The targeted inactivation of oncogenes may be a specific and effective treatment for cancer. However, because human cancers are the consequence of multiple genetic changes, the inactivation of one oncogene may not be sufficient to cause sustained tumor regression. Moreover, cancers are genomically unstable and may readily compensate for the inactivation of a single oncogene. Here we confirm by spectral karyotypic analysis that MYC-induced hematopoietic tumors are highly genetically complex and genomically unstable. Nevertheless, the inactivation of MYC alone was found to be sufficient to induce sustained tumor regression. After prolongedMYC inactivation, some tumors exhibited a distinct propensity to relapse. When tumors relapsed, they no longer required the overexpression of MYC but instead acquired novel chromosomal translocations. We conclude that even highly genetically complex cancers are reversible on the inactivation of MYC, unless they acquire novel genetic alterations that can sustain a neoplastic phenotype.
Introduction
Tumorigenesis is associated with the activation of oncogenes and the inactivation of tumor-suppressor genes.1The pharmacologic inactivation of these mutant genes may be an effective therapy for cancer. Human cancer cells generally possess many genetic abnormalities, so that the repair or inactivation of a single mutant gene may not be sufficient to induce sustained tumor regression. Moreover, because cancers are genomically unstable, they may readily acquire genetic lesions that could compensate for any pharmacologically inactivated gene product.
The MYC proto-oncogene may be a good pharmacologic target for the treatment of human cancer. MYC has been implicated in the pathogenesis of many types of human neoplasia, in particular hematopoietic tumors such as Burkitt lymphoma, large B-cell lymphoma, lymphoblastic lymphoma, and multiple myeloma.2-4 A consistent feature of these tumors is that MYC expression levels are up-regulated through several different mechanisms, including chromosomal translocation, genomic amplification, and mutation.2,4,5MYC normally regulates cellular growth and proliferation and, under some circumstances, induces apoptosis. MYC overexpression is thought to contribute to tumorigenesis by causing unrestrained cellular proliferation, blocking differentiation, and perhaps promoting genomic destabilization.2
Many oncogenes are known to cooperate with MYC during tumorigenesis, including Ras,6Bcl-2,7Bmi-1,8Pim-1,9 andPim-2.10 Similarly, the loss of function of many tumor-suppressor genes, including p53 andp19ARF, has been shown to cooperate with MYC to induce tumorigenesis.11 Hence, MYC is thought to induce tumorigenesis in concert with multiple genetic factors. More recently, chromosomal aberrations in tumors associated with MYC overexpression have been identified by spectral karyotypic analysis (SKY). SKY is a multicolor chromosome painting method that allows for visualization of chromosomal abnormalities otherwise difficult to identify in mouse metaphases.12Examples of abnormalities detected using SKY include translocations in myeloma,13 chromosomal alterations in mammary tumors,14 and recurrent translocations inMYC-induced BALB/c plasmacytomas.15
To determine whether MYC may be an effective target for cancer therapy, we developed a conditional transgenic model system for the regulation of MYC expression in murine hematopoietic tumors.16 We reported that with MYCinactivation, hematopoietic tumors undergo sustained regression. After prolonged MYC inactivation, we found that 20% of the tumors relapsed, apparently bypassing the requirement for continuedMYC overexpression. We concluded that MYC-induced hematopoietic tumorigenesis is reversible, but that there are mechanisms by which tumors can become independent of MYC. We postulated that there might be genetic changes that can functionally replace the requirement of MYC overexpression to sustain a neoplastic phenotype.
Here we have characterized the ability of individualMYC-induced tumors to undergo sustained regression uponMYC inactivation. We found that individual tumors exhibit a distinct propensity to relapse with prolonged MYCinactivation and that when tumors relapse, they become independent ofMYC to maintain their neoplastic features. Moreover, through the use of SKY, we examined a panel of primary MYC-induced hematopoietic tumors and variants of these tumors that had relapsed after prolonged MYC inactivation. We found that primary transgenic tumors exhibited complex patterns of karyotypic abnormalities, including aneuploidy, polyploidy, homogeneously stained regions (HSRs), and chromosomal translocations. Relapsed tumor variants consistently acquired novel chromosomal translocations. Our results confirm that MYC inactivation is sufficient to induce sustained tumor regression even in genomically complex tumors, and they suggest that there may be genetic changes that can functionally compensate for the requirement of MYC overexpression to maintain a neoplastic phenotype.
Materials and methods
Tumor cell lines and tumorigenicity assays
Tumor-derived cell lines were generated, and tumorigenicity assays were performed exactly as previously described.16Tumor cell lines were transplanted into cohorts of at least 10 syngeneic mice by the intraperitoneal injection of 107tumor cells. When mice were moribund with tumor burden, MYCwas inactivated by doxycycline (100 μg/mL) treatment in the drinking water. Animals were monitored for tumor relapse for up to 30 weeks after inoculation.
Western blots
MYC protein levels were measured using conventional Western blot techniques for human c-MYC (9E10; Oncogene, San Diego, CA), mouse c-Myc (c19; Santa Cruz Biotechnology, Santa Cruz, CA), mouse N-Myc (OP13; Oncogene), and mouse L-Myc (c20; Santa Cruz Biotechnology). As a positive control for N-Myc and L-Myc, we used a lysate from BJAB cells (Santa Cruz) and NCI-H209 cells (ATCC Biotechnology, Manassas, VA), respectively.
Metaphase preparation and spectral karyotypic analysis
Metaphase spreads were prepared according to standard procedures. Briefly, 1 mg/mL colcemide was added to the cells for 30 minutes in early passage. Cells were incubated with 0.075 M KCl for 20 minutes and then were fixed using methanol/acetic acid fixative. Spectral karyotyping was performed using the Applied Spectral Imaging (ASI; Carlsbad, CA) skypaint kit for mouse chromosomes, according to the manufacturer's protocol. SKY images were acquired with a Spectracube-2 (ASI) mounted on a Leica DMR microscope. Acquisition and analysis were performed using the ASI image capturing (SI 2.2) and analysis software (SkyView 1.6.1). At least 15 metaphase spreads of good spectral hybridization quality and banding morphology were analyzed for each preparation. Every karyotype was confirmed by cytogenetic G-banding analysis.
Results
Tumor regression and relapse upon MYC inactivation
Previously, we described the use of the tetracycline regulatory system to generate transgenic mice that conditionally overexpress theMYC proto-oncogene in hematopoietic cells.16 In this system, the tetracycline-transactivating protein (tTA) mediates the transcription of MYC placed under the control of the tetracycline-responsive promoter (tet-o). The presence of doxycycline inactivates the transcription mediated by tTA, and MYC is turned off. In the absence of doxycycline, mice develop hematopoietic tumors. When treated with doxycycline, 100% of the tumors initially regressed and 80% of the tumors showed sustained tumor regression with continuous doxycycline treatment. Approximately 20% of the tumors relapsed after prolonged MYC inactivation. We concluded thatMYC-induced tumorigenesis is reversible but that some tumors can become independent of MYC for their neoplastic properties.
We considered there are 2 possible explanations for the relapse of some tumors after prolonged MYC inactivation. First, all tumors may have the ability to give rise to rare cells that have acquired the ability to become independent of MYC for their neoplastic properties. Second, only some of the tumors may have an inherent ability to relapse following prolonged MYC inactivation. To distinguish between these 2 possibilities, we generated a panel of tumor-derived cell lines. We reasoned that if the ability of a tumor to relapse after MYC inactivation is a property of all tumors, then each tumor should exhibit a similar ability to relapse. Previously, we verified that tumor-derived cell lines faithfully continue to exhibit a conditional malignant phenotype in vitro and in vivo.16 We confirmed that each of these cell lines had lost its neoplastic features upon MYC inactivation in vitro, as demonstrated by the loss of cellular proliferation and the induction of apoptosis, observed by phase microscopy, trypan blue staining, and fluorescence-activated cell sorter (FACS) analysis for DNA content (data not shown). These results are similar to what we described previously in detail.16
To directly investigate the ability of each tumor to relapse afterMYC inactivation, we inoculated 12 primary tumor lines into at least 10 syngeneic mice. When mice were moribund with tumor burden, we investigated the ability of tumors to regress on the inactivation ofMYC (Table 1, Figure 1). Representative mice were killed and analyzed grossly and histologically for evidence of tumors before and after MYC inactivation (Figure 1A-C). Initially, tumors invaded and effaced the healthy architecture of the thymus, spleen, and lymph nodes (Figure 1A). After MYCinactivation, all tumors initially regressed; this was associated with the restoration of healthy hematopoietic architecture (Figure 1B). Mice were observed to determine the frequency at which tumors relapsed after prolonged MYC inactivation. Some tumors consistently relapsed after prolonged MYC inactivation (Figure 1C). In summary, tumors behaved in 1 of 3 different manners. Many tumors exhibited sustained tumor regression and were not observed to relapse despite prolonged MYC inactivation (mouse nos. 1137, 1979, 2263, 5719, 5720). Other tumors relapsed in 10% to 50% of the mice (mouse nos. 6780, 6021, 1232, 966, 967), and some tumors relapsed in 100% of the mice (mouse nos. 6814, 6816) after prolongedMYC inactivation (Table 1, Figure2). It was only from the same tumors that had relapsed in vivo that we could isolate tumor cells that were able to continue to proliferate in vitro after 2 to 8 weeks of doxycycline treatment (Table 1). We conclude that although allMYC-induced tumors initially regress upon MYCinactivation, some tumors have the intrinsic ability to relapse and become independent of MYC.
Summary of tumorigenesis experiments showing different abilities of MYC-induced tumors to relapse after prolonged MYC inactivation
| Tumor . | No. experiments . | Relapsed tumors/ total tumors . | Relapse in vitro . |
|---|---|---|---|
| 1137 | 2 | 0/20 | No |
| 1979 | 1 | 0/10 | No |
| 2263 | 1 | 0/10 | No |
| 5719 | 1 | 0/10 | No |
| 5720 | 1 | 0/10 | No |
| 6780 | 2 | 2/20 | Yes |
| 6021 | 1 | 1/10 | ND |
| 1232 | 2 | 3/15 | ND |
| 966 | 1 | 5/10 | ND |
| 967 | 1 | 3/10 | ND |
| 6814 | 2 | 10/10 | Yes |
| 6816 | 3 | 15/15 | Yes |
| Tumor . | No. experiments . | Relapsed tumors/ total tumors . | Relapse in vitro . |
|---|---|---|---|
| 1137 | 2 | 0/20 | No |
| 1979 | 1 | 0/10 | No |
| 2263 | 1 | 0/10 | No |
| 5719 | 1 | 0/10 | No |
| 5720 | 1 | 0/10 | No |
| 6780 | 2 | 2/20 | Yes |
| 6021 | 1 | 1/10 | ND |
| 1232 | 2 | 3/15 | ND |
| 966 | 1 | 5/10 | ND |
| 967 | 1 | 3/10 | ND |
| 6814 | 2 | 10/10 | Yes |
| 6816 | 3 | 15/15 | Yes |
ND indicates not determined.
Tumor regression and relapse on prolonged
MYC inactivation. (A) Transplantation of tumor cells into a syngeneic recipient results in the animal becoming moribund with tumor burden, as demonstrated grossly by tumor invasion into thymus, lymph nodes, spleen, liver, and intestine and histologically by the effacement of spleen architecture. (B) AfterMYC inactivation with doxycycline treatment for 2 weeks, autopsy and histologic examination show evidence of complete tumor regression and the restoration of healthy spleen architecture. (C) After prolonged MYC inactivation, tumors can relapse, again invading the thymus, spleen, lymph nodes, liver, and intestine. Histologic specimens were stained with hematoxylin and eosin. Bottom panels show × 10 and × 60 (inset) images of the spleen. LN indicates lymph node; SC, superficial cervical; P, popliteal; M, mesenteric.
Tumor regression and relapse on prolonged
MYC inactivation. (A) Transplantation of tumor cells into a syngeneic recipient results in the animal becoming moribund with tumor burden, as demonstrated grossly by tumor invasion into thymus, lymph nodes, spleen, liver, and intestine and histologically by the effacement of spleen architecture. (B) AfterMYC inactivation with doxycycline treatment for 2 weeks, autopsy and histologic examination show evidence of complete tumor regression and the restoration of healthy spleen architecture. (C) After prolonged MYC inactivation, tumors can relapse, again invading the thymus, spleen, lymph nodes, liver, and intestine. Histologic specimens were stained with hematoxylin and eosin. Bottom panels show × 10 and × 60 (inset) images of the spleen. LN indicates lymph node; SC, superficial cervical; P, popliteal; M, mesenteric.
Each tumor exhibits a distinct propensity to relapse after prolonged
MYC inactivation. Survival graph of mice receiving transplants of tumors 1137, 967, or 6814. Mice at tumor onset were treated with doxycycline (closed figures) or were not treated (■). All the nontreated mice died within 2 weeks.
Each tumor exhibits a distinct propensity to relapse after prolonged
MYC inactivation. Survival graph of mice receiving transplants of tumors 1137, 967, or 6814. Mice at tumor onset were treated with doxycycline (closed figures) or were not treated (■). All the nontreated mice died within 2 weeks.
Relapsed tumors escape dependence on MYCoverexpression
We considered that there were 2 likely explanations for tumor relapse after prolonged MYC inactivation. First, tumors may find a means to restore MYC overexpression through bypassing the tetracycline regulatory system or activating the expression of endogenous Myc. Second, tumors may acquire novel genetic alterations that can functionally replace the ability of MYCto maintain a tumorigenic phenotype. To address the first possibility, we examined the protein expression of transgenic human c-MYC and endogenous mouse c-, N-, and L-Myc proteins in primary tumors and in tumors that had relapsed after doxycycline treatment. We used antibodies specific for human c-MYC (9E10) for human and mouse c-Myc (c19) or mouse N-Myc (OP13) and L-Myc (c20) (Figure3). As expected, we found that primary tumors expressed the MYC protein in the absence of doxycycline but failed to exhibit expression of the human MYC transgene in the presence of doxycycline (Figure 3). Furthermore, we found that relapsed tumors similarly did not express the MYC transgene in the presence of doxycycline treatment. Some of the primary tumors and relapsed tumors exhibited low levels of expression of endogenous c-Myc, but all failed to express N- or L-Myc in the presence of doxycycline treatment. Note that because c-Myc is located on chromosome (Chr) 15, the small relative increase of c-Myc seen in some of the primary and relapsed tumors could be related to the gain of Chr 15 (see below). However, none of the relapsed tumors exhibited levels of c-Myc higher than those detected in the primary tumors. We conclude that relapsed tumors became independent of MYCoverexpression for the maintenance of their tumorigenic properties.
Relapsed
MYC-induced tumors are independent ofMYC expression. Western blot analysis showed the protein levels of human c-MYC and mouse endogenous c-Myc, N-Myc, and L-Myc in primary and relapsed MYC-induced tumors. Anti-MYC 9E10 antibody detects only the human c-MYC transgene. Anti-Myc c19 antibody detects the human c-MYC transgene and endogenous murine c-Myc. Anti-N-Myc OP13 and anti-L-Myc c20 antibodies detect only N-Myc and L-Myc, respectively. Results from 3 primary tumors are shown in the left panel, and results from the relapsed tumors are shown in the right panel.
Relapsed
MYC-induced tumors are independent ofMYC expression. Western blot analysis showed the protein levels of human c-MYC and mouse endogenous c-Myc, N-Myc, and L-Myc in primary and relapsed MYC-induced tumors. Anti-MYC 9E10 antibody detects only the human c-MYC transgene. Anti-Myc c19 antibody detects the human c-MYC transgene and endogenous murine c-Myc. Anti-N-Myc OP13 and anti-L-Myc c20 antibodies detect only N-Myc and L-Myc, respectively. Results from 3 primary tumors are shown in the left panel, and results from the relapsed tumors are shown in the right panel.
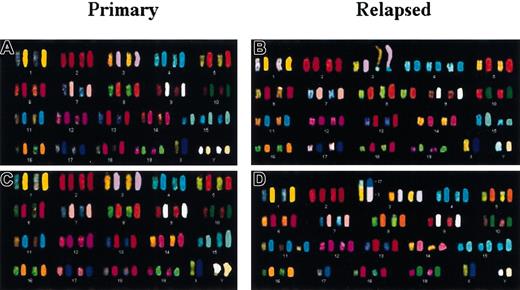
Primary tumors exhibit complex and unique patterns of genomic changes
We considered that the propensity of individual tumors to relapse might reflect the particular combination of genomic alterations in a given tumor. To characterize the patterns of genomic alterations in primary and relapsed transgenic tumors, we performed cytogenetic analysis by inverted DAPI banding and SKY (summarized in Table2). Representative SKY results and examples of the karyotypic abnormalities are shown in Figures4 and 5. In the 16 primary tumors analyzed, clonal chromosomal abnormalities were identified in 14 (88%) cases. In 12 (75%) tumors, we noted numerical chromosomal aberrations. The most common numerical aberrations were gain of Chr 15 in 7 (44%) cases, gain of the Y chromosome in 4 (25%) cases, trisomy 4 or 11 in 5 (31%) cases, and trisomy 11 together with trisomy 15 in 3 (19%) cases. Loss of chromosomes occurred less frequently. The most common of these was the loss of the X chromosome in 2 (13%) cases.
Summary of cytogenetic and SKY findings in 16 primary and 11 relapsed MYC-induced hematopoietic tumors
| Tumor . | Phenotype . | 2n (range) . | Structural aberrations . | Chromosome losses . | Chromosome gains . |
|---|---|---|---|---|---|
| 820 | Primary | 42 (40-45) | — | 14, 17, X | 3, 7, 15, Y |
| 6021 | Primary | 41 | iso(15) | — | 15 |
| 7525 | Primary | 42 (41-43) | t(3;2), 3hsr | — | 11p+, 15 |
| 1137 | Primary | 40 (32-65) | 3, 15hsr | — | — |
| 1979 | Primary | 41 (41-42) | — | — | 17 |
| 2263 | Primary | 42 (39-43) | t(3;17), 3, 15hsr, del(4) | — | 3q+, 11p+, 15 |
| 5719 | Primary | 41 (39-43) | 3, 15hsr | — | 1, 4 |
| 5720 | Primary | 41 (40-41) | t(Y;15), 3, 15, Yhsr | X | 5, Y |
| 1936 | Primary | 40 (40-45) | — | — | — |
| 1936 R1 | Relapsed | 41 (40-42) | — | — | 15 |
| 1937 | Primary | 40 (37-41) | — | — | — |
| 1937 R1 | Relapsed | 41 (37-41) | iso(15), 3, 15hsr | — | 15 |
| 6780 | Primary | 40 (36-44) | — | 16, Y | 4, 5, 11 |
| 6780 R1 | Relapsed | 42 (41-65) | t(3;17), 3hsr | — | 3, 15, Y |
| 1232 | Primary | 41 (41-45) | iso(4), 3, 15hsr | — | 4, 16 |
| 1232 R2 | Relapsed | 41 (38-51) | t(1; 18), 11hsr | 14 | 1q+, 13, 14, 15 |
| 1232 R3 | Relapsed | 42 (41-43) | t(1;18), Rob(3;10), 11hsr | 2 | 1q+, 3, 4, 11p+, 15 |
| 966 | Primary | 41 (38-43) | — | 14, 18 | 15 |
| 966 R1 | Relapsed | 40 (38-56) | iso(4), t(13;3) | — | 2, 4, 10, 19 |
| 967 | Primary | 42 | — | — | 15, Y |
| 967 R1 | Relapsed | 39 (37-40) | 11hsr | 2, 4 | — |
| 967 R2 | Relapsed | 40 (38-41) | t(3;4), iso(4), 3, 15, Yhsr | 8, 11, X | 1, 3, 4, Y |
| 6814 | Primary | 40 (38-43) | t(3;4), t(3;15), 1, 3hsr | 7 | 15, Y |
| 6814 R1 | Relapsed | 42 (39-62) | t(3;1), 3hsr | — | 3, 5, 11p+, 15 |
| 6816 | Primary | 40 (40-41) | t(Y;15), 3, 15, Yhsr | — | — |
| 6816 R3 | Relapsed | 41 (39-43) | t(3;17), t(15;5), 3, 15hsr | 18 | 3, 5, 7, 11p+, 15 |
| 2453 R | Relapsed | 42 (40-44) | t(3;4), t(3;7), t(3;15), 3hsr | 11 | 15, Y |
| Tumor . | Phenotype . | 2n (range) . | Structural aberrations . | Chromosome losses . | Chromosome gains . |
|---|---|---|---|---|---|
| 820 | Primary | 42 (40-45) | — | 14, 17, X | 3, 7, 15, Y |
| 6021 | Primary | 41 | iso(15) | — | 15 |
| 7525 | Primary | 42 (41-43) | t(3;2), 3hsr | — | 11p+, 15 |
| 1137 | Primary | 40 (32-65) | 3, 15hsr | — | — |
| 1979 | Primary | 41 (41-42) | — | — | 17 |
| 2263 | Primary | 42 (39-43) | t(3;17), 3, 15hsr, del(4) | — | 3q+, 11p+, 15 |
| 5719 | Primary | 41 (39-43) | 3, 15hsr | — | 1, 4 |
| 5720 | Primary | 41 (40-41) | t(Y;15), 3, 15, Yhsr | X | 5, Y |
| 1936 | Primary | 40 (40-45) | — | — | — |
| 1936 R1 | Relapsed | 41 (40-42) | — | — | 15 |
| 1937 | Primary | 40 (37-41) | — | — | — |
| 1937 R1 | Relapsed | 41 (37-41) | iso(15), 3, 15hsr | — | 15 |
| 6780 | Primary | 40 (36-44) | — | 16, Y | 4, 5, 11 |
| 6780 R1 | Relapsed | 42 (41-65) | t(3;17), 3hsr | — | 3, 15, Y |
| 1232 | Primary | 41 (41-45) | iso(4), 3, 15hsr | — | 4, 16 |
| 1232 R2 | Relapsed | 41 (38-51) | t(1; 18), 11hsr | 14 | 1q+, 13, 14, 15 |
| 1232 R3 | Relapsed | 42 (41-43) | t(1;18), Rob(3;10), 11hsr | 2 | 1q+, 3, 4, 11p+, 15 |
| 966 | Primary | 41 (38-43) | — | 14, 18 | 15 |
| 966 R1 | Relapsed | 40 (38-56) | iso(4), t(13;3) | — | 2, 4, 10, 19 |
| 967 | Primary | 42 | — | — | 15, Y |
| 967 R1 | Relapsed | 39 (37-40) | 11hsr | 2, 4 | — |
| 967 R2 | Relapsed | 40 (38-41) | t(3;4), iso(4), 3, 15, Yhsr | 8, 11, X | 1, 3, 4, Y |
| 6814 | Primary | 40 (38-43) | t(3;4), t(3;15), 1, 3hsr | 7 | 15, Y |
| 6814 R1 | Relapsed | 42 (39-62) | t(3;1), 3hsr | — | 3, 5, 11p+, 15 |
| 6816 | Primary | 40 (40-41) | t(Y;15), 3, 15, Yhsr | — | — |
| 6816 R3 | Relapsed | 41 (39-43) | t(3;17), t(15;5), 3, 15hsr | 18 | 3, 5, 7, 11p+, 15 |
| 2453 R | Relapsed | 42 (40-44) | t(3;4), t(3;7), t(3;15), 3hsr | 11 | 15, Y |
— indicates none detected.
Different chromosomal abnormalities in primary versus relapsed
MYC-induced tumors. (A-D) SKY showing the complete karyotypes and chromosomal abnormalities in 2 primary and corresponding relapsed tumors. (A) Primary tumor 967 has trisomy 15 and 2 Y. (B) Relapsed tumor 967 R2 contains trisomy 4 and 8, HSRs on Chr 8 and 15, and nonreciprocal translocation t(3;4). Note that most of the metaphases from 967 R2 exhibited a loss of chromosome 8. (C) Primary tumor 6816 displays HSRs on Chr 15 and Y. (D) Relapsed tumor 6816 R3 exhibits trisomy 3, 11p+ and 15, and nonreciprocal translocation t(3;17).
Different chromosomal abnormalities in primary versus relapsed
MYC-induced tumors. (A-D) SKY showing the complete karyotypes and chromosomal abnormalities in 2 primary and corresponding relapsed tumors. (A) Primary tumor 967 has trisomy 15 and 2 Y. (B) Relapsed tumor 967 R2 contains trisomy 4 and 8, HSRs on Chr 8 and 15, and nonreciprocal translocation t(3;4). Note that most of the metaphases from 967 R2 exhibited a loss of chromosome 8. (C) Primary tumor 6816 displays HSRs on Chr 15 and Y. (D) Relapsed tumor 6816 R3 exhibits trisomy 3, 11p+ and 15, and nonreciprocal translocation t(3;17).
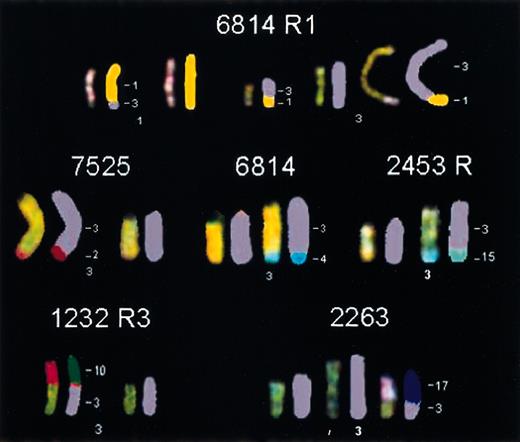
Novel translocations involving chromosome 3 in different
MYC-induced lymphomas. In tumor 6814 R1, a reciprocal translocation, t(3;1), was found. Tumors 7525, 6814, and 2453 R all contained a nonreciprocal translocation in which part of chromosome 2, 4, or 15, respectively, had been translocated to the telomeric end of Chr 3. Tumor 1232 R3 and tumor 2263 harbored a translocation in which the entire chromosome 10 or 17, respectively, was translocated to the proximal part of Chr 3.
Novel translocations involving chromosome 3 in different
MYC-induced lymphomas. In tumor 6814 R1, a reciprocal translocation, t(3;1), was found. Tumors 7525, 6814, and 2453 R all contained a nonreciprocal translocation in which part of chromosome 2, 4, or 15, respectively, had been translocated to the telomeric end of Chr 3. Tumor 1232 R3 and tumor 2263 harbored a translocation in which the entire chromosome 10 or 17, respectively, was translocated to the proximal part of Chr 3.
In addition to the numerical changes, we noted that 4 (25%) tumors exhibited structural aberrations in chromosomes. We observed a nonbalanced translocation involving Chr 3 in 3 (19%) tumors, with the translocation partner originating from different chromosomes in different tumors. One tumor (6816) showed a translocation of Chr Y (t(Y;15)). Notably, only 2 (1936 and 1937) of the 16 (12%) primary tumors had no clonal structural or numerical chromosomal rearrangements, but they had several nonclonal alterations in individual metaphase cells. Tumor 1936 had numerical changes in 3 of 15 metaphases, whereas tumor 1937 exhibited numerical aberrations in 7 of 15 metaphases. The numerical changes observed were all random gains and losses of different chromosomes. Hence, the MYC-induced tumors contain complex unique patterns of genomic abnormalities and evidence for genomic destabilization. Our results underscore that genomically complex and unstable tumors can regress on the inactivation of a single oncogene.
We attempted to identify genomic events that correlated with the propensity of tumors to relapse. We were unable to correlate the specific genomic abnormalities we observed with the ability of tumors to relapse after MYC inactivation (Figure6A). There are several possible explanations. The sample size was likely too small to have enabled discernment of the patterns of gross chromosomal changes associated with a propensity to relapse. Alternatively, the genetic events responsible might not have been detectable by karyotypic analysis, or a nongenetic mechanism might have been responsible.
Different chromosomal aberrations in primary versus relapsed
MYC-induced tumors. (A) Illustration of chromosomal changes in primary tumors organized into groups that never (0%), sometimes (10%-50%), or always (100%) relapse. (B) Illustration of chromosomal abnormalities in primary versus relapsed tumors. Each primary tumor is compared with its relapsed counterpart(s). Gain of chromosome is illustrated in green, loss of chromosome in red, HSR in orange, and translocation in blue. ND indicates not determined
Different chromosomal aberrations in primary versus relapsed
MYC-induced tumors. (A) Illustration of chromosomal changes in primary tumors organized into groups that never (0%), sometimes (10%-50%), or always (100%) relapse. (B) Illustration of chromosomal abnormalities in primary versus relapsed tumors. Each primary tumor is compared with its relapsed counterpart(s). Gain of chromosome is illustrated in green, loss of chromosome in red, HSR in orange, and translocation in blue. ND indicates not determined
Relapsed tumors exhibit novel chromosomal translocations
We noted that all the relapsed tumors exhibited additional chromosomal alterations when compared with their parent primary tumor (Table 2; Figures 4 and 6B). Structural chromosomal abnormalities were observed in 10 (91%) of the relapsed tumors. The most common abnormality was a nonreciprocal translocation involving Chr 3 (6 cases) (Figure 4), with the translocation partner varying between different tumors. In one case part of Chr 1, t(3;1)(H4;?), and in 2 cases part of chromosome 4, t(3;4)(H4;?), had translocated to the distal end of Chr 3, with the breakpoint at 3(H4). The translocation was always accompanied by an amplification of the distal part of Chr 3. An entire Chr 17 had translocated to a partial Chr 3 [+der(3)t(3;17)(G;A-D)] in 2 cases, and Chr 10 had translocated to a partial Chr 3 [+der(3)t(3;10)(G;A-D)] in one case. Two (18%) tumors showed an unbalanced translocation of Chr 1 and 18 [+der(1)t(1;18)(G;A-D)]. Therefore, the ability of a relapsed tumor to become independent ofMYC to maintain its neoplastic properties correlated with the acquisition of a novel chromosomal translocation.
Relapsed tumors also exhibited aneuploidy. The gain of Chr 15 in 8 (73%) cases was the most common numerical aberration. Trisomy 15 was the sole aberration in 1 case. Gain of the Y chromosome occurred in 3 (27%) cases (Table 2). Gain of Chr 4 or 11 was found in 5 (45%) cases, and trisomy 11 was observed with trisomy 15 in all those cases. Partial trisomy of Chr 3 was present in 5 (45%) tumors and was always associated with a translocation involving this chromosome. However, no specific chromosome was found to be consistently lost in the relapsed tumors.
Thus, every relapsed tumor exhibited additional chromosomal rearrangements, both numerical and structural, compared with the primary tumor of origin (Figures 5-6). In 9 of 10 of the relapsed tumors, this structural rearrangement was a previously unidentified translocation. For 2 of the primary tumors (1232, 967), we analyzed 2 independently derived relapsed tumor variants. In both cases, the relapsed tumors originating from the same primary tumor exhibited different chromosomal abnormalities. Tumor 967 R1 contained an amplification of Chr 11, whereas tumor 967 R2 displayed a translocation of Chr 3 and an isochromosome (iso) 4. Tumor 1232 R3 had acquired an additional translocation of Chr 3 compared with tumor 1232 R2 (Table 1, Figure 5). We infer that even in a given primary tumor, more than one genetic event can functionally replace the ability of MYC to maintain tumorigenesis.
Discussion
Many reports document that oncogene-induced tumorigenesis is reversible.16-22 Even brief oncogene inactivation can be sufficient to result in sustained tumor regression.20 Less clear is whether human tumors, which are generally presumed to be more genetically complicated than tumors arising in transgenic mice, would similarly regress with the inactivation of a single oncogene. Further complicating this issue is the recent suggestion that MYCcan induce tumorigenesis in transgenic mice without associated genomic instability.21 23 On the contrary, we found thatMYC-induced hematopoietic tumors are highly genetically complex and genomically unstable. Nevertheless, the inactivation ofMYC alone was sufficient to induce sustained tumor regression. Moreover, each hematopoietic tumor exhibited different complex patterns of chromosomal aberration, suggesting that regardless of the combination of genomic disturbances, tumorigenesis induced byMYC overexpression is reversible with MYCinactivation. We infer from these results that it is likely that even in human tumors the inactivation of individual oncogenes may be effective in inducing sustained tumor regression.
Collectively, our results raise 3 questions. First, why does inactivation of a single oncogene in a cancer that consists of multiple genomic abnormalities induce tumor regression? Second, can we predict when tumors will escape dependence on a given oncogene? Third, what are the mechanisms by which tumors become independent of a given oncogene? To address the first question, we consider there are at least 3 nonmutually exclusive explanations for why oncogene inactivation could cause tumor regression in genetically complex tumors. First, oncogene inactivation can restore the physiologic program of differentiation,16,20 and in this context genetic alterations could be silenced. We have recently shown that by even briefly inactivating MYC, we can induce the differentiation of tumor cells. In this new epigenetic context, the reactivation ofMYC fails to restore the neoplastic features.20Second, sustained oncogene activity may be required to provide for host factors for sustained growth, such as angiogenesis.17,21Although this appears to be true under at least some circumstances, this would not appear to account for our observation that oncogene inactivation can cause the loss of a neoplastic phenotype in vitro.16,20 Third, oncogene inactivation may restore the function of endogenous DNA damage checkpoint mechanisms that initiate senescence or apoptotic programs. Previously, we proposed thatMYC could abrogate the ability of healthy cells to appropriately respond to DNA damage.24 The inactivation ofMYC may restore this function in tumor cells. Perhaps this would permit tumor cells to recognize that they are genomically damaged, resulting in the induction of differentiation, apoptosis, or both. We suspect that the specific mechanisms by which oncogene inactivation induces tumor regression are likely to be influenced by the particular oncogene involved, the combination of genetic events in a given tumor, and the specific type of cancer.
Although MYC inactivation was sufficient to induce tumor regression, we found that tumors occasionally relapse. The tendency of a given hematopoietic tumor to relapse appears to be distinct for each tumor. Some tumors never or rarely relapse, whereas some tumors always relapse after prolonged MYC inactivation. Tumor relapse was associated with the loss of expression of the MYC transgene, without evidence that tumors activated the expression of endogenousMyc genes. We conclude that tumors can acquire the ability to recover their neoplastic properties independent of the expression of even the oncogene that initiated tumorigenesis. Our results suggest mutant genes exist that can functionally substitute for MYCto maintain a neoplastic phenotype.
We were unable to find specific chromosomal abnormalities that correlated with the propensity for a given tumor to relapse uponMYC inactivation. This may reflect that we did not analyze enough primary tumors to discern the specific abnormalities. Alternatively, the genetic alterations responsible for relapse may not be readily detectable by karyotypic analysis and instead may be a consequence of small genetic deletions or mutations. Finally, it is possible that the ability of a tumor to relapse reflects epigenetic features of a tumor, such as its particular differentiation state, which may influence either the consequences of MYCinactivation or the propensity of tumors cells to acquire novel genomic aberrations. Our observations that all relapsed tumors exhibited additional chromosomal abnormalities and that almost all exhibited novel clonal chromosomal translocations strongly suggest that tumors relapse through a genetic mechanism.
The most common abnormality we observed in relapsed tumors was a nonbalanced translocation involving Chr 3. The translocating partner and breakpoint varied among different tumors, suggesting that more than one gene product was involved. Nonreciprocal translocations are considered a hallmark of human cancer25 but are rarely found in human lymphomas and leukemias. Telomeres are generally protected from chromosome fusions.26 This could indicate that in our model system, the telomere of Chr 3 might have been shortened and was unable to protect the end from fusing to another chromosome. The distal part of Chr 3 is homologous to human chromosome 1p22-p31. According to the chromosomal abnormality database,27 this chromosomal region has been reported to undergo allelic loss in different types of tumors, and it also shows balanced chromosomal abnormalities in lymphomas and leukemias. Based on these observations, we propose the existence of an important tumor progression locus in the distal region of Chr 3, which presumably becomes involved in tumor development by promiscuous chromosomal translocations. These translocations may result in the disruption of the healthy function of a gene product, which can at least partially phenocopy the effects of MYC overexpression.
The activation of oncogenes and the loss of function of tumor-suppressor genes are known to cooperate with MYC in tumorigenesis or to enhance tumor formation. These genetic events would be potential candidates for the mechanism by which tumors exhibit the propensity to relapse or to acquire the ability to become independent of MYC overexpression.2,4 Specifically, the loss of p19ARF and p53 function are known to cooperate with MYC in the initial steps of tumorigenesis.11 We examined whether any of the genes known to cooperate with MYC are located on the chromosomes that were altered. The mouse p19ARF locus is located on MMU4. This chromosome was frequently involved in a Robertsonian translocation (Rob).4 This kind of translocation is not thought to cause any genetic alterations because the chromosomal material remains intact. In addition, the p19ARF gene is situated in the middle of the chromosome and is therefore not likely to be altered. Trp53 (p53) is located on Chr 11. In 3 of the relapsed tumors, 2 of which were derived from the same primary tumor, this chromosome was involved in a structural rearrangement that resembled an interstitial C-band together with an amplification of part of the chromosome. Because p53 is located on the middle of the chromosome, it should not be affected by this rearrangement. Thus, it does not appear that p19ARF or p53 function is disrupted through gross chromosomal rearrangements or deletions. We presume that these functions are likely disrupted through other genetic or epigenetic mechanisms, as has been frequently observed inMYC-induced tumors. We are investigating whetherp53 function is disrupted in our tumors.
Some of the chromosomal abnormalities are proximal to oncogenes known to cooperate with MYC. In 2 of the relapsed tumors, the entire Chr 18 had translocated to part of Chr 1. The oncogeneBcl-2 is located on the middle of Chr 1 at 59.8 cM. Overexpression of Bcl-2 blocks many cell death pathways, andBcl-2 is known to cooperate with Myc to promote tumorigenesis.7 The position of Bcl-2corresponds to band D. The breakpoint of the translocation occurred at band D, which might be close to the location of Bcl-2. Because of the imprecision of cytogenetic position, we cannot exclude that Bcl-2 was altered in these 2 tumors and that this alteration contributed to tumorigenesis. However, we failed to observe the increased expression of Bcl-2 by Western blot analysis, making it unlikely that this gene product has been activated (data not shown). In 1 primary and 2 relapsed tumors, there was a translocation involving the fusion of Chr 17 to part of Chr 3 [t(3;17)]. The oncogenes Pim-1 and Pim-2, which have been described earlier to cooperate with Myc,9 10are both located on Chr 17. None of the Pim genes are located near the fusion point, which indicates that their expression is not altered by the translocation in these tumors. Thus, we do not have evidence that a particular oncogene is associated with the chromosomal aberrations we observed.
We considered that because hematopoietic tumors frequently exhibit the activation of oncogenes through their translocation to immunoglobulin or T-cell receptor loci, the chromosomal deletions or translocations we observed could be associated with one of these loci. However, none of the chromosomes, or even regions of chromosomes, in which immunoglobulin or T-cell receptor genes are located were significantly altered or involved in any of the observed translocations. We conclude that the chromosomal translocations observed in the relapsed tumors are likely to be associated with the activation or inactivation of previously uncharacterized genes.
We have shown that tumorigenesis is reversible with the inactivation of a single oncogene, even in genetically highly complex and genomically unstable tumors. These observations make it more likely that the targeted inactivation of oncogenes may be effective in the treatment of human cancers. We demonstrate that the propensity for a given tumor to relapse appears to be an intrinsic property of each tumor. Moreover, the ability of a cancer to become independent of an oncogene was found to be associated with the acquisition of specific chromosomal translocations. Because MYC overexpression can cause genomic destabilization, we speculate that MYC may directly contribute to the generation of these chromosomal translocations. Indeed, we recently found that MYC overexpression may contribute to genomic instability by disrupting double-strand DNA break repair (A.K. et al, manuscript submitted). Thus, although MYC inactivation induces tumor regression, the ability of MYC to cause genomic destabilization may be responsible for the ability of some tumors cells to acquire the compensatory genetic lesions required to avert dependence on MYC for their neoplastic properties. We hope to be able to define the specific genetic contexts in which the inactivation of MYC is sufficient to induce sustained tumor regression, the mechanisms by which MYC facilitates tumor relapse, and the genetic changes responsible for the ability of tumors to escape dependence on MYC.
We thank the members of the Felsher laboratory and Drs Michael Cleary and Joseph Lipsick for helpful suggestions.
Prepublished online as Blood First Edition Paper, November 27, 2002; DOI 10.1182/blood-2002-10-3091.
Supported by National Cancer Institute grants K08-CA75967-01 and R01-CA89305-01, the Lymphoma Research Foundation of America, the Leukemia Research Foundation, the V Foundation, the Esther Ehrman Lazard Faculty Scholar, and the Elsa Pardee Foundation (D.W.F).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dean Felsher, Division of Oncology, CCSR 1105, Stanford University, 269 Campus Dr, Stanford, CA 94305-5151; e-mail: dfelsher@stanford.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal