Abstract
JunB is a component of the Jun family genes of the activating protein-1 transcription factors that are important in the control of cell growth and differentiation and neoplastic transformation. Recently, it was demonstrated that transgenic mice specifically lacking JunB expression in the myeloid lineage developed a myeloproliferative disease, eventually progressing to blast crisis that resembled human chronic myeloid leukemia (CML). To gain further insights into the role of JunB in human CML, we examined peripheral blood from 17 healthy individuals and CML patients (11 in blastic crisis and 21 in chronic phase) by real-time quantitative reverse transcription–polymerase chain reaction analysis for the expression of JunB. The results showed the expression levels of JunB were significantly impaired in CML cases (blastic crisis < chronic phase < normal). Mutational analysis of the whole gene and methylation analysis of cytosine-phosphate guanosine (CpG) sites at the promoter area were further performed to investigate the possible mechanisms. However, no mutation was found within the coding region or the 9 flanking evolutionarily conserved regions in all CML cases. Interestingly, in the promoter area of JunB gene, most of the CpG sites were methylated in CML cases; in contrast, none of these CpG sites were methylated in normal cases. Demethylation by treatment of hypermethylated K562 cells with 5′-aza-2′-deoxycytidine resulted in partial reactivation of JunB expression. Our results suggest that the down-regulated JunB expression in CML was due to the inactivation of JunB gene by methylation and the differential expression was correlated to the ratio of cells being methylated.
Introduction
JunB is a component of the activator protein 1 (AP-1), which consists of Jun (JunB, c-Jun, and JunD), Fos (c-Fos, FosB, Fra-1, Fra-2), and activating transcription factor (ATF) families. The 3 Jun proteins form dimers with each other or with members of related Fos and ATF families to constitute the AP-1 transcription factor, which converts extracellular signals into changes in the transcription of specific target genes.1JunB gene is constitutively expressed in human mature granulocytes2 and its expression promotes myeloid differentiation.3JunB gene is an immediate early or primary response gene that is induced by polypeptide growth factors, cytokines, and chemical agents in various cell types, including myeloid,4 lymphoid,5liver,6 neuronal,7 fibroblast,8and epidermal9 lineages. Initially, JunB was suggested to function as general repressor of AP-1–mediated transactivation.10 However, it was later shown that depending on the interacting partner and the promoter context, JunB also selectively regulates transcription either in a positive or negative way.11 12
Targeted disruption of the junB locus in mice demonstrated that lack of JunB causes early embryonic lethality with defects in placentagenesis and vascularization.13 In contrast, constitutive JunB overexpression from the human ubiquitin C promoter has no significant effects in Ubi-junB transgenic mice.14 JunB has also been shown to negatively regulate cell proliferation by activating the p16INK4a inhibitor15 and decreasing cyclin D1 expression.16 The absence of JunB has also been shown to result in down-regulation of p16 and in increased levels of c-Jun, a potent activator of cell proliferation that regulates the p53 pathway.17
Recently, Passegué et al18 have developed transgenic mice that uniformly expressed the junB gene (Ubi-junB) to rescue the lethality of JunB−/− knockouts. These JunB−/−Ubi-junB mice appear normal, but develop a myeloproliferative disease by 11 months of age. These animals showed progressive myeloid hyperplasia and some mice developed blast crisis. Passegué et al suggest that these JunB−/−Ubi-junB mice recapitulate the natural course of human chronic myeloid leukemia (CML).19 The disease is associated with age-dependent and myeloid-specific silencing of the junB transgene, although the mechanism for this phenomenon is still unclear. This interesting study by Passegué et al defines a cellular and molecular mechanism by which JunB might regulate the process of myelopoiesis. It also further confirmed the role of JunB as a negative growth regulator and a potential tumor suppressor.
To explore the possible role of JunB in human CML, we have used the real-time quantitative reverse transcription–polymerase chain reaction (RT-PCR) to examine if the JunB expression is also down-regulated in human CML. We have also investigated the possible mechanisms that cause the JunB down-regulation in patients with CML.
Materials and methods
Samples
Peripheral blood (PB) samples were collected from healthy individuals and CML patients from June 1998 throughout December 2001. Patients who carried more than 30% blast cells in bone marrow were included in the group of blastic crisis (52.98% ± 11.96%, n = 11) and those who carried less than 30% blast cells in bone marrow were included in the group of chronic phase (11.40% ± 1.62%, n = 21). RNA and gDNA were extracted by standard procedures as described previously.20 21
Real-time quantitative RT-PCR analysis
The cDNA sequence of JunB gene was evaluated and the specific forward and reverse primers and TaqMan probe were designed using Primer Express software (Applied Biosystems, Foster City, CA). The probe designed by the software was synthesized and labeled with appropriate fluorescent dyes (Applied Biosystems). Sequences of the forward and reverse primers and probe are as follows: 5′-GCA CTA AAA TGG AAC AGC CCT T-3′, 5′-GGC TCG GTT TCA GGA GTT TG-3′, and 5′-FAM-ACG ACG ACT CAT ACA CAG CTA CGG GAT ACG-TAMRA-3′. For internal control of the RNA, expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was also examined by RT-PCR. The amount of JunB was normalized to the endogenous reference GAPDH to obtain the relative threshold cycle (ΔCT) and then related to the ΔCT of normal cases to obtain the relative expression level (2−ΔΔCT) of JunB.
All reactions were performed in an ABI-7700 sequence detector (Applied Biosystems) and TaqMan EZ RT-PCR Core Kit (Applied Biosystems) and protocol were used. RT-PCR was performed in a 25-μL final volume containing 400 nM each primer, 200 nM probe, 300 μM each deoxynucleoside triphosphate (dNTP), 3.0 mM manganese acetate, 2.5 U rTth DNA polymerase, and 1 × PCR buffer. The RT-PCR cycling parameters were set as follows: the RT reaction at 50°C for 2 minutes, 60°C for 30 minutes, and 95°C for 5 minutes followed by 40 cycles of PCR reactions at 94°C for 20 seconds and 62°C for 1 minute.
To determine the precision of the assay, 4 replicates of total RNA of each sample were run on 4 separate days. Intra-assay (within-run) precision was determined by calculating mean, standard deviation (SD), and coefficient of variance (CV) of the CT values for each sample and for each set of primers and probe on each day.
Immunocytochemistry
For cytospin preparation, 5 × 105 cells were cytocentrifuged onto glass slides and fixed in 1% formaldehyde/phosphate-buffered saline (PBS), permeabilized with 0.5% Triton X-100/PBS, and blocked for nonspecific binding with 10% bovine serum albumin (BSA)/PBS. Cells were then incubated with JunB antibody (1:200 dilution; Santa Cruz Biotechnology, Santa Cruz, CA), peroxidase-conjugated IgG (1:500 dilution; Jackson Immunoresearch Laboratories, West Grove, PA), and then visualized by DAB Substrate Kit (Vector Laboratories, Burlingame, CA). Finally, cells were counterstained with Mayer hematoxylin (Merck, Darmstadt, Germany).
Gene mutational analysis
PCR was performed using primers for amplification of coding region and 9 flanking evolutionarily conserved regions ofJunB gene as listed in Table1. PCR was performed in a 25-μL final volume containing 200 nM each primer, 200 μM each dNTP, 3.5 mM MgCl2, 2 U Taq DNA polymerase (Promega, Madison, WI), and 1 × PCR buffer. The amplification procedure was carried out as follows: 35 cycles of PCR reactions at 95°C for 1 minute, 58 to 62°C (depending on the melting temperature [Tm] of each primer set) for 1 minute, and 72°C for 2 minutes. For single-strand conformational polymorphism (SSCP) analysis, PCR products (4 μL) of each of the amplicons was denatured in 2 μL denaturing solution (95% formamide, 10 mM EDTA [ethylenediaminetetraacetic acid], 0.05% bromophenol blue, 0.05% xylenol cyanol) at 95°C for 5 minutes and thereafter directly placed on ice. A 4-μL aliquot of each sample was loaded, and electrophoresis was carried out in a GenePhor Electrophoresis Unit using the GeneGel Excel 12.5/24 Kit (Pharmacia Biotech, Uppsala, Sweden). Staining was performed in a Gene Stain Automated Gel Stainer using PlusOne Silver Staining Kit (Pharmacia Biotech).
Oligonucleotide primers for amplification of coding region and 9 FECSs of JunB gene
| . | Forward primer (5′→3′) . | Location . | Reverse primer (5′→3′) . | Location . | Annealing temperature, °C . |
|---|---|---|---|---|---|
| Coding region-1 | cgacggccaatcgga | 5639-5653 | ggctcggtttcaggagtttg | 6112-6193 | 60 |
| Coding region-2 | tggctgctacccgcc | 5854-5868 | gagacgcgccggtgt | 6235-6221 | 62 |
| Coding region-3 | gctcggacaccggcg | 6215-6229 | gttggtgtaaacggg | 6549-6528 | 62 |
| Coding region-4 | cccgtttacaccaactca | 6535-6553 | cttcaccttgtcctccagg | 6900-6882 | 64 |
| Coding region-5 | gctgtcgagtaccgccg | 6924-6940 | ccaccaacagcactgagct | 7624-7606 | 60 |
| FECS I | accccgaggtcctttgagc | 5485-5503 | gctgtgcgcaaaagccct | 5846-5828 | 62 |
| FECS II | gaaccattgcttgaggg | 4374-4390 | ggtaacctcttagtcatctgtattc | 4752-4728 | 58 |
| FECS III | gcagccgccgttccagc | 3880-3896 | ggaacccgactatctgcc | 4155-4138 | 56 |
| FECS IV | tgcaagttagcttcccaa | 2960-2978 | caccaagtgtgggaaagag | 3351-3333 | 58 |
| FECS V | catgctctctggttgtcattattca | 2436-2460 | cgcagtgactcagaaacactt | 2760-2736 | 60 |
| FECS VI | ggcctggaaggccctatac | 1056-1074 | tacaggtgtgagccaccgc | 1272-1254 | 58 |
| FECS VII | tgcgatttgtgggttctcg | 633-651 | atacccggttccaggcct | 1072-1055 | 62 |
| FECS VIII | ccagtcacacgcttgcc | 126-142 | cgagaacccacaaatcgc | 651-634 | 60 |
| FECS IX | cgagacttggagtgcg | 7785-7801 | cgccaaacgagcgga | 8181-8167 | 58 |
| . | Forward primer (5′→3′) . | Location . | Reverse primer (5′→3′) . | Location . | Annealing temperature, °C . |
|---|---|---|---|---|---|
| Coding region-1 | cgacggccaatcgga | 5639-5653 | ggctcggtttcaggagtttg | 6112-6193 | 60 |
| Coding region-2 | tggctgctacccgcc | 5854-5868 | gagacgcgccggtgt | 6235-6221 | 62 |
| Coding region-3 | gctcggacaccggcg | 6215-6229 | gttggtgtaaacggg | 6549-6528 | 62 |
| Coding region-4 | cccgtttacaccaactca | 6535-6553 | cttcaccttgtcctccagg | 6900-6882 | 64 |
| Coding region-5 | gctgtcgagtaccgccg | 6924-6940 | ccaccaacagcactgagct | 7624-7606 | 60 |
| FECS I | accccgaggtcctttgagc | 5485-5503 | gctgtgcgcaaaagccct | 5846-5828 | 62 |
| FECS II | gaaccattgcttgaggg | 4374-4390 | ggtaacctcttagtcatctgtattc | 4752-4728 | 58 |
| FECS III | gcagccgccgttccagc | 3880-3896 | ggaacccgactatctgcc | 4155-4138 | 56 |
| FECS IV | tgcaagttagcttcccaa | 2960-2978 | caccaagtgtgggaaagag | 3351-3333 | 58 |
| FECS V | catgctctctggttgtcattattca | 2436-2460 | cgcagtgactcagaaacactt | 2760-2736 | 60 |
| FECS VI | ggcctggaaggccctatac | 1056-1074 | tacaggtgtgagccaccgc | 1272-1254 | 58 |
| FECS VII | tgcgatttgtgggttctcg | 633-651 | atacccggttccaggcct | 1072-1055 | 62 |
| FECS VIII | ccagtcacacgcttgcc | 126-142 | cgagaacccacaaatcgc | 651-634 | 60 |
| FECS IX | cgagacttggagtgcg | 7785-7801 | cgccaaacgagcgga | 8181-8167 | 58 |
The locations indicated are base numbers according to the human JunB gene sequence (GenBank accession no.U20734).
For further confirmation, direct sequencing of the PCR products was carried out for at least 5 cases of each amplicon for each group of samples. DNA sequencing was performed on ABI Prism 310 Genetic Analyzer and BigDye Terminator Cycle Sequencing Kit (Applied Biosystems) and protocol was followed for the reactions.
Methylation-specific PCR
Bisulfite treatment of gDNA was done as described previously.22 23 gDNA, 2 μg in a volume of 100 μL, was denatured in 0.2 N NaOH at 37°C for 10 minutes and incubated with 3 M sodium bisulfite at 50°C for 16 hours. After incubation, modified DNA was purified using Wizard DNA Clean-Up System (Promega) according to the manufacturer's instructions and was desulfonated in 0.3 N NaOH at room temperature for 5 minutes. DNA was then ethanol precipitated, washed, and resuspended in 20 μL water and used immediately or stored at −20°C until use.
Bisulfite-modified DNA was amplified by PCR using primer sets designed specifically for the promoter region of JunB genes as follows: JunB-M (forward) 5′-GAC GTT AGG AAA GTT ATC GC-3′ (−224 to −205 to the transcriptional start site of human JunB gene, according to the human JunB gene sequence of GenBank accession no.U20734) and (reverse) 5′-CGA ACT AAA TAC CTA ATC GCG-3′ (−89 to −109 to the transcriptional start site of human JunB); JunB-U (forward) 5′-TTG GGG GAA ATG ATG TTA GGA AAG TTA TTG T-3′ (−235 to −205 to the transcriptional start site of human JunB gene) and (reverse) 5′-ACT ACA ACA AAC AAC AAA CTC TCC ACT ACA-3′ (−54 to −83 to the transcriptional start site of human JunB gene). The amplification areas correspond to the −89 to −224 (JunB-M) and −54 to −235 (JunB-U) to the transcriptional start site of humanJunB gene. PCR was performed in a 50-μL final volume containing 200 nM each primer, 200 μM each dNTP, 1.5 mM MgCl2, 2 U of Taq DNA polymerase (Promega), and 1 × PCR buffer. The amplification procedure was carried out as follows: 95°C for 10 minutes followed by 40 cycles of PCR reactions at 95°C for 90 seconds, 50°C for 1 minute, and 72°C for 2 minutes. The PCR products were analyzed by agarose gel electrophoresis and visualized by ethidium bromide staining with a 100-bp ladder as a reference.
CpG methylase (Sss I)–treated gDNA was used as a positive control for methylation-specific primers JunB-M, because CpG methylase methylates all cytosine within the double-stranded dinucleotide CG. DNA samples from healthy individuals that were negative for JunB-M primer set and positive for JunB-U primer set were used as positive controls for JunB-U primers. To ensure the specificity of JunB-M and JunB-U primer sets for bisulfite-modified DNA, amplification using unmodified gDNA samples from healthy individuals and patients with CML was also carried out.
To calculate the numbers of methylated CpG sites, direct sequencing using both forward and reverse primers was carried out for each sample. DNA sequencing was performed on ABI Prism 310 Genetic Analyzer and BigDye or dRhodamine Terminator Cycle Sequencing Kits (Applied Biosystems) and protocols followed for the reactions.
Subcloning of methylation-specific PCR products
Bisulfite-modified DNA was amplified by PCR using a primer set designed for detection of both methylated and unmethylated promoter region of JunB genes as follows: JunB-MU (forward) 5′-GGG ATT TTG AGA GTG GTT AGG-3′ (−286 to −266 to the transcriptional start site of human JunB gene) and (reverse) 5′-CCA TAT CCC ATA ACT ATA TAT AAA CT-3′ (+62 to +36 to the transcriptional start site of human JunB gene). The amplification area corresponds to the +62 to −286 to the transcriptional start site of human JunBgene. PCR was performed as described for methylation-specific PCR. Cloning reactions were carried out using TOPO TA cloning kit version K2 (Invitrogen, Groningen, the Netherlands). Briefly, JunB-MU PCR products were first purified and added 3′ A-overhangs by incubation in 1 UTaq and 2.5 mM deoxyadenosine triphosphate (dATP) at 72°C for 10 minutes and then put on ice. PCR products with A-overhangs were then incubated with vector at room temperature for 30 minutes and then put on ice. The reaction mixture was then added to TOPO-competent cells and incubated on ice for 20 minutes and then 42°C for 30 seconds to heat shock the competent cells and then put on ice immediately. The mixture was then added to 250 μL SOC (2% bacto-tryptone, 0.5% bacto-yeast extract, 0.05% NaCl, and 20 mM glucose) medium and shaken horizontally at 37°C for 1 hour. After shaking, the mixture was plated out on agar plates containing X-gal and 50 μg/mL ampicillin and incubated at 37°C overnight. The next day, colonies were picked and cultured in Luria-Bertani (LB) medium containing 50 μg/mL ampicillin overnight. After overnight incubation, the pDNA was extracted from competent cells using QIApre Spin Miniprep Kit (Qiagen, Hilden, Germany) and digested with EcoRI to examine if the pDNA contained the correct insert. The pDNA containing the correct insert was then sequenced using SP6, T7, or JunB-MU primers on ABI Prism 310 Genetic Analyzer.
Tissue culture and demethylation studies
K562 cells were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin, and grown at 37°C with 5% CO2. Cells were plated at 106 cells/100-mm dishes and treated the next day with 5-aza-2′-deoxycytidine (Sigma Chemical, St Louis, MO) at a final concentration of 10−6 M for 24 hours. The cells were then washed with PBS and fresh medium was added. The treated cells were harvested 1, 2, 3, 4, and 5 days after treatment for immunocytochemical, real-time quantitative RT-PCR, and methylation-specific PCR analysis as described.
Statistics
Comparisons were made with t test using SPSS for Windows Release 9.0 (SPSS, Chicago, IL).
Results
Determination of JunB expression level by real-time quantitative RT-PCR
To investigate whether the expression of the JunB gene was down-regulated in patients with CML, we have analyzed PB from 17 healthy individuals and 32 patients with CML (11 in blastic crisis and 21 in chronic phase) using real-time quantitative RT-PCR. For internal control of the RNA, expression of GAPDH was also examined and the amount of JunB was normalized to the endogenous reference GAPDH. The normalized JunB expression (ΔCT) of CML cases was then related to the ΔCT of healthy cases for their relative expression levels. The results showed that the expression levels of JunB in CML patients were significantly impaired compared with those in healthy individuals. The relative expression levels (2−ΔΔCT) of JunB for patients in blastic crisis and chronic phase were 1:31.12 and 1:4.72 of healthy individuals, respectively. Interestingly, the degrees of down-regulation of JunB gene expression were correlated to the clinical phases in the patients with CML, that is, in blastic crisis it is less than chronic phase which is less than normal (Table 2).
JunB gene expression levels and methylated CpG frequencies at the promoter area in healthy individuals and patients with CML
| . | No. . | Real-time quantitative RT-PCR . | Methylation study . | ||||
|---|---|---|---|---|---|---|---|
| ΔCT. JunB-GAPDH . | ΔΔCT. CML-normal . | 2−ΔΔCT . | Methylation rate . | No. of CpG . | No. of T/Cp G . | ||
| Healthy | 17 | 0.98 ± 0.32* | 0/17 (0%) | 0 | 0 | ||
| CML | |||||||
| Blastic crisis | 11 | 5.94 ± 0.78† | 4.96 | 1/31.12 | 11/11 (100%) | 12.64 ± 0.31 | 3.36 ± 0.31 |
| Chronic phase | 21 | 3.22 ± 0.47† | 2.24 | 1/4.72 | 21/21 (100%) | 12.50 ± 0.39 | 3.50 ± 0.39 |
| . | No. . | Real-time quantitative RT-PCR . | Methylation study . | ||||
|---|---|---|---|---|---|---|---|
| ΔCT. JunB-GAPDH . | ΔΔCT. CML-normal . | 2−ΔΔCT . | Methylation rate . | No. of CpG . | No. of T/Cp G . | ||
| Healthy | 17 | 0.98 ± 0.32* | 0/17 (0%) | 0 | 0 | ||
| CML | |||||||
| Blastic crisis | 11 | 5.94 ± 0.78† | 4.96 | 1/31.12 | 11/11 (100%) | 12.64 ± 0.31 | 3.36 ± 0.31 |
| Chronic phase | 21 | 3.22 ± 0.47† | 2.24 | 1/4.72 | 21/21 (100%) | 12.50 ± 0.39 | 3.50 ± 0.39 |
JunB gene expression was determined by real-time quantitative RT-PCR. The amount of JunB was normalized to the endogenous reference GAPDH to obtain the ΔCT value for each sample. The normalized JunB expression (ΔCT) of CML cases was first related to the ΔCT of normal cases to obtain the relative threshold cycle (ΔΔCT) and then the relative expression levels (2−ΔΔCT) were calculated. In the methylation study, all the healthy cases are positive for the nonmethylation-specific PCR and all the CML cases are positive for methylation-specific PCR. The numbers of CpG and T/Cp G were calculated from direct sequencing results of JunB-U or JunB-M PCR products of bisulfite-modified DNA.
Results are the means ± SEs.
P < .001 when compared with normal.
JunB protein was also down-regulated in cells from CML
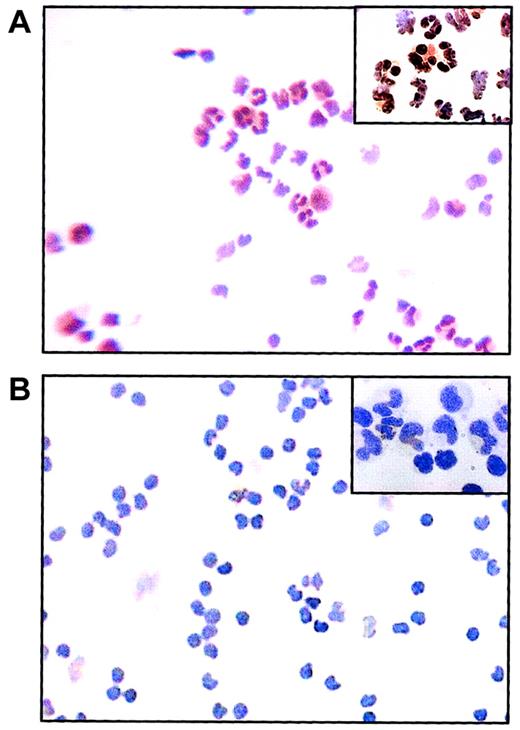
We further investigated the JunB protein expression using immunocytochemistry staining. As shown in Figure1A, most of the cells in healthy individuals were positively stained by the JunB antibody. In contrast, only a few cells were positively stained in CML cases (Figure 1B). These results confirmed the observation obtained from real-time quantitative RT-PCR that JunB expression was indeed down-regulated in CML.
Immunocytochemical stained cytospins for JunB.
(A) In cells from healthy individuals, most cells are positively stained (brown) for JunB antibody. (B) In cells from CML patients, few cells are positively stained and most cells are negatively stained for JunB antibody. JunB antibody staining was detected using peroxidase with DAB substrate. Cells were counterstained with nematoxylin. Original magnifications × 100, × 1000 for insets.
Immunocytochemical stained cytospins for JunB.
(A) In cells from healthy individuals, most cells are positively stained (brown) for JunB antibody. (B) In cells from CML patients, few cells are positively stained and most cells are negatively stained for JunB antibody. JunB antibody staining was detected using peroxidase with DAB substrate. Cells were counterstained with nematoxylin. Original magnifications × 100, × 1000 for insets.
JunB gene expression was not down-regulated by mutations within coding region and 9 FECSs in CML
Comparison of the murine and human junB loci reveals 9 flanking evolutionarily conserved sequences (FECSs) at distal 5′- and 3′-flanking DNA that share 72% to 91% sequence identity.24 These FECSs have been shown to be necessary for maximal mitogenic induction of junB and have been suggested to be required for effecting the proper transcriptional regulation of this gene.25 To elucidate the mechanism for the down-regulation of the JunB gene in CML, we first screened mutations in the coding region and 9 FECSs of JunB gene using PCR-SSCP and confirmed by direct sequencing. However, no mutation was found in these regions in all CML cases (data not shown). Results of gene mutational analysis suggested that mutations in these regions were not responsible for the down-regulation of JunB gene expression in CML.
JunB gene expression was inactivated by methylation in CML
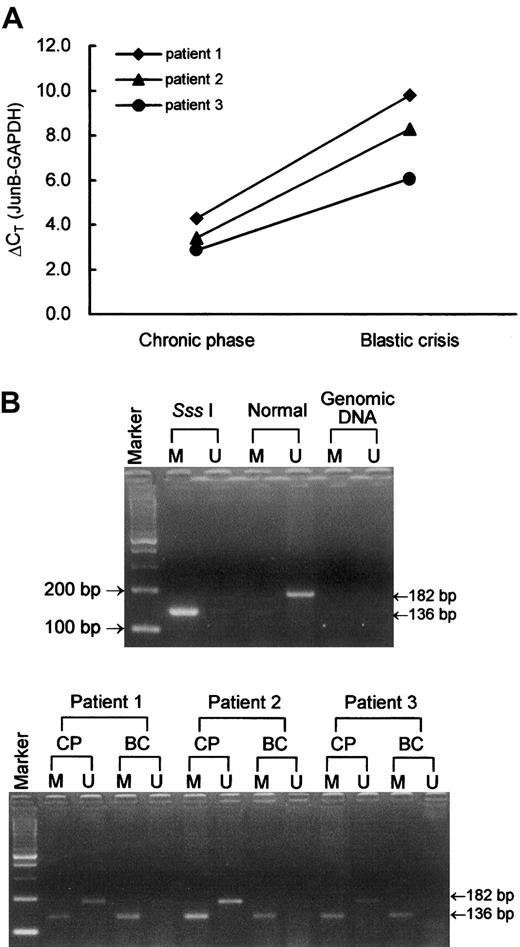
Second, to investigate whether the aberrant CpG island methylation was the possible mechanism for the down-regulation of theJunB gene in CML, we analyzed the methylation status of the promoter area of the JunB gene. We designed 2 pairs of methylation-specific primers designated JunB-U and JunB-M to discriminate between unmethylated and methylated alleles and to discriminate between bisulfite-modified and unmodified DNA. In Figure2, we show 3 representative cases in which JunB expression levels were higher (lower ΔCT) at chronic phase and lower (higher ΔCT) at blastic crisis (Figure 2A). The methylation-specific PCR results demonstrated that they are positive for both JunB-M and JunB-U at chronic phase and are positive for JunB-M but negative for JunB-U at blastic crisis (Figure2B). The JunB-M and JunB-U primer sets are specific for methylated and unmethylated alleles, respectively; in addition, both primer sets are specific for bisulfite-modified DNA but not for unmodified gDNA (Figure2B). The JunB-U primer set covers 23 CpG sites and JunB-M primer set covers 16 CpG sites. PCR products of these primer sets were direct-sequenced to calculate the numbers of methylated CpG sites (examples in Figure 3). As shown in Table2, no methylation was observed in healthy individuals, whereas methylation was observed in all CML cases. However, the methylated CpG frequencies of JunB gene did not differ in patients at blastic crisis (12.64 ± 0.31) or at chronic phase (12.50 ± 0.39) of CML. In addition, all CML cases carried some partial methylated sites (3.44 ± 0.26). We also designed a universal primer designated JunB-MU that covers 34 CpG sites and can detect both methylated and unmethylated alleles. JunB-MU PCR products of the same healthy and CML cases were used for subcloning analysis and similar results as those gained from methylation-specific PCR and sequencing were obtained. Figure 4 shows the allelic patterns of CpG island methylation of chronic phase and blastic crisis of a representative CML case (patient 3 in Figure 2). In this representative case, several clones from chronic phase were devoid of methylated CpG sites, whereas clones from blastic crisis were methylated at most CpG sites. The allelic patterns of CpG island methylation are consistent with the results observed by methylation-specific PCR in this patient.
JunB gene expression and methylation-specific PCR.
(A) JunB gene expression levels of 3 representative CML cases as measured by real-time quantitative RT-PCR. In each CML patient, JunB expression level was higher (lower ΔCT) at chronic phase and was lower (higher ΔCT) at blastic crisis. (B) Methylation-specific PCR of JunB. Sss I is Sss I–treated, bisulfite-modified DNA of a healthy individual. This DNA sample was used as a positive control for methylation-specific primers. Normal is bisulfite-modifed DNA of the same healthy individual as in lane Sss I Tx. Genomic DNA is unmodified gDNA of a CML patient. M and U indicate methylation-specific PCR using JunB-M and JunB-U primer sets, respectively. CP and BC indicate chronic phase and blastic crisis of CML, respectively. Marker represents the 100-bp ladder DNA marker. Patients 1, 2, and 3 correspond to the same patients in panel A.
JunB gene expression and methylation-specific PCR.
(A) JunB gene expression levels of 3 representative CML cases as measured by real-time quantitative RT-PCR. In each CML patient, JunB expression level was higher (lower ΔCT) at chronic phase and was lower (higher ΔCT) at blastic crisis. (B) Methylation-specific PCR of JunB. Sss I is Sss I–treated, bisulfite-modified DNA of a healthy individual. This DNA sample was used as a positive control for methylation-specific primers. Normal is bisulfite-modifed DNA of the same healthy individual as in lane Sss I Tx. Genomic DNA is unmodified gDNA of a CML patient. M and U indicate methylation-specific PCR using JunB-M and JunB-U primer sets, respectively. CP and BC indicate chronic phase and blastic crisis of CML, respectively. Marker represents the 100-bp ladder DNA marker. Patients 1, 2, and 3 correspond to the same patients in panel A.
Examples of methylation analysis of CpG islands at the promoter area of JunB gene.
The sequence shown corresponds to the −77 to −116 to the transcriptional start site of JunB gene. (A) Unmodified DNA with all the cytokines remains intact. (B) Modified DNA of an unmethylated sample with all the cytokines converted to thymidine. (C) Modified DNA of a methylated sample with all Cs in CpG dinucleotides remaining as C. (D) Modified DNA of a partially methylated sample with part of the Cs in CpG dinucleotides showing C/T heterozygosity. Blue indicates cytosine (C); black, guanine (G); red, thymidine (T); and green, adenosine (A).
Examples of methylation analysis of CpG islands at the promoter area of JunB gene.
The sequence shown corresponds to the −77 to −116 to the transcriptional start site of JunB gene. (A) Unmodified DNA with all the cytokines remains intact. (B) Modified DNA of an unmethylated sample with all the cytokines converted to thymidine. (C) Modified DNA of a methylated sample with all Cs in CpG dinucleotides remaining as C. (D) Modified DNA of a partially methylated sample with part of the Cs in CpG dinucleotides showing C/T heterozygosity. Blue indicates cytosine (C); black, guanine (G); red, thymidine (T); and green, adenosine (A).
Allelic patterns of CpG site methylation at the promoter area of JunB gene.
Methylation of the JunB CpG island located between positions −89 to −224 corresponding to the transcriptional starting site was examined by genomic bisulfite sequence analysis. Ten clones were randomly selected and sequenced. Methylated CpG sites are marked as filled circles (●) and unmethylated sites as open circles (○). (A) The allelic patterns of chronic phase of a representative CML case. (B) The allelic patterns of blastic crisis of a representative CML case.
Allelic patterns of CpG site methylation at the promoter area of JunB gene.
Methylation of the JunB CpG island located between positions −89 to −224 corresponding to the transcriptional starting site was examined by genomic bisulfite sequence analysis. Ten clones were randomly selected and sequenced. Methylated CpG sites are marked as filled circles (●) and unmethylated sites as open circles (○). (A) The allelic patterns of chronic phase of a representative CML case. (B) The allelic patterns of blastic crisis of a representative CML case.
Demethylation-associated reactivation of JunB expression in CML cell line K562
Because reversibility is a hallmark of epigenetic transcriptional repression, demethylation studies were carried out. We treated hypermethylated CML cells, K562, with 5-aza-2′-deoxycytidine to attempt demethylation of the JunB locus over a 24-hour exposure period. As shown in Figure 5A, JunB expression was increased after 24-hour incubation in 1 μM 5-aza-2′-deoxycytidine and persisted to 5 days after treatment as determined by real-time quantitative RT-PCR. Methylation-specific PCR results also showed a time-dependent decrease of intensities of JunB-M PCR products and a slight increase of intensities of JunB-U PCR products. (Figure 5B). Figure 5C shows the allelic patterns of CpG island methylation of K562 cell before and 5 days after 5-aza-2′-deoxycytidine treatment. Nearly all the CpG sites were methylated in most of the K562 cells without treatment. The addition of 5-aza-2′-deoxycytidine abandoned the methylation of some of the CpG sites. However, the increase in JunB protein expression was insignificant as analyzed by immunocytochemical staining using JunB antibody (data not shown). The results demonstrated that 5-aza-2′-deoxycytidine treatment partially reactivated JunB expression in K562 cells.
Demethylation studies using K562 cells treated with 5-aza-2′-deoxycytidine.
Cells (106) were treated with 1 μM 5-aza-2′-deoxycytidine for 24 hours and cells were then washed and fresh medium was added. The treated cells were harvested at days 1, 2, 3, 4, and 5 after 5-aza-2′-deoxycytidine treatment for JunB expression and methylation-specific PCR analysis. (A) JunB gene expression levels in K562 cells were increased (decreased ΔCT) after 5-aza-2′-deoxycytidine treatment for 24 hours and persisted for 5 days as determined by real-time quantitative RT-PCR. Data are presented as mean ± SE calculated from 5 duplicate studies. (B) Methylation-specific PCR of JunB-M and JunB-U of K562 cells after 5-aza-2′-deoxycytidine treatment. JunB-M PCR products showed a time-dependent decrease of intensities and JunB-U PCR products showed a slight increase of intensities. Internal controls are PCR products of universal primer set JunB-MU. (C) Allelic patterns of CpG site methylation at the promoter area of JunB gene of K562 cells before and 5 days after 5-aza-2′-deoxycytidine treatment. Methylated CpG sites are marked as filled circles (●) and unmethylated sites as open circles (○).
Demethylation studies using K562 cells treated with 5-aza-2′-deoxycytidine.
Cells (106) were treated with 1 μM 5-aza-2′-deoxycytidine for 24 hours and cells were then washed and fresh medium was added. The treated cells were harvested at days 1, 2, 3, 4, and 5 after 5-aza-2′-deoxycytidine treatment for JunB expression and methylation-specific PCR analysis. (A) JunB gene expression levels in K562 cells were increased (decreased ΔCT) after 5-aza-2′-deoxycytidine treatment for 24 hours and persisted for 5 days as determined by real-time quantitative RT-PCR. Data are presented as mean ± SE calculated from 5 duplicate studies. (B) Methylation-specific PCR of JunB-M and JunB-U of K562 cells after 5-aza-2′-deoxycytidine treatment. JunB-M PCR products showed a time-dependent decrease of intensities and JunB-U PCR products showed a slight increase of intensities. Internal controls are PCR products of universal primer set JunB-MU. (C) Allelic patterns of CpG site methylation at the promoter area of JunB gene of K562 cells before and 5 days after 5-aza-2′-deoxycytidine treatment. Methylated CpG sites are marked as filled circles (●) and unmethylated sites as open circles (○).
Discussion
Recent studies have made it ever more clear that JunB, at least in mice, can act as a tumor suppressor gene. However, the JunBgene has not yet been connected with any human leukemia. In the present study, we examined the JunB expression levels in CML patients using real-time quantitative RT-PCR and explored its role in CML. Interestingly, we found JunB gene expression was not only significantly down-regulated in cells from CML patients but also correlated to the patients' clinical phases, that is, in blastic crisis it is less than in chronic phase which is less than normal. Results from immunocytochemical staining further confirmed our observation that JunB expression was indeed down-regulated in CML.
Promoter region hypermethylation is an important mechanism in abolishing gene transcription in cancer. Numerous studies have reported the inactivation of tumor suppressor genes by point mutation or chromosomal deletion in the development of cancers. However, epigenetic silencing of tumor suppressor gene by promoter region hypermethylation is also common in human cancer.26,27 Aberrant methylation of normally unmethylated CpG islands has been associated with transcriptional inactivation of defined tumor suppressor genes in human cancers, such as renal carcinoma,28 esophageal adenocarcinoma,29 carcinoma of the uterine cervix,30 acute lymphoblastic leukemia,31 and so forth. In this situation, promoter region hypermethylation stands as an alternative to coding region mutations in eliminating tumor suppressor gene function. Here, we first analyzed the coding region and 9 FECSs of JunB gene for mutations but no mutation was found in these regions. Because the primers used for mutational analysis cover the whole coding region of JunB and PCR products were obtained from all the CML cases, we can also exclude the possibility that homozygous deletion accounts for the loss of JunB expression. Therefore, we further analyzed the methylation of CpG sites at the promoter area of the JunB gene using methylation-specific PCR and direct sequencing and we found at promoter CpG sites hypermethylation of JunB gene related to the human CML. Although we did not find a strong association of methylation frequency with clinical phases of CML disease, we did notice differences in the allelic patterns of CpG island methylation in some cases. In combination with the mutational analysis and methylation analysis, our data suggested that mutation or deletion was irrelevant to this down-regulation; instead, the CpG-site methylation at the promoter area is responsible for this JunB gene down-regulation in patients with CML. Furthermore, the epigenetic alterations found in CML patients were absent in healthy individuals, suggesting that it may also be useful as a cancer-specific marker.
There are 2 possible reasons for the differential expression ofJunB gene in chronic phase and in blastic crisis. First, the differential CpG-island methylation frequencies may cause differential expression of JunB gene. Second, the proportions of normal cells may determine the degree of JunB gene expression. Because the CpG methylation frequencies of JunB gene were not different in cells from patients in chronic phase or in blastic crisis of CML, it is obvious that normal cells contributed the expression of JunB gene, because patients in chronic phase carry more normal cells than those in blastic phase. The immunocytochemical staining patterns have also provided additional evidence for the assumption that the differential JunB expression in patients in chronic phase and in blastic crisis was due to differential distribution of normal cells in these 2 phases of CML. In addition, the partial methylation (T/Cp G) and different methylation number of CpG island may also explain the differential JunB antibody staining observed in cells from patients with CML.
Because reversibility is a hallmark of epigenetic transcriptional suppression, we treated hypermethylated CML cells K562 with 5-aza-2′-deoxycytidine to attempt demethylation of CpG sites at the promoter area of the JunB gene. In the treated cells, increased JunB gene expression was observed after 24 hours and carried on for at least 5 days. In addition, decreased intensities of JunB-M PCR products and slightly increased intensities of JunB-U PCR products were also observed. The allelic patterns of CpG island methylation also revealed an increase in unmethylated sites. Results from demethylation studies demonstrated that methylation of CpG sites at the promoter area of JunB did cause the loss of JunB gene expression. However, the JunB gene expression level in cells treated with 5-aza-2′-deoxycytidine cells was not restored to the normal level. Consistent with the JunB gene expression, the JunB protein was increased insignificantly, which may be because the method is not sensitive enough to detect the change, and the JunB-M PCR products were slightly diminished but not fully vanished in treated cells. Previous study has demonstrated that transcription from densely methylated promoters can only be partially restored by 5-aza-2′-deoxycytidine treatment but can obviously be enhanced by cotreatment with an inhibitor of histone deacetylase.34Therefore, another mechanism, such as deacetylation change ofJunB gene, may play some role in the inactivation of the CpG island of JunB gene in K562 cells.
The t(9;22) chromosomal aberration is believed to be the initiating event in human CML. This aberration results in fusion ofABL and BCR genes and synthesis of a chimeric BCR-ABL p210 protein with deregulated tyrosine kinase activity. The direct effects of BCR-ABL in leukemogenesis have been proven in several studies.32 33 However, the increased number of myeloid cells in the chronic phase of CML could result not only from the reduced dependence of progenitor cells on growth factors but also from a partial loss of negative growth responses to proliferation. This may happen if BCR-ABL antagonizes some of the pathways that are triggered by negative growth regulators, leading to an increase in the number of cell divisions. The finding that the down-regulation of JunB in human CML was correlated with patients' clinical phases implies that JunB dysfunction may be a downstream event of BCR-ABL activity. It is possible that BCR-ABL antagonizes the negative regulatory function on cell proliferation of JunB and the progression to blast crisis is due to the aberrant proliferation caused by the inactivation ofJunB gene. Therefore, JunB expression may provide a novel and useful marker for clinical follow-up for CML and ultimately to design effective target therapies in blast crisis.
JunB has been demonstrated to control the number of granulocyte progenitors by inhibiting proliferation and promoting apoptosis in mice.17 The fact that humans with CML exhibit decreased JunB levels by methylation of the CpG sites at the JunB promoter area further demonstrated that JunB can act as a tumor suppressor gene, which is consistent with previous studies in mice. Impaired apoptosis has also been suggested to contribute to leukemogenesis.35 However, whether the role of JunB in promoting apoptosis observed in mice can be extended to human leukemia remains to be explored. Meanwhile, the balance of Jun proteins with opposing effects appear to determine whether cells progress through the cell cycle or die. Therefore, it also remains to be investigated whether the observed JunB-related leukemogenesis is dependent on expression of other Jun proteins and which Fos dimerization partner is involved in regulating granulocyte proliferation and apoptosis.
Prepublished online as Blood First Edition Paper, December 27, 2002; DOI 10.1182/blood-2002-05-1598.
Supported by grant NSC91-2314-B-037-248 from the National Science Council; Taiwan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sheng-Fung Lin, Division of Hematology-Oncology, Department of Internal Medicine, Kaohsiung Medical University Hospital, 100 Shih-Chuan 1st Rd, Kaohsiung 80706, Taiwan; e-mail: shlin@cc.kmu.edu.tw.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal