still fully capable of transmitting differentiation signals in 32D cells.24 Strikingly, cells expressing mutant Tyr764Phe showed significantly accelerated differentiation with a concomitant reduction in proliferation, suggesting that Tyr764 plays an essential role in controlling the balance between proliferation and differentiation. Recently, similar observations have been made in primary bone marrow cells transduced with chimeric epidermal growth factor receptor (EGFR)/G-CSF-R Tyr764Phe.28G-CSF-R lacking all tyrosines (m0) fails to elicit proliferation and differentiation in 32D cells, although survival signals are still transduced.25 The presence of Tyr704 or Tyr744, which serve as major docking sites for STAT3, restored G-CSF–induced proliferation and differentiation to a significant extent.13,25 Introduction of Tyr764, involved in p21Ras activation and signaling via ERK and p38MAPK pathways,19,24,25 29 generated strong proliferative signals resulting in exponential growth without neutrophilic differentiation. A specific function and signaling mechanism linked to Tyr729 did not emerge from these studies.
The mechanisms by which G-CSF signaling is negatively regulated have not been elucidated. In contrast to, for example, the erythropoietin (EPO) receptor or granulocyte-macrophage colony-stimulating factor (GM-CSF)/interleukin 3 (IL-3)/IL-5 receptor common β chain, G-CSF-R tyrosines do not serve as docking sites for the protein tyrosine phosphatase SHP-1, although negative effects of SHP-1 on G-CSF signaling have been reported.20,30,31Because STAT3 and STAT5 are prominently activated by G-CSF, it is conceivable that suppressor of cytokine signaling (SOCS) proteins, which are under the direct transcriptional control of STATs, are involved in down-modulation of G-CSF responses.32-34 The SH-2 domain of SOCS1 has a high affinity for JAK kinases and interferes directly with JAK activity.35 On the other hand, other SOCS proteins, such as SOCS3, are recruited to phosphotyrosines in activated receptors and exert their negative activity either by blocking positively acting signaling substrates docking to the same receptor tyrosine, by inhibiting the activity of receptor-associated kinases, or by proteosomal targeting of signaling molecules.32-34
Although myeloid cell lines have provided useful models for studying G-CSF signaling, these cells are transformed and immortalized and therefore do not fully recapitulate the physiologic features of normal myeloid progenitor cells. In the present study, we have used retroviral transduction of G-CSF-R mutants into bone marrow cells of G-CSF-R–deficient mice to investigate how signals emanating from the cytoplasmic tyrosine residues in the G-CSF-R contribute to the clonogenic abilities of primary murine myeloid progenitor cells. We show that tyrosines are dispensable for G-CSF–induced colony formation per se, but individually contribute significantly to both G-CSF–induced colony growth and the maintenance of clonogenicity after sequential replatings. Prominent negative regulatory effects on colony growth were projected by Tyr729, which we found to be associated with recruitment of SOCS3. Conversely, Tyr764 greatly enhanced proliferative signals through activation of the Erk kinases.
Materials and methods
Cells and culture
Embryonic stem (ES) cells (ES-E14), a gift from M. Hooper (Edinburgh, United Kingdom), were cultured as described.36Briefly, cells were grown in culture medium consisting of Dulbecco modified Eagle medium (DMEM, Gibco BRL, Breda, The Netherlands), 50% Buffalo rat liver–conditioned medium, 10% fetal calf serum (FCS, ES qualified; Gibco-BRL) supplemented with 1% nonessential amino acids (Gibco BRL), 0.1 mM 2-mercaptoethanol (Sigma Chemical, St Louis, MO), 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco BRL), and 1000 U/mL leukemia inhibitory factor (LIF; Gibco BRL) in dishes coated with 0.1% gelatin (G-1890; Sigma). The cells were passaged every 2 to 3 days.
Targeting construct and probe
The targeting strategy used to inactivate the gcsfrgene is shown in Figure 1. Isolation, cloning, and sequencing of gDNA were done according to standard procedures.37 gDNA was isolated from a mouse 129SV/Cosmid library (Cosmid SC1-6 SuperCos 1, Stratagene Cloning Systems, La Jolla, CA) as described.38 A region of 10 kb including exon 7 to 17 of the gcsfr gene was replaced by a Neo gene driven by the PGK promoter. To construct the targeting vector, a 2.9-kbSau3AI-PstI fragment containing exon 5 and 6 and a 4.5-kb XbaI fragment, 3-prime of the gcsfrgene, including the noncoding region of exon 17, were cloned into pBluescript yielding pEUR11 and pEUR5, respectively. pEUR5 was opened by SpeI, and PGK-Neo was inserted in reverse orientation, yielding pEUR17. pEUR17 was opened by NotI, blunted, and subsequently cut by XhoI. The resulting 6.5-kbNotI-XhoI fragment containing the 4.5-kbXbaI fragment and the PGK-Neo gene was ligated into pEUR11 after ApaI opening, blunting, and subsequentXhoI digestion, resulting in pEUR18. A uniqueNotI site in the vector backbone was used for linearization prior to transfection. A 0.6-kb Sau3A fragment 5-prime of exon 4 (probe A) was used to screen for homologous recombination, yielding a 9.6-kb band in germ line configuration, and a 7-kb band after homologous recombination.
Targeting strategy to inactivate gcsfrgene.
(A) Targeting strategy to delete exons 7 to 16 of the gcsfrgene in ES cells. The position of the probe used to screen recombinant colonies is shown. Southern analysis of HindIII digests of gDNA detected a 9.6-kb band from the WT allele and a 7-kb band from the targeted allele (not shown). pBKS indicates pBluescript KS (Stratagene, La Jolla, CA). (B) Specific binding of G-CSF to bone marrow cells from gcsfr+/+,gcsfr+/−, and gcsf-−/−mice analyzed by flow cytometry. Cells were incubated with biotinylated G-CSF in the absence (solid line) or presence (dotted line) of a 100-fold molar excess of nonlabeled G-CSF followed by incubation with PE-conjugated streptavidin.
Targeting strategy to inactivate gcsfrgene.
(A) Targeting strategy to delete exons 7 to 16 of the gcsfrgene in ES cells. The position of the probe used to screen recombinant colonies is shown. Southern analysis of HindIII digests of gDNA detected a 9.6-kb band from the WT allele and a 7-kb band from the targeted allele (not shown). pBKS indicates pBluescript KS (Stratagene, La Jolla, CA). (B) Specific binding of G-CSF to bone marrow cells from gcsfr+/+,gcsfr+/−, and gcsf-−/−mice analyzed by flow cytometry. Cells were incubated with biotinylated G-CSF in the absence (solid line) or presence (dotted line) of a 100-fold molar excess of nonlabeled G-CSF followed by incubation with PE-conjugated streptavidin.
Disruption of the gcsfr gene by homologous recombination
ES-E14 cells (107) were transfected with 25 μg linearized pEUR18 by electroporation using a Progenetor II, PG200 Hoefer Gene pulser set at 350 V/cm, 1200 μF, 10 msec. The next day, cells were transferred to culture medium containing 200 μg/mL Gly418 (Gibco BRL), with Gly418-resistant colonies picked on day 8 or 9 after electroporation. gDNA of these colonies was digested withHindIII, transferred to nylon membranes, and hybridized to probe A and a neomycin probe. Correctly targeted clones were subjected to cytogenetic analysis and clones with a normal karyotype were used for blastocyst injections.
Generation of gcsfr knock-out mice
Two ES cell clones were injected into blastocysts of C57BL/6 mice. The resulting male chimeras were mated to FVB females to generategcsfr+/− F1 mice. Heterozygousgcsfr+/− mice were intercrossed to obtaingcsfr−/− mice. DNA was isolated from tail segments and analyzed by polymerase chain reaction (PCR) using primers for exon 17 (5′-GTATATCCCTGTGTTCAGGAAACC and 5′-GGCAGGGTCTTCAAGATACAAGG) and primers for the neo gene (5′-TACTCGGATGGAAGCCGGTC and 5′-AGTCGATGAATCCAGAAAAG).
Flow cytometric analysis of G-CSF-R expression
Expression levels of G-CSF-R on neutrophilic cells were measured by flow cytometry. To this end, G-CSF was biotinylated using D-biotinoyl-ε-aminocaproic acid-N-hydroxysuccinimide ester (Biotin-7-NHS; Boehringer, Mannheim, Germany). Free biotin was removed by gel filtration on Sephadex G-25. Bone marrow cells (106) were incubated in 96-well plates for 60 minutes at room temperature in 25 μL PBA (phosphate-buffered saline with 1% bovine serum albumin (BSA) and 0.1% NaN3) and 0.2 μg/mL biotinylated G-CSF, either in the absence or the presence of a 100-fold molar excess of nonbiotinylated G-CSF. Subsequently, cells were incubated for 30 minutes at 4°C with phycoerythrin-conjugated streptavidin (SA-PE; Caltag Laboratories, Burlingame, CA). To determine G-CSF-R expression on transduced bone marrow cells, cells were labeled for 30 minutes at 4°C with biotinylated antihuman G-CSF-R antibody (LMM741; Pharmingen, San Diego, CA) and subsequently with SA-PE. Cells were subjected to flow cytometric analysis on a FACScan (Becton Dickinson, Sunnyvale, CA).
Construction of G-CSF-R retroviral vectors and virus production
Vectors containing cDNA encoding human G-CSF-R WT and tyrosine substitution mutants have been described previously.13Inserts were recloned into the retroviral vector pBabe, containing a puromycin-resistance gene. Correct insertion was verified by nucleotide sequencing. Phoenix E virus producer cells (a gift from G. Nolan, Stanford, CA) were transfected with these constructs using Promega Profection Mammalian Transfection Systems (Promega, Madison, WI). Supernatants containing high-titer, helper-free recombinant viruses were harvested after culturing approximately 80% confluent producer cells for 16 to 20 hours in DMEM medium (with 5% FCS and penicillin/streptomycin) and passed through a 45-μm filter before use.
Retroviral infection of hematopoietic progenitor cells
Bone marrow cells were harvested from the femurs and tibiae of 8- to 12-week-old G-CSF-R–deficient mice as described.38After depletion of adherent cells, the remaining cells were fractionated on a Percoll density gradient (Amersham Pharmacia Biotech, Uppsala, Sweden) as described.39 Cells were washed twice in Hanks balanced salt solution (HBSS)/5% FCS/0.5% BSA, and prestimulated for 2 days in Cell Gro (Boehringer Ingelheim Bioproducts Partnership, Heidelberg, Germany) supplemented with a cytokine cocktail containing murine IL-3 (10 ng/mL), human FLT3 ligand, human thrombopoietin (hTPO), murine stem cell factor (mSCF), and GM-CSF (all 100 ng/mL) at a final density of 5 × 105cells/mL. Cells where then transferred to 35-mm culture dishes (Becton Dickinson, Lincoln Park, NJ) coated with 12 μg/mL recombinant fibronectin fragment CH-296 (Takara Shuzo, Otsu, Japan) and preincubated with the appropriate virus supernatant for 30 minutes at 37°C. Subsequently, bone marrow cells (106 cells/mL) were mixed with fresh virus supernatant in a 1:1 ratio, supplemented with a fresh cytokine cocktail, and cultured overnight at 37°C and 5% CO2. Virus supernatant and cytokine cocktail were once again refreshed the next day and the cells cultured for an additional 24 hours.
Progenitor cell assays and suspension culture
Bone marrow cells were plated in triplicate at densities of 1 × 105/mL in methyl cellulose medium supplemented with 30% FBS, 1% BSA, 0.1 mM 2-mercaptoethanol, 2 mMl-glutamine, and G-CSF (100 ng/mL). To calculate infection efficiencies for the different receptor mutants, cells were also plated in GM-CSF (20 U/mL) containing colony assays, with or without 2.5 μg/mL puromycin (Sigma, Zwijndrecht, The Netherlands). Colonies (30 cells or more) were counted on day 7 of culture. For cytologic analysis and replating experiments, colony cells were mass harvested and washed twice in HBSS. Suspension cultures were performed in RPMI (Gibco BRL) supplemented with 10% FCS and 100 ng/mL G-CSF. Every 3 to 4 days, culture medium was renewed and cells were counted on a Casy R-1 cell counter (Scharfe System, Reutlinger, Germany). Cell densities were kept between 0.3 × 106 and 1 × 106/mL. For inhibitor studies, cells were grown as described, in the presence of either 10 μM SB203580 or U0126 (Calbiochem, San Diego, CA) dissolved in dimethyl sulfoxide (DMSO) or 0.1% (vol/vol) DMSO as a solvent control. Viable cells were counted daily and every second day cells were spun down and resuspended in fresh media with fresh inhibitor. Cell densities of proliferating cells were kept between 0.5 and 1.5 × 106 cells/mL. Cell viability was assessed by flow cytometric analysis (FACScan, Becton Dickinson, Sunnyvale, CA) using 7-amino actinomycin D (7-AAD; Molecular Probes, Eugene, OR).
Reporter assay for SOCS3 effects on G-CSF-R activity
To determine the effects of SOCS3 on the activity of G-CSF-R and mutants, we used a STAT5 luciferase assay essentially as described previously.40 In brief, HEK 293 cells, seeded in 24-well dishes at 0.2 × 106 cells/well in 1 mL DMEM/10% FCS, were cultured overnight and transfected by means of standard Ca-PO4 precipitation with a mixture of the following plasmids: pME18S-STAT5 for expression of STAT5, a STAT5 luciferase reporter plasmid consisting of 5 repeats of the β-casein sequence upstream of a SV40 promoter in the pGL-3-promotor vector (Promega), a β-galactosidase expression plasmid pRSVLacZ, derived from pCH110,41 pcDNA3-SOCS3,42 or empty pcDNA3 (Invitrogen, Breda, The Netherlands) and pBabe with the different G-CSF-R mutants. A volume of 100 μL precipitate with a total of 2 μg DNA (400 ng DNA for each construct) was added to each well. For SOCS3, 12.5 ng SOCS3 supplemented with 387.5 ng pcDNA3 (empty vector) was added. On day 4, the cells were stimulated for 6 hours with 100 ng/mL G-CSF and subsequently lysed in 100 μL lysis buffer (25 mM Tris [tris(hydroxymethyl)aminomethane] phosphate, pH 7.8, 15% glycerol, 1% Triton X-100, 1 mM dithiothreitol [DTT], 8 mM MgCl2). To measure luciferase activity, cell lysates (25 μL) were transferred to 96-well flat bottom plates (Costar, Corning, Corning, NY) and 25 μL of a 16 mg/mL luciferase substrate-containing buffer (Steady-Glo luciferase assay System; Promega) was added to each well. Emitted light was measured in a TopCount luminometer (Packard, Meriden, CT). To correct luciferase activity levels for variations in transfection efficiencies, 25 μL cell lysate was incubated in parallel with 75 μL β-galactosidase substrate buffer (100 mM Na-PO4 buffer, pH 7.8, 10 mM KCl, 1 mM MgSO4, 2.7 mM DTT) and 0.56 mg/mL O-nitrophenyl β-d-galactopyranoside (oNPG; Sigma) for 15 minutes at 37°C. Absorption was measured in a microplate reader (Biorad 450; Veenendaal, The Netherlands) at 450 nm. All experiments were performed in duplicate.
Results
Generation of gcsfr-deficient mice
To inactivate the murine gcsfr gene, we constructed a targeting vector in which the genomic sequence spanning exon 7 to 17 was replaced by a pgk-Neo selection cassette (Figure 1A). Two independently isolated ES cell clones were injected into blastocysts and the resulting chimeras were crossed with FVB mice. Germ line transmission of the knock-out allele was achieved for both clones. Flow cytometric analysis of bone marrow neutrophils using biotinylated G-CSF confirmed the absence of G-CSF-R in gcsfr−/−mice and a 50% reduced expression in gcsfr+/−mice (Figure 1B). In agreement with a previously reportedgcsfr knock-out line,5 peripheral neutrophil counts in this gcsfr−/− strain are 15% to 20% of levels found in WT littermates.
Role of receptor tyrosines in G-CSF–induced colony formation
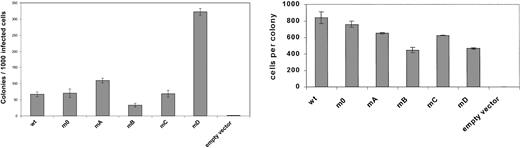
To study the involvement of receptor tyrosines in G-CSF-R–mediated signaling in primary hematopoietic cells, we introduced single tyrosine add-back mutants, m0, G-CSF-R WT, or Babe control vector (Figure 2), intogcsf−/− bone marrow cells and determined G-CSF responses in colony assays. As predicted, no colonies were formed by gcsf−/− cells transduced with Babe vector (Figure 3, left panel). Cells expressing m0 produced colonies at numbers equivalent to cells transduced with G-CSF-R WT. At first glance, these data would suggest that the receptor tyrosines are fully dispensable for G-CSF–controlled colony growth. However, experiments with the add-back mutants unveiled a more subtle scenario. Expression of mA (Tyr704) slightly increased colony formation, whereas colony numbers obtained with mC (Tyr744) were similar to m0. Colony formation induced by mB (Tyr729) was reduced by approximately 50%, indicating that Tyr729 has a negative influence on G-CSF–induced colony growth. The presence of Tyr764 (mD) resulted in approximately 6-fold increase in cloning efficiency. Assessment of the mean number of cells per colony did not show a correlation between colony number and size, except for mB (Figure 3, right panel). The latter observation suggests that negative signals emanating from Tyr729 affect both clonogenicity as well as proliferative potential of the transduced progenitor cells. Morphologic analysis revealed no differences in the composition of colonies induced by the various receptor forms, suggesting that the receptor tyrosines are dispensable for G-CSF–induced differentiation (data not shown).
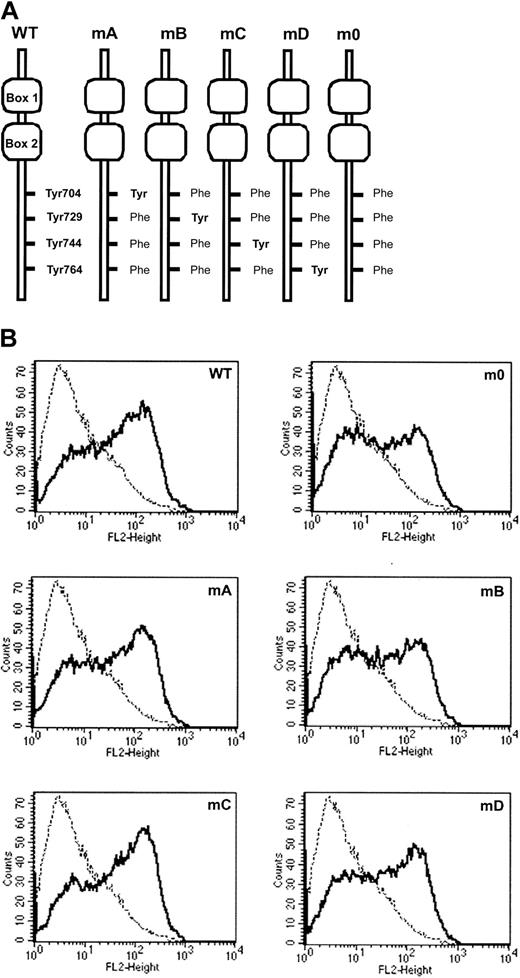
Expression of G-CSF-R mutants.
(A) Schematic representation of cytoplasmic domains of human G-CSF-R wild-type and tyrosine substitution mutants showing positioning of conserved tyrosines relative to membrane proximal Box1 and Box2. (B) Flow cytometric analysis of G-CSF-R expression ongcsfr−/− bone marrow cells retrovirally transduced with G-CSF-R WT and tyrosine mutants shown in panel A. Bold histograms indicate cells stained with biotinylated G-CSF-R antibodies and SA-PE; dotted histograms, cells stained with SA-PE only.
Expression of G-CSF-R mutants.
(A) Schematic representation of cytoplasmic domains of human G-CSF-R wild-type and tyrosine substitution mutants showing positioning of conserved tyrosines relative to membrane proximal Box1 and Box2. (B) Flow cytometric analysis of G-CSF-R expression ongcsfr−/− bone marrow cells retrovirally transduced with G-CSF-R WT and tyrosine mutants shown in panel A. Bold histograms indicate cells stained with biotinylated G-CSF-R antibodies and SA-PE; dotted histograms, cells stained with SA-PE only.
Primary colony formation by gcsfr−/−bone marrow progenitors transduced with different G-CSF-R constructs.
Colonies were grown in the presence of 100 ng/mL G-CSF. Left panel shows mean colony numbers ± SD per 1000 infected bone marrow cells from triplicate colony dishes; data are representative of 4 independent experiments. Colony numbers were normalized to the numbers of infected cells based on puromycin resistance of CFU-GMs (responsive to GM-CSF) to correct for differences due to variations in transduction efficiencies. Right panel shows mean numbers of cells per colony ± SD from triplicate dishes.
Primary colony formation by gcsfr−/−bone marrow progenitors transduced with different G-CSF-R constructs.
Colonies were grown in the presence of 100 ng/mL G-CSF. Left panel shows mean colony numbers ± SD per 1000 infected bone marrow cells from triplicate colony dishes; data are representative of 4 independent experiments. Colony numbers were normalized to the numbers of infected cells based on puromycin resistance of CFU-GMs (responsive to GM-CSF) to correct for differences due to variations in transduction efficiencies. Right panel shows mean numbers of cells per colony ± SD from triplicate dishes.
G-CSF-R tyrosines are involved in G-CSF–dependent maintenance of myeloid progenitor cell levels
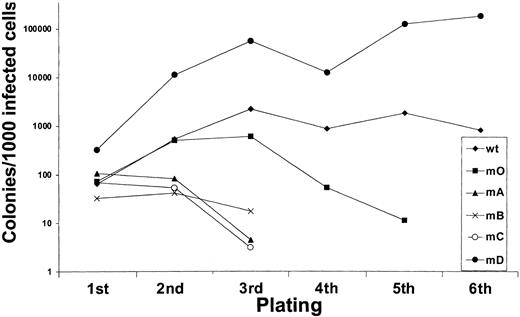
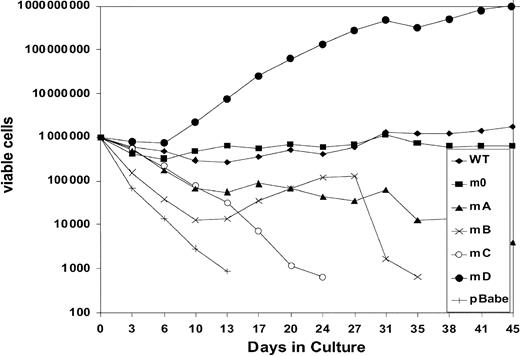
We next investigated to what extent G-CSF-R tyrosines contribute to the expansion of the progenitor cells in serial replatings. As shown in Figure 4, progenitor cells transduced with WT G-CSF-R maintained recloning abilities at a relatively constant level up to the sixth replating. In contrast, replating abilities of m0-expressing cells gradually declined after the third replating, suggesting that receptor tyrosines are required for sustained G-CSF–dependent maintenance and expansion of myeloid progenitor cells. The presence of Tyr729 (mB), Tyr704 (mA), or Tyr744 (mC) further suppressed the recloning potential of progenitors. In contrast, the presence of Tyr764 (mD) alone greatly enhanced recloning potential, resulting in colony numbers after the fifth replating that were 100-fold higher than WT and 10 000-fold higher than m0-expressing cells cultured in parallel. The sustained expansion of progenitor cells mediated via Tyr764 also translated into exponential cell proliferation in long-term suspension culture (Figure5). These expanded cells did not express mainly features of immature blast cells, but rather represented a mixture of myeloid cell types at various stages of differentiation. Notably, the cells remained fully dependent on G-CSF for proliferation (data not shown). Cell numbers were maintained at relatively stable levels in cultures from cells expressing WT and m0 G-CSF-R, whereas cells expressing mA, mB, or mC progressively lost proliferative abilities.
Serial replatings of progenitor cells from
gcsfr−/− bone marrow transduced with G-CSF-R expression constructs.
Serial replatings of progenitor cells from
gcsfr−/− bone marrow transduced with G-CSF-R expression constructs.
Expansion of gcsfr−/−bone marrow cells transduced with G-CSF-R expression constructs in suspension culture.
Cells were cultured in the presence of 100 ng/mL G-CSF. Viable cell counts and replenishment of culture media were performed at 3- to 4-day intervals.
Expansion of gcsfr−/−bone marrow cells transduced with G-CSF-R expression constructs in suspension culture.
Cells were cultured in the presence of 100 ng/mL G-CSF. Viable cell counts and replenishment of culture media were performed at 3- to 4-day intervals.
Proliferative signals from Tyr764 are mediated via Erk
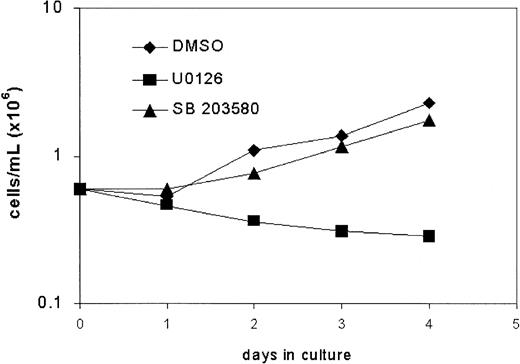
Tyr764 is a docking site for connector proteins implicated in p21Ras/MAPK signaling and has been shown to play a prominent role in the activation of Erk as well as p38.17,24 29 We therefore studied the effects of the MEK1 inhibitor U0126, which blocks activation of Erk1 and Erk2, and the p38 inhibitor SB203580 on G-CSF–induced proliferation of bone marrow cells expressing mD in suspension culture. As shown in Figure6, addition of U0126 to the cultures inhibited proliferation, whereas the effects of SB203580 were minimal. These findings establish that Erk kinases are the principle mediators of proliferative signaling via Tyr764 of the G-CSF-R.
Effects of inhibitors of MEK (U0126) and p38MAPK (SB203580) on proliferation of mD-expressing cells.
Effects of inhibitors of MEK (U0126) and p38MAPK (SB203580) on proliferation of mD-expressing cells.
SOCS3 inhibits G-CSF responses via Tyr729 of G-CSF-R
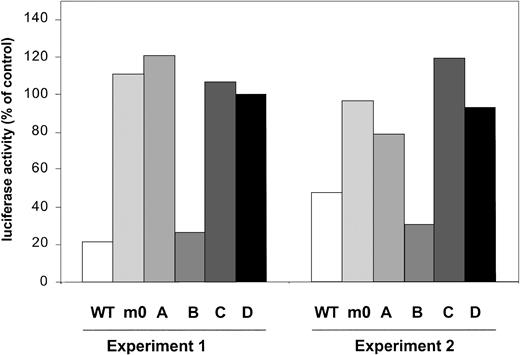
Recently, several studies have demonstrated that SOCS3 mediates its inhibitory activity on a variety of cytokine receptors, for example, leptin receptor and gp130, via binding to receptor tyrosines.42,43 Based on structural similarities between G-CSF-R and gp130, we performed a G-CSF-R activity assay based on the onset of STAT5-mediated gene expression, which is activated via the membrane proximal region of G-CSF-R.27Introduction of SOCS3 severely interfered with activity of G-CSF-R WT (Figure 7). In contrast, under similar conditions, activity of m0 was not affected, confirming that one or more of the receptor tyrosines are involved in SOCS3 recruitment. Experiments with single tyrosine add-back mutants subsequently showed inhibition only with mB, suggesting that Tyr729 is the major binding site for SOCS3. We suggest on the basis of these data that the negative effects generated by Tyr729 on G-CSF-R signaling are mediated by SOCS3.
STAT5-luciferase reporter assay showing prominent inhibitory effects of cotransfection of pcDNA-SOCS3 on G-CSF-R WT and mB (Tyr729).
Data are expressed as the percentage of activity measured after cotransfection of pcDNA3 empty vector.
STAT5-luciferase reporter assay showing prominent inhibitory effects of cotransfection of pcDNA-SOCS3 on G-CSF-R WT and mB (Tyr729).
Data are expressed as the percentage of activity measured after cotransfection of pcDNA3 empty vector.
Discussion
The aim of this work was to investigate to what extent signals from G-CSF-R tyrosines contribute to the G-CSF–induced responses of primary myeloid progenitor cells. Previous studies in myeloid factor–dependent cell lines have revealed that these tyrosines contribute to G-CSF–dependent proliferation, differentiation, and survival via signaling mechanisms involving the activation of STAT3 and p21Ras.17,21,24,25,29,44 Although these models have provided useful information, it has also become clear that major discrepancies in signaling requirements exist between cell lines and primary cells. For instance, STAT3 was shown to induce growth arrest and neutrophilic differentiation in cell lines, whereas a recent study in a transgenic mouse model demonstrated that STAT3 is essential for G-CSF–induced proliferation of primary myeloid progenitors.40,45 46 The fact that certain mechanisms underlying growth factor–induced proliferation of primary progenitor cells are bypassed or constitutively activated in cell lines is likely to contribute to such differences.
Some of the findings reported here are consistent with certain observations in cell lines. For instance, Tyr764 confers hyperproliferative responses to G-CSF in both primary progenitor cells and cell lines.25 However, m0, which was unable to transduce proliferation signals in 32D cells, fully supported G-CSF–induced colony formation of primary progenitors at plating efficiencies comparable to G-CSF-R WT. Thus, coupling of signaling mechanisms to G-CSF-R tyrosines in primary myeloid progenitor cells is redundant for G-CSF–induced colony growth. This might be attributed to either alternative activation of the pathways linked to the tyrosines or to compensatory influences of other signaling pathways.
Although G-CSF-R tyrosines were not required per se for G-CSF–induced colony formation, the experiments with the add-back mutants clearly suggested that the individual tyrosines exert regulatory functions. In particular, this applies to the growth inhibitory role of Tyr729 and the growth-promoting role of Tyr764. No specific inhibitory pathway had previously been assigned to Tyr729. We have identified Tyr729 here as the single tyrosine involved in SOCS3-mediated inhibition of G-CSF signaling. In similar experiments we could not functionally link SOCS1 or SOCS2 to Tyr729 (G.-J. van de G. and I.P.T., unpublished results, 2002). We hypothesize that SOCS3 binds directly to Tyr729 via its SH2 domain. Tyr729 is located in a motif (VLYGQLLGS) that shows striking homology with the SOCS3-SH2–binding sites within gp130 and the leptin receptor. Characteristics of this motif are the valine at pY − 2, a hydrophobic residue at Y+343 and the serine at Y+6 (or Y+5).42,43 While this paper was under review, Hörtner et al published data supporting the notion that the Tyr729-containing motif of G-CSF-R indeed forms a direct binding site for SOCS3.47 Notably, G-CSF-R deletion mutants in patients with severe congenital neutropenia that progresses to acute myeloid leukemia lack this motif,48-50 which may contribute to the hyperproliferative signaling properties of these receptor forms.51 52
The G-CSF–induced colonies grown from the gcsfr−/−bone marrow cells transduced with G-CSF-R constructs were of granulocyte, granulocyte-macrophage, macrophage, or mast cell origin and contained fully mature cells. We did not observe differences in the composition of the colonies grown from cells transduced with G-CSF-R WT, tyrosine add-back, or tyrosine null mutants. This argues against a major role of the receptor tyrosines in controlling myeloid differentiation. A similar conclusion was recently drawn by Akbarzadeh et al,28 who further demonstrated that expression of myeloperoxidase and gelatinase, enzymatic markers of granulocytic differentiation, was not affected by substitution of the tyrosines. Interestingly, these authors observed a slight, but significant, increase in the numbers of macrophage colonies and reduction of granulocyte colonies with mutant Tyr729Phe, but not with their Tyr null mutant. This suggests that Tyr729, possibly via recruitment of SOCS3, influences the balance between granulocyte and macrophage colony growth only when pathways activated via one or more of the remaining tyrosines remain intact.
Both G-CSF– and G-CSF-R–deficient mice have reduced numbers of granulocyte-macrophage colony-forming units (CFU-GMs) in the bone marrow. Thus, G-CSF not only stimulates myeloid progenitors to proliferate and differentiate toward neutrophils, but also controls the size of the progenitor cell compartment in the bone marrow. The data from the sequential platings suggest that signaling pathways emanating from the G-CSF-R tyrosines contribute significantly to this control. The prominent stimulatory influence of Tyr764 suggests that activation of the p21Ras/Erk pathway contributes to progenitor cell expansion, whereas the inhibitory signal provided by Tyr729, most likely involving SOCS3, has the opposite effect. Strikingly, Tyr704 and Tyr744, while exerting no inhibitory effect on primary colony growth, suppressed progenitor cell expansion in the replating experiments. The signaling pathways responsible for this inhibition are not known. Tyr704 and Tyr744 both function as direct docking sites for STAT3.25 STAT3 activation by G-CSF-R can also occur in the absence of receptor tyrosines.13,28 Still, we think that STAT3 activation via Tyr704 and Tyr744 may play an important role because depending on the levels of activation in conjunction with other signaling pathways, STAT3 can exert variable functions in myeloid progenitor cells. The effects of STAT3 on cell proliferation and differentiation are diverse and depend on cell type and stage of differentiation.40,46,53 Even within one cell type, unexpected variations in the effects of STAT3, depending on the status of activation of other signaling pathways, have been reported. Based on these findings, a model was proposed in which STAT3 orchestrates conflicting signals during G1 to S transition in the cell cycle.54 In view of this more complex role of STAT3 in the regulation of cell growth, we propose that STAT3 mediates stimulatory effects on primary colony growth and neutrophilic differentiation, thereby negatively affecting the expansion of myeloid progenitors controlled by G-CSF. Studies in which STAT3 activation in bone marrow cells can inducibly be inactivated are in progress to unravel the full spectrum of activities of STAT3 in myeloid cell development.
We thank Drs Marieke von Lindern and Joanna Prasher for their critical evaluation of this manuscript.
Prepublished online as Blood First Edition Paper, December 5, 2002; DOI 10.1182/blood-2002-07-2062.
Supported by grants from the Netherlands Organization for Scientific Research and the Dutch Cancer Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ivo Touw, Institute of Hematology, Erasmus University Rotterdam, PO Box 1738, 3000 DR Rotterdam, the Netherlands; e-mail: touw@hema.fgg.eur.nl.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal