Abstract
We have analyzed the phenotype, cytokine profile, and mitotic history (telomere length) of monoclonal T-cell expansions in 5 CD3+ T-cell large granular lymphocyte (TLGL) leukemia patients by fluorescence activated cell sorting (FACS) and single-cell polymerase chain reaction (PCR). We confirm that the common phenotype of TLGL leukemia is CD3+CD8+CD45RA+CD27−CD94+(CD57+). Interestingly, the C-type lectin-like type killer cell receptor CD94 was invariably associated with the activating form of its signal-transducing molecule NKG2. Furthermore, when judged by criteria such as interferon gamma (IFN-γ)/tumor necrosis factor (TNF) production, expression of granzyme, FasL, and NKG2D, the TLGL cells had all the features of a cytotoxic effector T cell. Telomere shortening in TLGL cells was in the normal range for CD8+ T cells, indicating that they had not divided significantly more than chronically stimulated CD8+ T cells in healthy individuals. In 25 of 27 controls, cells with a TLGL phenotype occurred at low (1%-3%) frequencies. However, in the other 2 individuals (ages 28-36 years), large stable (> 3 years) monoclonal expansions of CD3+CD8+CD45RA+CD27−CD57+CD94+ NKG2C+ were found which rendered these controls phenotypically indistinguishable from TLGL leukemia patients. We believe that the TLGL clonopathy, rather than being of a neoplastic nature, is more likely an extreme manifestation of the large and stable clonal size characteristic of CD8+ effector cells. Such a TLGL clone consisting of cells without any particular pathologic trait might exist in a considerable number of individuals. Clinical symptoms may occur in individuals in whom the TLGL clone encounters antigen and is triggered to produce large amounts of effector molecules that dysregulate the immune system, which could manifest itself as autoimmunity or as a FasL-mediated neutropenia.
Introduction
Lymphoproliferative disorders of large granular lymphocytes (LGLs) are classified as lymphoid neoplasms.1,2 They are divided into 2 distinct entities: approximately 10% to 20% are of the natural killer (NK) lineage (CD3−CD16+), whereas the others consist of mature CD3+ T cells expressing either an αβ or a γδ T-cell receptor (TCR). NK-LGL leukemia is by far the more aggressive form. Most patients have more than 10 × 109/L LGLs in their blood and a significant percentage dies within 2 months after diagnosis.3 By contrast, CD3+ T-cell LGL (TLGL) leukemia that affects mainly elderly patients progresses in a much milder way. Lymphocytosis is usually modest4 and many patients may even remain asymptomatic.3,5-7 Moreover, clinical complications consist primarily of neutropenia or autoimmune features.8 Therefore, it remains under debate whether this T-cell clonopathy of variable significance9 should be considered as a malignancy or rather as a secondary reactive phenomenon.6
Apart from the low malignancy of TLGL leukemia, there are more indications that the clonal expansion is the result of a perhaps rather common dysregulation of CD8+ T cells. In fact, large oligoclonal expansions of CD8+ T cells are quite frequent in elderly people.10 Such clones that may be maintained by cytokines rather than by antigen11 are of a phenotype similar to that of normal CD8+ effector T cells of which the clonal size is still controlled through apoptosis. The latter is also true for the TLGL clone. In the majority of patients, these cells are CD8+CD45RA+CD27−,6,7,12,13express Fas and FasL,14,15 and may or may not express CD57.6,7 Therefore, they share many features with their normal CD8+ effector T-cell counterparts.16 In fact, except for their typical monoclonality, no marker or combination of markers has been found to date that distinguishes these cells from a subpopulation of normal effector T cells. Moreover, the original diagnostic criterion of a granular lymphocytosis greater than 2000/μL and lasting for more than 6 months5 had also to be revised7 because the size of the TLGL clone may remain limited. Thus, owing to the lack of either an aberrant phenotype or a typical leukemia-like clonal progression, most of the current criteria for TLGL leukemia are based on the presence of a monoclonal T-cell proliferation and/or on clinical symptoms such as neutropenia or autoimmune phenomena.
Recently, it has been reported that TLGL cells express killer cell receptors specific for class I HLA antigens.7,17-19 These receptors, either of the C-type lectin-like type or members of the immunoglobulin (Ig) superfamily have been initially characterized on NK cells as receptors that inhibit their effector function when engaged by their major histocompatibility complex (MHC) ligand on the target cell.20-22 Killer cell immunoglobulin-like receptors (KIRs) with a long cytoplasmic tail (KIRLs)23abrogate the activation pathway initiated by other receptors by recruiting tyrosine phosphatases through an immunoreceptor tyrosine-based inhibition motif (ITIM) in their intracytoplasmic domain. C-type lectin-like inhibitory killer cell receptors consist of heterodimers of CD94 and NKG2A (CD94:NKG2A) of which the latter molecule also harbors an ITIM motive. The inhibitory function of killer cell receptors has been studied extensively and their detection on lymphocytes by fluorescence activated cell sorting (FACS) is often perceived as evidence that the effector function of the cell is down-regulated when these receptors are engaged. However, only a few monoclonal antibodies discriminate the ITIM-bearing killer cell receptors from their activating (ITAM) counterparts that are either very homologous with respect to their extracellular domain (Ig superfamily), or differ only because they are associated with another member of the NKG2 family (CD94:NKG2C, CD94:NKG2E, or CD94:NKG2F, respectively). Hence, little is known about the incidence or the physiologic role of this perhaps more rare activating form of the killer cell receptor.
In this study, we characterized the monoclonal expansion in 5 patients diagnosed with TLGL leukemia. We confirm that the expression of markers such as CD16, CD56, and CD57, once presumed to be characteristic for LGL leukemia, is indeed heterogeneous and that these cells express an array of killer cell receptors. Furthermore, we show that most of these killer cell receptors are activating receptors. As a result, the CD8+ LGL T cell might be readily activated, which could be the cause of the clinical symptoms characteristic of the disease.
Materials and methods
Blood samples and flow cytometry
Peripheral blood mononuclear cells (PBMCs) were obtained from blood samples by Ficoll-Hypaque (Ficoll-Hypaque; Pharmacia, Uppsala, Sweden) density gradient centrifugation and kept in liquid nitrogen until use. Either a FacsCalibur or a FacsStarPlus (both from Becton Dickinson, Mountain View, CA) was used to perform 4-color immune fluorescence and cell sorting, using the following reagents: CD4–allophycocyanin (APC), CD8-APC, CD45RA-biotin, CD94–phycoerythrin (PE) (Pharmingen, Basel, Switzerland), CD16-PE, CD27–fluorescein isothiocyanate (FITC) (Dako, Zug, Swizerland), CD56-PE (Becton Dickinson), and streptavidin-red 670 (Gibco BRL, Basel, Switzerland). In addition, we used 3 monoclonal antibodies, CD57-FITC, OKT8-biotin (ATCC, Manassas, VA), and CD94-Cy5 (39C10), prepared from hybridoma supernatants and coupled with the respective reagents.
Preparation of cDNA
cDNA was obtained by lysing the (single) cells in 10 μL to 15 μL of buffer provided with the Moloney murine leukemia virus (M-MLV) reverse transcriptase kit (Gibco BRL) supplemented with 0.12% Triton X-100 (Fluka, Buchs, Switzerland), 80 U/mL rRNAsin (Promega, Madison, WI), 200 ng/mL oligo-dT (T)20 (Amplimmun, Madulain, Switzerland), 50 nM deoxynucleotide triphosphate (dNTP) (Gibco BRL), and 3 μg t-RNA (Boehringer-Mannheim, Rotkreuz, Switzerland) followed by a 1-hour incubation at 37°C. For further processing, the cDNA was precipitated for 2 hours at −80°C after the addition of 5 μL C2H7NO2 7.5 M (Fluka), 30 μL ethanol 100% (Fluka) using 2 μL acryl as carrier, washed in 150 μL ethanol 70% and dried for 1 hour at room temperature.
cDNA amplification
The procedures are based on methods published by Brady24 and modified by Sauvageau et al.25 In brief, sufficient quantities of cDNA from a single cell were obtained by addition of a 3′ oligo(dA) tail to the cDNA by an incubation of 30 minutes at 37°C with 0.5 mM dATP (2′-deoxyadenosine 5′-triphosphate; Gibco BRL) and 1 U terminal deoxynucleotidyl transferase (Promega) in 5 μL of buffer prepared following the manufacturer's protocol. After denaturation (95°C for 3 minutes), 45 μL polymerase chain reaction (PCR) buffer (10 mM Tris-HCl, pH 8.8, 2 mM MgCl2, 100 nM dNTP [2′-deoxynucleoside 5′-triphosphate; Gibco BRL], 0.005% Triton X-100) containing 1 ng oligo dT(60) (CATGTCGTCCAGGCCGCTCTGGGACAAAATATGAATTCT23) and 5 U Taq DNA recombinant polymerase (Gibco BRL) was added, followed by 5 cycles of PCR (50 seconds at 94°C, 2 minutes at 37°C, 9 minutes at 71°C) and 35 cycles (50 seconds at 94°C, 1.5 minutes at 60°C, and 8 minutes at 71 °C).
PCR
For amplification (35-40 cycles; 30 seconds at 94°C, 45 seconds at 58°C, 60 seconds at 72°C) of the respective genes from 0.5 μL to 1 μL of the amplified cDNA, we used the following primers (40 ng, 1.5 mM MgCl2): glyceraldehyde phosphate dehydrogenase (GAPDH): 5′-GGACCTGACCTGCCGTCTAG-3′, rev-5′-GGCCATGTGGGCCATGAGGTC-3′; CD3: 5′-CGTTCAGTTCCCTCCTTTTCTT-3′, rev-5′-GATTAGGGGGTTGGTAGGGAGTG-3′; CD94: 5′-AGAAATCCAGCCTGCTTCAGCTTC-3′, rev-5′-CACCTTCTCTGCCCCA- AGAAAC-3′; NKG2A(B): 5′-ACTGAACAGGAAATAACCTATGCG-3′, rev-5′-GTCACCCATGGATGATGACTGCTG-3′; NKG2C(E,H): 5′-ACCGAACAGGAAATATTCCAAGTA-3′, rev-5′-GTCACCCATGGATGATGACTGCTG-3′; NKG2D: 5′-CGCTGTAGCCATGGGAATC-3′, rev-5′-AATGTG- TACTAGTCCCATCCAATGA-3′; NKG2F: 5′-ACCGAACAGGAAATATTCCAAGTA-3′, rev-5′-CAGATCAGAGTTCTTCGAAG-3′; 1.5 mM MgCl2, KIR2DL1,2,3 KIR3DL12: 5′-GTGACCTTGTCCTG(CT)AGCTCC-3′, rev-5′-GATGAAGAGGA(AT)GATGACCACTG-3′; KIR2DL4: 5′-GTGACCTTGTCCTG(CT)AGCTCC-3′, rev-5′-GCCACTGAGTACCTAATCACAG-3′; KIR2DS1,2,3,4 KIR3DS1: 5′-TACAGATGCTTCGGCTCTTTC-3′, rev-5′-GGAGGATGGTGAAAGGGATTT-3′; tumor necrosis factor alpha (TNF-α): 5′-GTGGACCTTAGGCCTTCCTC-3′, rev-5′-ACGGAAAACATGTCT- GAGCC-3′; interferon gamma (IFN-γ): 5′-GCAGAGCCAAATTGTCTCCT-3′, rev-5′-ATGCTCTTCGACCTCGAAAC-3′; FasL: 5′-GAGCCAGACAAATGGAGGAA-3′, rev-5′-GAAGTGAAGATGCTGCCAGTG-3′; granzyme B: 5′-CCTGGGAAAACACTCACACA-3′, rev-5′-GCCATTGTTTCGTCCATAGG-3′; IL-15secreted: 5′-CATGTCTTCATTTTGGGCTGT-3′, rev-5′-TGCATCTCCGGACTCAAGTG-3′; CD14: 5′-AGCTCAGAGGTTCGGAAGACTTA-3′, rev-5′-ATCTCCACCTCTACTGCAGACACA-3′. For the T-cell receptor (TCR) analysis (spectratyping), all conditions as well as the sequences of the 6-FAM, HEX, and TET dyes 5′-labeled primers (Amplimmun) corresponding to the 21 variable segments of the TCR beta chain used in this study have been published previously.26 Data analysis was performed using the Genescan analysis software.
Telomere fluorescence in situ hybridization and flow cytometry (flow FISH)
All procedures have been described before.27,28FACS analysis was performed with a gate on lymphocytes and telomere fluorescence was calculated by subtracting the mean fluorescence of the background control (no probe) from the mean telomere fluorescence obtained from cells hybridized with the telomere probe after calibration with FITC-labeled fluorescent calibration beads (Quantum-24 Premixed; Flow Cytometry Standards, San Juan, Puerto Rico) and conversion into molecules of equivalent soluble fluorochrome (MESF) units. To calculate the telomere length (in base pairs) from the telomere fluorescence in MESF units, the slope (y = 0.019x) of the calibration curve previously described27 was used in the equation bP = MESF × 0.026 × 0.019 × 103.
Results
Phenotypes of TLGL cells
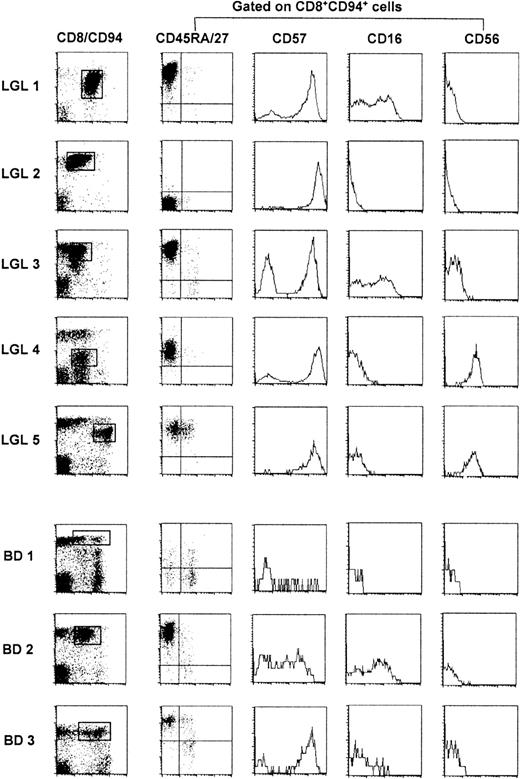
We studied 5 patients diagnosed with TLGL leukemia based on the invariable presence of high numbers of CD3+ LGLs in their blood. During the study, their disease course remained indolent and only one patient (LGL 1) needed treatment with antibiotics for recurrent infections. Lymphocytosis (7 ± 0.9, range 5.9-7.5 G/L) was observed in all but one patient (LGL 5). The left panel of Figure 1 shows that the majority of lymphocytes were CD8+ T cells of which at least 30% to 90% expressed the C-type lectin-like killer cell receptor CD94.18
FACS analysis of TLGL cells and of their CD8+CD94+ counterparts in 3 blood donors.
BD 1 is representative for most healthy blood donors whereas BD 2 and BD 3 are examples of blood donors with high numbers of CD8+CD94+ T cells. Cells of patients with TLGL leukemia are marked LGL 1-5; blood donor cells are marked BD 1-3.
FACS analysis of TLGL cells and of their CD8+CD94+ counterparts in 3 blood donors.
BD 1 is representative for most healthy blood donors whereas BD 2 and BD 3 are examples of blood donors with high numbers of CD8+CD94+ T cells. Cells of patients with TLGL leukemia are marked LGL 1-5; blood donor cells are marked BD 1-3.
CD8 expression was either normal (LGL 2, LGL 3), slightly reduced (LGL 1, LGL 5), or dim (LGL 4). In the latter patient, these cells coexpressed the CD4 molecule. To study other phenotypic markers of LGL cells, we gated on the virtually monoclonal population (Figure2) of CD8+CD94+ cells in LGL 1, LGL 2, LGL 3, and LGL 5 or on the CD8dimCD94+ cells in LGL 4. As shown in the second panel of Figure 1, these cells were almost homogeneous with respect to the expression of the CD45R-isoforms and of CD27. The TLGLs in LGL 1, LGL 3, and LGL 5 were CD8+CD45RAbrightCD27−, whereas in LGL 2 and LGL 4, the T cells were CD45RA−CD27− or CD45RAintermCD27−, phenotypes that are reminiscent of genuine effector T cells.29,30 As reported by others,7 the expression of CD57, CD16, and CD56 was heterogeneous. With some previously observed31 intraclonal variation in LGL 1, LGL 3, and LGL 4, the LGLs of all 5 patients expressed CD57. LGL 4 and LGL 5 were CD56+ while a part of the TLGL in LGL 1 and LGL 3 expressed CD16. Hence, based on our data and those in the literature,7 the most universal TLGL phenotype appears to be CD3+CD8+CD45RA+CD27−CD94+( CD57+).13 18
Spectratype analysis of the T-cell repertoire in LGL 1-5 and in the 2 blood donors with large expansions of CD8+CD94+ T cells.
The analysis has been performed for 21 BVs in 8 pools containing the specific primers for 2-4 BV families. Only the data of the pool with the BV depicted in the figure containing the BV of the dominant clone are shown. For each graph, the number in parentheses represents the size of the amplified fragment by Genescan analysis. The double peak in BD 3 has been authenticated by analysis with BJ-specific primers. The clone BV3197 uses the BJ2.5 segment, whereas the clone BV3201 uses the BJ2.7 segment.
Spectratype analysis of the T-cell repertoire in LGL 1-5 and in the 2 blood donors with large expansions of CD8+CD94+ T cells.
The analysis has been performed for 21 BVs in 8 pools containing the specific primers for 2-4 BV families. Only the data of the pool with the BV depicted in the figure containing the BV of the dominant clone are shown. For each graph, the number in parentheses represents the size of the amplified fragment by Genescan analysis. The double peak in BD 3 has been authenticated by analysis with BJ-specific primers. The clone BV3197 uses the BJ2.5 segment, whereas the clone BV3201 uses the BJ2.7 segment.
To study the prevalence of T cells with a TLGL phenotype in healthy controls, we analyzed the expression of the respective surface markers in a panel of 27 healthy blood donors (BD). In 25 of 27 of the individuals, we detected low numbers (1%-5%, see BD 1, Figure 1, for a representative example) of CD8+CD94+ T cells that coexpressed the various combinations of CD45RA, CD45RO, CD27, and CD57 that can be expressed by effector/memory CD8+ T cells.16 Among these cells, a minor part was indeed CD45RA+CD27− (effector cells), and thus of the TLGL phenotype. However, this population was clearly polyclonal with respect to its TCR usage (data not shown). Rather unexpectedly, in 2 of the 27 healthy blood donors (BD 2, age 28 years and BD 3, age 36 years) we found a significant expansion of CD8+CD94+ T cells that did express a homogeneous CD45RA+CD27− effector phenotype. Moreover, the cells expressing CD16 as well as CD57 persisted in sequential samples over a period of more than 3 years, which makes these blood donors phenotypically indistinguishable from TLGL leukemia patients.
Large expansions of CD8+CD94+ T cells are monoclonal in nature
Figure 2 shows the spectratype analysis of the TCR BV families in the LGL leukemia patients as well as in BD 2 and BD 3. Analysis was performed on mononuclear cells (MNCs; left panel) as well as on the cells with the TLGL phenotype (right panel) that were FACS-sorted using the windows shown in Figure 1. Spectratyping of the MNC samples of LGL 1 and LGL 2 with a pool of primers specific for BV9, BV15, BV17, and BV21 generated a peak that entirely dominated the 4 Gaussian curves of 7 to 9 peaks that these primers generate in healthy controls.26 Furthermore, the CD16+ and CD16− cells in LGL 1 used the same TCR, showing that this surface marker can be expressed with a similar intraclonal variation as has been reported for CD57.6 Also in the other 3 patients, who still had cells with a heterogeneous phenotype (Figure 1) and with a normal polyclonal TCR repertoire (Figure 2, left panel), the cells sorted on the basis of their CD8+CD94+ phenotype were also practically monoclonal. This was in sharp contrast to the individuals with only minor populations of T cells with a TLGL phenotype where the TCR repertoire of these cells was polyclonal (data not shown). Interestingly, the CD8+CD94+ cells in the 2 blood donors with the persisting high numbers of CD8+CD94+ T cells were also monoclonal, or in the case of BD 3, biclonal. Altogether, this suggests that high numbers of CD8+CD94+ T cells are usually caused by the expansion of only one or 2 cells. Although such a massive expansion might be rare, it still occurs frequently enough to cause a monoclonal, or as has been showed in 20% of the cases,32 a biclonal expansion in a relatively high number of individuals.
The telomere lengths of TLGL cells are comparable to those of chronically stimulated CD8 effector cells
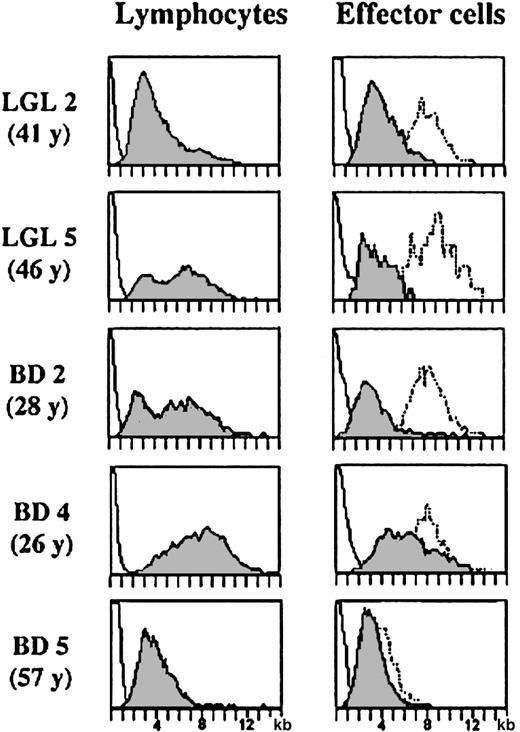
Telomeres are specialized structures at the end of chromosomes that shorten with each cell division. Therefore, the telomere length in T cells reflects their mitotic history and the number of divisions of a memory/effector cell can be estimated by comparing its telomere length with that of a naive T cell. CD8 effector cells are terminally differentiated cells that undergo very few cell divisions after being generated from central memory cells. Expansion of the antigen-specific clone occurs in the pre-effector/central memory stage and therefore the telomere length of memory and effector cells is quite similar.27 We argued that if a TLGL cell were leukemic, the constant ratio between the telomere length of normal CD8 effector cells and that of the other leukocytes in the blood of the same individual33 would be lost. Interestingly, the telomere data of LGL 2, LGL 5, and BD 2 (similar results were obtained for LGL 1, LGL 3, LGL 4, and BD 3) in Figure3 show that this is not the case.
Comparison of telomere length in lymphocytes and CD8+CD94+ effector T cells in TLGL leukemia patients and in lymphocytes and CD8+ effector cells in healthy blood donors.
Telomere length in lymphocytes (left panel) and in CD8+(CD94+) effector T cells (right panel) in the individuals with large expansions of TLGL cells (LGL 2, LGL 5, and BD 2) as well as in a young (BD 4, 26 years of age) and in an elderly (BD 5, 57 years of age) blood donor. For comparison, the telomere lengths in the monocytes (dotted line, right panel) of the same individual are shown.
Comparison of telomere length in lymphocytes and CD8+CD94+ effector T cells in TLGL leukemia patients and in lymphocytes and CD8+ effector cells in healthy blood donors.
Telomere length in lymphocytes (left panel) and in CD8+(CD94+) effector T cells (right panel) in the individuals with large expansions of TLGL cells (LGL 2, LGL 5, and BD 2) as well as in a young (BD 4, 26 years of age) and in an elderly (BD 5, 57 years of age) blood donor. For comparison, the telomere lengths in the monocytes (dotted line, right panel) of the same individual are shown.
In all 5 patients as well as in the 2 blood donors with the large TLGL-like expansions, the telomere length of the lymphocytes showed a bimodal distribution with population sizes of cells with short telomeres being proportional to population sizes of monoclonal cells found by spectratyping. Importantly, the difference in telomere length between the expanded clone and the other lymphocytes or monocytes in the same individual was comparable (Figure 3). In LGL 2, representative of a patient with a dominant monoclonal population of CD8+ cells, the bimodal distribution was completely biased toward the TLGL CD8+ effector cells with short telomeres, whereas in LGL 5 and BD 2, the number of cells with short telomeres was less. However, in all individuals, the difference between the TLGL cells and the monocytes (dotted line, right panel) was 4 kb to 5 kb, which is comparable to the difference between effector cells and monocytes in healthy controls.33 Furthermore, in all 7 individuals, the length of the telomeres of the expanded clone was similar—4.3 kb ± 0.7 kb, which was nearly identical to the CD8 effector cells with short telomeres accumulating in healthy individuals through life. This is shown in the 2 lower panels in Figure 3. In the younger individual, BD 4, the lymphocytes as well as the CD8+ effector cells showed a considerable variation in telomere length. However, some of the CD8+ effector cells had already shortened their telomeres to a size comparable to that of the TLGL cells in the patients. Furthermore, in the elderly blood donor, BD 5, who had high numbers of CD8 effector cells in his blood, most cells had telomeres of a size in the range of the TLGL cells. Therefore, we think that with respect to their mitotic history, TLGL cells are not very different from normal effector T cells that have considerably shortened their telomeres after several rounds of antigenic stimulation.
TLGL cells express ITAM-harboring killer cell receptors and share many features with functionally active CD8+ effector cells
Clinical symptoms of LGL leukemia point to a dysregulation of the immune system. Moreover, the neutropenia might be a result of the high levels of soluble Fas ligand that are produced by the patient's TLGL cells.34 Because these phenomena are hardly compatible with cells expressing CD94 inhibitory receptors with ubiquitous ligands (HLA-E), we determined which NKG2 molecules were expressed by the TLGL cells. In addition, we measured whether the KIRs that had been detected through FACS analysis in LGL 3, LGL 5, and in the 2 blood donors were inhibitory or activating receptors. Because a sort of CD8+CD94+ cells would not yield a 100% pure TLGL population and CD8+CD94+ non-LGL cells might effect the results, we analyzed the killer cell receptors with a PCR method24 that allows the detection of several genes in a single cell. To ascertain that the cell analyzed was a member of the TLGL clone, we determined the TCR BV usage of the single cells FACS-sorted into separate tubes on the basis of the windows of Figure 1. Subsequent analysis was performed on the first 10 cells in which we detected the TCR BV–spectratype band of the dominant clone as the sole TCR transcript. The lower panel (controls) of Table1 shows that by this method we detected a CD3ε transcript in 60% to 90% and a CD94 transcript in approximately 50% of the cells expressing the clonotypic BV. Obviously, this indicates the method's lack of sensitivity in detecting minute amounts of RNA, because cells expressing a TCR should also transcribe the CD3 molecule that is required for its expression, while CD94 is very likely transcribed in CD94+ cells. Therefore, for the killer cell receptor genes for some of which no transcripts were detected in any of the single cells analyzed, we also analyzed the transcript in 10 cells sorted with the windows of Figure 1as controls. Table 1 shows that in 6 of 7 TLGL-like cells, transcripts for one of the ITAM-associated forms of the NKG2 molecules (NKG2C/D/E/F) were found, while only LGL 5 coexpressed NKG2A. Furthermore, all TLGL cells expressed NKG2D, the activating killer cell receptor not associated with CD94. As expected, no KIR transcripts were found in LGL 1, LGL 2, and LGL 4, who were negative by FACS analysis. In the 2 FACS-positive patients, LGL 3 and LGL 5, as well in BD 2 and BD 3, transcripts for KIRL and for the activating KIRs with a short cytoplasmic tail (KIRS) were found. Interestingly, the coexpression of the inhibitory KIRL in LGL 3 was without functional consequences because the ligands (HLA-Bw4 and HLA-A*030120) for the KIR3D2 receptor detected by FACS were not expressed by the patient. We do not know whether this was also the case in LGL 5 and in the 2 blood donors. Here, the cells expressed an array of different KIRs, which made it impossible by our methods to determine whether the different receptors that would see their HLA ligands harbored an ITIM or an ITAM motive.
Killer cell receptor and effector cell marker transcripts in single CD8+CD94+ T cells expressing the same TCR as the respective dominant clones in LGL 1-5 and BD 2-3
| . | LGL 1 . | LGL 2 . | LGL 3 . | LGL 4 . | LGL 5 . | BD 1 . | BD 2(1)* . | BD 2(2)† . |
|---|---|---|---|---|---|---|---|---|
| T-cell receptor‡ . | BV17 . | BV9 . | BV13 . | BV12 . | BV8 . | BV7 . | BV3 . | BV3 . |
| (163)1-153 . | (154) . | (176) . | (196) . | (164) . | (227) . | (197) . | (201) . | |
| Killer cell receptors | ||||||||
| NKG2A | 0/101-155 | 0/10 | 0/10 | 0/10 | 3/10 | 0/10 | 0/5 | 0/6 |
| NKG2A 101-154 | − | − | − | − | + | − | − | − |
| NKG2C/E/F# | 5/10 | 2/10 | 5/10 | 0/10 | 1/10 | 4/10 | 2/5 | 3/6 |
| NKG2C/E/F 101-154 | + | + | + | − | + | + | + | + |
| NKG2D | 6/10 | 4/10 | 4/10 | 2/10 | 1/10 | 3/10 | 0/5 | 3/6 |
| KIRL | 0/10 | 0/10 | 6/10 | 0/10 | 7/10 | 2/10 | 0/5 | 3/6 |
| KIRL 101-154 | − | − | + | − | + | + | + | + |
| KIRS | 0/10 | 0/10 | 4/10 | 0/10 | 1/10 | 4/10 | 0/5 | 3/6 |
| KIRS 101-154 | − | − | + | − | + | + | + | + |
| Effector cell markers | ||||||||
| Granz B | 10/10 | 8/10 | 10/10 | 6/10 | 7/10 | 9/10 | 4/5 | 5/6 |
| IFNγ | 7/10 | 5/10 | 7/10 | 4/10 | 0/10 | 4/10 | 0/5 | 0/6 |
| TNFα | 4/10 | 6/10 | 5/10 | 3/10 | 6/10 | 4/10 | 3/5 | 4/6 |
| FasL | 1/10 | 2/10 | 4/10 | 1/10 | 2/10 | 2/10 | 1/5 | 0/6 |
| Controls | ||||||||
| CD3 | 6/10 | 9/10 | 8/10 | 8/10 | 9/10 | 10/10 | 3/5 | 3/6 |
| CD94 | 9/10 | 6/10 | 5/10 | 7/10 | 3/10 | 7/10 | 1/5 | 1/6 |
| . | LGL 1 . | LGL 2 . | LGL 3 . | LGL 4 . | LGL 5 . | BD 1 . | BD 2(1)* . | BD 2(2)† . |
|---|---|---|---|---|---|---|---|---|
| T-cell receptor‡ . | BV17 . | BV9 . | BV13 . | BV12 . | BV8 . | BV7 . | BV3 . | BV3 . |
| (163)1-153 . | (154) . | (176) . | (196) . | (164) . | (227) . | (197) . | (201) . | |
| Killer cell receptors | ||||||||
| NKG2A | 0/101-155 | 0/10 | 0/10 | 0/10 | 3/10 | 0/10 | 0/5 | 0/6 |
| NKG2A 101-154 | − | − | − | − | + | − | − | − |
| NKG2C/E/F# | 5/10 | 2/10 | 5/10 | 0/10 | 1/10 | 4/10 | 2/5 | 3/6 |
| NKG2C/E/F 101-154 | + | + | + | − | + | + | + | + |
| NKG2D | 6/10 | 4/10 | 4/10 | 2/10 | 1/10 | 3/10 | 0/5 | 3/6 |
| KIRL | 0/10 | 0/10 | 6/10 | 0/10 | 7/10 | 2/10 | 0/5 | 3/6 |
| KIRL 101-154 | − | − | + | − | + | + | + | + |
| KIRS | 0/10 | 0/10 | 4/10 | 0/10 | 1/10 | 4/10 | 0/5 | 3/6 |
| KIRS 101-154 | − | − | + | − | + | + | + | + |
| Effector cell markers | ||||||||
| Granz B | 10/10 | 8/10 | 10/10 | 6/10 | 7/10 | 9/10 | 4/5 | 5/6 |
| IFNγ | 7/10 | 5/10 | 7/10 | 4/10 | 0/10 | 4/10 | 0/5 | 0/6 |
| TNFα | 4/10 | 6/10 | 5/10 | 3/10 | 6/10 | 4/10 | 3/5 | 4/6 |
| FasL | 1/10 | 2/10 | 4/10 | 1/10 | 2/10 | 2/10 | 1/5 | 0/6 |
| Controls | ||||||||
| CD3 | 6/10 | 9/10 | 8/10 | 8/10 | 9/10 | 10/10 | 3/5 | 3/6 |
| CD94 | 9/10 | 6/10 | 5/10 | 7/10 | 3/10 | 7/10 | 1/5 | 1/6 |
Clone 1 of BD 2 (TCR: BV3BJ2.5).
Clone 2 of BD 2 (TCR: BV3BJ2.7).
Only cDNAs from single cells sorted with windows of Figure 1containing the transcript of the LGL-TCR have been analyzed.
Characteristic migration by Genescan.
Number of positive cells per number of cells tested.
Presence (+) or absence (−) of the respective transcript in the cDNA prepared from 10 cells sorted with windows of Figure 1.
#PCR with primers for the 3 activating NKG2 molecules.
The middle panel of Table 1 shows that the same cells with most of their killer cell receptors in activating form expressed granzyme B, IFN-γ, and TNF-α, as well as FasL, all typical for TH1-type CD8+ effector cells. It should be noted that our analysis has been done directly on individual cells ex vivo. It is therefore likely that TLGL cells are highly active effector cells producing a plethora of cytokines that may be at the origin of the autoimmune phenomena that are associated with LGL leukemia.
Discussion
CD3+ LGL leukemia is a monoclonal lymphoproliferative disorder that is currently classified as a lymphoid neoplasm.1 However, whereas the neoplastic character of NK-LGL leukemia is evident, this is not so for monoclonal expansions of large granular T lymphocytes of which the clinical and hematologic characteristics are far from being typical for neoplastic disease. For instance, the number of TLGL cells may already stabilize below 2000/μL and spontaneous regressions do occur. Furthermore, in contrast to malignant cells, the TLGL cell is highly differentiated, usually without chromosomal abnormalities,9,35 and invasion of the bone marrow, which is characteristic for other leukemias, is rarely observed.36 In addition, benign clonal expansions of CD8+ T cells are rather common. Hence, the question is where to draw the line between the truly transformed T lymphocyte on the one hand and a normal T cell escaping from homeostatic control on the other.
In this study, we show that the most common phenotype of expanded monoclonal T-cell populations in patients diagnosed with LGL leukemia is CD3+CD8+CD45RA+CD27−CD94+, whereas the expression of CD57 is variable. This largely confirms other reports on a more restricted series of surface markers.7We extend these findings by showing that the TLGL cells express the activating receptor NKG2D and that the killer cell receptor CD94 is associated with the activating form of NKG2. As a result, the response of these cells will be boosted by HLA-E, the ubiquitous ligand of CD94, or by the stress-induced ligands for NKG2D such as MHC class I–related chains A (MICA)37 or the UL binding proteins (ULBPs).38 Thus, TLGL cells are T cells prone to activation and loaded with effector molecules such as granzyme and FasL with a high potential of dysregulating the immune system or inducing neutropenia through FasL-induced apoptosis.34 It should be noted however that none of the characteristics of TLGL cells or the combination thereof is unique. In fact, except for their clonal size, TLGL cells appear to be indistinguishable from common CD8+effector cells for all parameters studied to date. This also holds for the expression of the NKG2D receptor and of the association of CD94 with the activating form of NKG2 reported in this study. CD8 effector cells express NKG2D,37 and also, in healthy controls, many of them seem to express the activating form of CD94 (L.A., manuscript in preparation). To our knowledge, all features of TLGL cells reported to date are by no means anomalous on an individual cell basis. They only seem to be aberrant because in healthy individuals, TLGL-like CD8+ T cells coexpressing CD16, CD94:NKG2C, or a KIR such as ILT219 occur only at low frequencies.
The reason why TLGL is intuitively classified as a neoplasm is the presence of the large monoclonal population of T cells. Yet, this might be less conspicuous for CD8+ T cells than it is for other leukocytes. The count of cells with a TLGL phenotype in healthy individuals ranges from 0.2 × 109/L to 0.4 × 109/L and this population is considerably less diverse than other T-cell populations.39,40 Moreover, we noticed that higher numbers of cells with a TLGL phenotype in healthy controls were mainly owed to the expansion of only a few clones that persisted for many years. It has been shown that more than 50% of the CD8+CD27−CD28− effector T cells may be monoclonal,40 which matches the finding that in healthy controls single CD8+ T-cell clones often represent more than 10% of the total T-cell population.41 Hence, stable monoclonal expansions of 0.1 × 109/L to 0.5 × 109/L are probably not exceptional and, although the difference with the clonal size of a TLGL clone may appear critical to the hematologist, it might be less so for a T cell that needs only a couple of divisions to reach this size.
Several reports have suggested that the abnormal clonal size of TLGL is owed to a defect in apoptosis,3,15,42 the pathway of programmed cell death confining the clonal size of CD8+ T cells. However, TLGL cells are only refractory to apoptosis when tested directly ex vivo but become sensitive after a short culture in vitro,15,43 which again is not so different from normal CD8+ effector cells.43 Likewise, the expression of the α chain of the IL-15 receptor suggested to be implicated in the expansion of the TLGL clone44 is in fact very characteristic for CD8+ effector/memory T cells. One could even argue that the responsiveness of the TLGL clone to IL-15 is a strong indication that the cell is still under the control of the regulator of the survival of CD8+ effector cells45 and therefore is not malignant.
It is not clear why in the same individual only a few CD8+T cells escape from the homeostatic mechanisms. With age, a continuous spectrum of different sizes of persisting CD8+CD27−CD28− clones emerges.10 After bone marrow transplantation, a relatively high number of clones expand and become indistinguishable from TLGL cells with respect to phenotype and function.12 Such a clonal expansion is driven by antigen46 and the large clonal size is probably the result of the absence of other T cells competing for the available IL-7 and IL-15 that control homeostasis. Interestingly, these clones persist for years after transplantation even after the normal T-cell compartment has been restored. Hence, an abnormal clonal size may be maintained without a general defect in the homeostatic control of the T-cell compartment. This also holds for TLGL leukemia patients in whom the spectratypes of the BVs other than BV of the TLGL clone remain normally distributed (Figure 2). Although another as-yet-unknown intrinsic defect of TLGL clones cannot be excluded, we believe that the large clonal size in TLGL simply represents the extreme of the spectrum of clonal sizes occurring in healthy individuals. The latter is also suggested by the fact that in approximately 20% of the patients diagnosed with TLGL leukemia, 2 clones of comparable size are found,32 which would be highly unlikely if TLGL leukemia would be a genuine leukemia.
If TLGL cells resemble normal CD8+ effector cells, what could then be the difference between an apparently healthy individual such as BD 2 or BD 3 with a large expansion of CD8+ effector T cells and a TLGL leukemia patient? Before trying to answer this question, one should not forget that the current criteria for TLGL leukemia fit to many individuals without overt disease.3 5-7 For instance, in our group of patients, only LGL 1 (neutropenia, recurring perirectal abscesses) and LGL 5 (autoimmune polysynovitis) had symptoms that may have hinted at TLGL leukemia. The monoclonal expansions in LGL 2 (upper airway infection without neutropenia) and in LGL 3 (diabetes, chronic obstructive pulmonary disease) were detected only after diagnostic procedures of which the indication was unrelated to TLGL leukemia, whereas for LGL 4 (no clinical symptoms, routine control) the diagnosis was entirely fortuitous. Therefore, many TLGL leukemia patients without severe disease may in fact not be so different from the blood donors BD 2 and BD 3 who were “fortuitously diagnosed with TLGL leukemia” while serving as controls in our study. Hence, the question asked above should be rephrased into “why do so few individuals with large monoclonal T-cell populations suffer from severe disease?”
Lamy and Loughran have suggested3 that the clinical manifestations of TLGL leukemia are caused by overproduction of Fas-L or of proinflammatory cytokines, which may subsequently lead to neutropenia and autoimmunity. We believe that it is indeed likely that high numbers of activated and ubiquitous3 effector cells are able to disrupt immune processes or boost autoimmune phenomena that otherwise would remain subclinical for years. However, because most expansions of effector T cells do not cause clinical manifestations, the activation state of the TLGL clone, that is, the number of triggers received by its TCR and/or by its activating killer cell receptors, must also be critical. Whether large clonal expansions of CD8+ effector cells will cause clinical symptoms may thus simply depend on how frequently the antigen recognized by the clone is encountered. When a virus has been at the origin of the initial expansion, reinfection, or in the case of a chronic infection, reactivation of the same virus may trigger the clone. Under these circumstances, additional signals would be received by the activating killer cell receptors specific for stress-induced ligands. Owing to the extremely high numbers of virus-specific cells, the virus will be efficiently dealt with so that all classical symptoms of a viral infection may remain unnoticed. By contrast, the sequel of the activation of so many effector cells will be evident. In these patients, the clinical symptoms of TLGL leukemia would be nothing more than an excessive manifestation of the autoimmune phenomena and/or neutropenia that may occur during common viral infections.
We thank Bertrand Huard for stimulating discussions and critical reading of this manuscript.
Prepublished online as Blood First Edition Paper, December 12, 2002; DOI 10.1182/blood-2002-08-2408.
Supported by a grant from the Swiss National Science Foundation (no. 3100-65′357.01) by the Dr Henri Dubois-Ferrière-Dinu Lipatti Foundation and by the Fondation pour la Lutte contre le Cancer.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Eddy Roosnek, Unité d'Immunologie de Transplantation, Hôpital cantonal universitaire de Genève, 24 rue Micheli-du-Crest, CH-1211 Genève 14, Switzerland; e-mail:roosnek@medecine.unige.ch.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal