Leukocytes are classified as myelocytic or lymphocytic, and each class of leukocytes consists of several types of cells that have different phenotypes and different roles. To define the gene expression in these cells, we have performed serial analysis of gene expression (SAGE) using human leukocytes and have provided the gene database for these cells not only at the resting stage but also at the activated stage. A total of 709 990 tags from 17 libraries were analyzed for the manifestation of gene expression profiles in various types of human leukocytes. Types of leukocytes analyzed were as follows: peripheral blood monocytes, colony-stimulating factor–induced macrophages, monocyte-derived immature dendritic cells, mature/activated dendritic cells, granulocytes, natural killer (NK) cells, resting B cells, activated B cells, naive T cells, CCR4− memory T cells (resting TH1 cells), CCR4+ memory T cells (resting TH2 cells), activated TH1 cells, and activated TH2 cells. Among 38 961 distinct tags that appeared more than once in the combined total libraries, 27 323 tags were found to represent unique genes in certain type(s) of leukocytes. Using probability (P) and hierarchical clustering analysis, we identified the genes selectively expressed in each type of leukocytes. Identification of the genes specifically expressed in different types of leukocytes provides not only a novel molecular signature to define different subsets of resting and activated cells but also contributes to further understanding of the biologic function of leukocytes in the host defense system.
Introduction
Leukocytes can be divided into various types such as monocytes/macrophages, dendritic cells (DCs), T cells, B cells, and natural killer (NK) cells. These cells communicate with each other through various surface molecules such as CD markers and through secreted factors such as cytokines.
In the past several years, the accumulation of a gene database including cDNA and genome has accelerated the identification of the molecules involved in cell–cell interaction, cell activation, and cell differentiation. In addition, the functional genomic technology such as DNA microarray1 and serial analysis of gene expression (SAGE)2 allow the expression of thousands of genes to be analyzed. These analyses are useful to know the function of each cell type because the characterization of each cell type depends on the genes selectively expressed at various stages. Among gene expression analyses, the SAGE method is quantitative and can cover the expressed genes that are unequaled by any mammalian DNA microarray systems available.3
We have recently reported the results of SAGE in human monocytes, macrophages,4,5 DCs,6 helper T (TH1 and TH2) cells,7 and NK cells.8 In this study, we analyzed gene expression by means of SAGE in leukocytes, including phagocytes, T cells, and B cells, at various differentiation and activation stages, and we constructed a gene database for these. In addition, we analyzed the set of genes restricted to myeloid or lymphoid cells.
Materials and methods
Cell preparation
Monocytes, macrophage–colony-stimulating factor (M-CSF)– and granulocyte macrophage–colony-stimulating factor (GM-CSF)–induced macrophages, immature and mature DCs, and lipopolysaccharide (LPS)–stimulated monocytes were prepared as described before.4-6 Langerhans-like cells were obtained by culture of purified monocytes for 7 days at 37°C in RPMI 1640 medium containing 7.5% heat-inactivated fetal calf serum (FCS; Gibco/Life Technologies, Tokyo, Japan), recombinant human GM-CSF (rhGM-CSF; 500 U/mL), interleukin-4 (IL-4; 100 U/mL), and transforming growth factor-β (TGF-β; 10 ng/mL) (R&D Systems, Minneapolis, MN). Granulocytes were prepared from fresh peripheral blood after the depletion of red blood cells by sedimentation in 0.25% dextran in phosphate-buffered saline (PBS) for 40 minutes at room temperature, and the remaining cells were subjected to Ficoll-Hypaque gradient centrifugation to obtain the buffy coat. Red blood cells in the buffy coat were depleted by hypotonic lysis, and the remaining cells were used as granulocytes.
Peripheral blood mononuclear cells (PBMCs) were isolated from venous blood drawn from healthy volunteers by centrifugation on a Ficoll-Metrizoate density gradient (d = 1.077; Lymphoprep; Nycomed, Oslo, Norway). CD4+CD45RA+ naive T cells were purified using CD4 multisort. CCR4+ resting TH1 and CCR4-resting TH2 cells were obtained from PBMCs by incubation with anti-CCR4 monoclonal antibody (mAb) followed by sorting with an EPICS XL (Beckman Coulter). NK, CD8+ T, and CD8+ B cells were separated from PBMCs by incubation with anti-CD56 mAb-coated, anti-CD8 mAb-coated, and anti-CD19 mAb-coated microbeads, respectively. Activated B cells were obtained from purified B cells by stimulation with a membrane containing CD40 ligand for 3 hours at 37°C. Activated TH1 and activated TH2 cells were generated as described before.7
Generation of SAGE and cDNA libraries
A modification of the original SAGE and micro-SAGE methods was used to generate all SAGE libraries. In brief, mRNAs of monocytes and macrophages were purified from a mixture of total RNA of at least more than 5 donors. Monocytes were incubated for 7 days at 37°C with M-CSF (100 ng/mL) or GM-CSF (500 U/mL) in RPMI 1640 containing 7.5% FCS. Total RNA from these cells was isolated by direct lysis in RNAzol B. Poly(A)+ RNA was isolated using a FastTrac (Invitrogen, Carlsbad, CA) mRNA purification kit according to the manufacturer's instructions.
SAGE libraries were generated using 1.5 μg poly(A)+ RNA and were converted to double-stranded cDNA with the use of a BRL synthesis kit, including biotin-5′-T18-3′ primer as described in the manufacturer's protocol. cDNA was cleaved with the restriction enzymeNlaIII, and the 3′-terminal cDNA fragments were bound to streptavidin-coated magnetic beads (Dynal). After ligation with oligonucleotides containing recognition sites for BsmF1, the linked cDNAs were released from the beads by digestion with BsmF1. Released tags were ligated to one another, ligated to concatemers, and cloned into the SphI site of pZero 1.0 (Invitrogen). Colonies were screened with polymerase chain reaction (PCR) using M13 forward and M13 reverse primers. PCR products containing inserts of more than 600 bp were sequenced with the Big Dye terminator version 2 kit and were analyzed using a 377 ABI automated sequencer (Applied Biosystems, Tokyo, Japan).
SAGE software (version 1) was used to quantify the abundance of each tag. Correction for tags containing linker sequences and other potential artifacts was made as described.9 Gene identification and UniGene cluster assignment of each SAGE tag were performed by using the SAGE Tag to UniGene Maps fromhttp://www.ncbi.nlm.nih.gov/SAGE/?SAGEtag.cgi and the table (updated March 2001; UniGene cluster fromhttp://www.sagenet.org/SAGEDatabases/unigene.htm). The cluster and treeview programs (http://rana.lbl.gov/EisenSoftware.htm) were used for clustering SAGE data. Briefly, the frequency tables of the 17 libraries were first normalized to expression levels per 55 000 tags. SAGE tags appearing at least 5 times in all 17 libraries were subjected to clustering analysis. Data were adjusted through centering on the median and mean by sample. Noncentered Pearson correlation coefficient was used for distance calculations, and the weighted-complete linkage was used for clustering as described.10
Reverse transcription–polymerase chain reaction
Total RNA (200 ng) was prepared by the use of RNAzol B. RNA was reverse-transcribed in 50 μL of 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 10 mM dithiothreitol, 1 mM each dNTP, 2 μM random hexamer, and 2.4 U/μL Moloney murine leukemia virus reverse transcriptase for 1 hour at 42°C. Complementary DNA (cDNA) was obtained by treating total RNA corresponding to 40 ng in boiled water for 3 minutes and quenching on ice before amplification by PCR. Conditions for PCR were as follows: in a 50-μL reaction, 0.15 μM each primer, 1.25 μM each dGTP, dATP, dCTP, and dTTP (Toyobo), 50 mM KCl, 10 mM Tris-HCl, pH 8.3, 0.15 mM MgCl2, and AmpliTaq (Perkin-Elmer, Branchburg, NJ).
PCR cycle numbers for genes and primers used were as follows—hemoglobin α 1: sense, 5′-TCTGGTCCCCACAGACTCAGA-3′, antisense, 5′-TTAACCTGGGCA GAGCCG T-3′;EST (Hs. 103296): sense, 5′-CTGGGCAGGAAATTGAAGGA-3′, antisense, 5′-TTTGAGATGGAGTCTCGCTCTG-3′; GRO2 oncogene: sense, 5′-TCCAACTGACCAGAAGGAAGGA-3′, antisense, 5′-CGTCACATTGATCTTACTGGCC-3′; EST (Hs.192427): sense, 5′-ATC CTCATCTCCTTGATGGGC-3′, antisense, 5′-TGAAAACACCCATGCTTG CA-3′;CCL18: sense, 5′-CATCATGAAGGGCCTTGCA-3′, antisense 5′-CGAAGAGTTGAAGGGAAAGGG-3′; p21SNFT: sense, 5′-AGAGCCCTGAGG ATGATGACA-3′, antisense, 5′-TCCATGCTGGATCTGCACAA-3′; CD6: sense, 5′-AAATATGCCCTCCCCGTAATG-3′, antisense, 5′-AGCTTGCTTTTGGGACTATTGC-3′; granzyme B: sense, 5′-TCCCCCATCCAGCCTATAA-3′, antisense, 5′-TGAGACATAACCCCAGCCA-3′; EST (Hs. 98785): sense, 5′-TCTTCCCCAGAGTGCGTTTTT-3′, antisense, 5′-CATGGAACACCAAGTTGGTGAT-3′;FLJ20706: sense, 5′-CTGAAAGGCATGGTCACAAAGA-3′, antisense, 5′-TCCACCATTGTCCCT GGTAAG-3′; EST (Hs. 290825): sense, 5′-CATGAATGTGTTCGTAGGGCC-3′, antisense, 5′-TCTTCCAGGAAACCACAGGCT-3′;MHC class IB: sense, 5′-TCTACC CTGCGGAGATCACACT-3′, antisense, 5′-TTC AGG TGC CTT TGC AGA AA-3′. Reaction mixtures were incubated in a Perkin-Elmer DNA Thermal Cycler (denaturation at 60 seconds, 94°C; annealing at 60 seconds, 58°C; extension at 120 seconds, 72°C; 29-31 cycles).
Statistics
Statistical significance (P) between samples was calculated as described previously.11 Gene ranking, a framework for finding the genes expressed in specific samples, was used for statistical evaluation of differential expression of SAGE tags between samples. Statistical significance (P) in the evaluation is the expansion of the Audic and Claverie method12 to manage more than 2 samples.
Results
SAGE libraries in leukocytes
Seventeen independent SAGE libraries were summarized in Table1 (generated from human peripheral blood monocytes, CSF-derived macrophages, monocyte-derived immature DCs, mature/activated dendritic cells, granulocytes, NK cells, resting B cells, activated B cells, naive T cells, CCR4-negative memory T cells (resting TH1 cells), CCR4-positive memory T cells (resting TH2 cells), activated TH1 cells, and activated TH2 cells. CCR4-negative and -positive memory T cells are called resting TH1 and resting TH2 cells because, when these cells were stimulated with PMA (phorbol 12-myristate-13-acetate) and ionomycin, the cells typically showed TH1 and TH2 phenotypes as described in Imai et al.13
SAGE in human leukocytes and transcript tags
| Cells . | No. tags . | Unique transcripts . | Unique genes . |
|---|---|---|---|
| Myeloid | |||
| Monocytes* | 58 700 | 10 391 | 8 141 |
| LPS-stimulated monocytes* | 35 991 | 8 172 | 6 710 |
| M-CSF-induced macrophages* | 54 047 | 10 629 | 8 295 |
| GM-CSF-induced macrophages* | 57 525 | 10 722 | 8 434 |
| Immature dendritic cells* | 58 700 | 12 577 | 9 844 |
| Mature dendritic cells* | 31 862 | 8 017 | 6 583 |
| Langerhans-like cells | 57 717 | 13 630 | 10 874 |
| Granulocytes | 31 466 | 8 007 | 6 821 |
| Lymphoid | |||
| CD4 T cells (naive) | 50 433 | 11 290 | 9 124 |
| CD4 T cells (memory, CCR4-negative) | 31 919 | 7 572 | 6 220 |
| CD4 T cells (memory, CCR4-positive) | 30 700 | 7 820 | 6 521 |
| Activated T cells (TH1)* | 32 219 | 8 111 | 6 676 |
| Activated T cells (TH2)* | 32 288 | 9 047 | 7 382 |
| CD8 T cells* | 51 017 | 11 789 | 9 380 |
| NK cells* | 34 831 | 8 187 | 6 569 |
| B cells | 53 236 | 10 903 | 8 499 |
| Activated B cells | 7 339 | 2 798 | 2 439 |
| Total | 709 990 | 38 961 | 27 323 |
| Cells . | No. tags . | Unique transcripts . | Unique genes . |
|---|---|---|---|
| Myeloid | |||
| Monocytes* | 58 700 | 10 391 | 8 141 |
| LPS-stimulated monocytes* | 35 991 | 8 172 | 6 710 |
| M-CSF-induced macrophages* | 54 047 | 10 629 | 8 295 |
| GM-CSF-induced macrophages* | 57 525 | 10 722 | 8 434 |
| Immature dendritic cells* | 58 700 | 12 577 | 9 844 |
| Mature dendritic cells* | 31 862 | 8 017 | 6 583 |
| Langerhans-like cells | 57 717 | 13 630 | 10 874 |
| Granulocytes | 31 466 | 8 007 | 6 821 |
| Lymphoid | |||
| CD4 T cells (naive) | 50 433 | 11 290 | 9 124 |
| CD4 T cells (memory, CCR4-negative) | 31 919 | 7 572 | 6 220 |
| CD4 T cells (memory, CCR4-positive) | 30 700 | 7 820 | 6 521 |
| Activated T cells (TH1)* | 32 219 | 8 111 | 6 676 |
| Activated T cells (TH2)* | 32 288 | 9 047 | 7 382 |
| CD8 T cells* | 51 017 | 11 789 | 9 380 |
| NK cells* | 34 831 | 8 187 | 6 569 |
| B cells | 53 236 | 10 903 | 8 499 |
| Activated B cells | 7 339 | 2 798 | 2 439 |
| Total | 709 990 | 38 961 | 27 323 |
Number of unique libraries is 17. Each cell was purified as described in “Materials and methods.” No. unique tags refers to number of tags observed in each cell. Unique transcripts representing the tags appeared more than once in all 17 libraries. Unique genes were counted using the UniGene database.
Published data.
We identified 709 990 SAGE tags that represent 112 555 distinct transcripts. Among them, to provide an accurate estimation of unique genes and to avoid sequencing errors, we omitted the tags that appeared only once in the data set. In 38 961 distinct tags that appeared more than once in the total libraries, 27 323 tags were represented as unique genes. Of the 27 323 genes, 14 557 tags had at least one match to a UniGene cluster, whereas 12 766 tags had no match. A list of all tags found is available at our Web site (http://bloodsage.gi.k.u-tokyo.ac.jp/). Among the 38 961 identified unique transcripts, 0.2% of these had more than 501 copies, 1.6% had between 500 and 51 copies, 16.8% had between 50 and 6 copies, and 81.4% had fewer than 5 copies (Table 2). Most of the unique transcripts were expressed at low levels; however, the mass of mRNAs with more than 5 copies per cell accounted for more than 80%. These categorized copies per cell in leukocytes were similar to those of other tissues from 3 496 829 tags, as described before. The gene abundance at moderate to high copies (more than 50 copies/cell) in activating cells such as LPS-stimulated monocytes, activated TH1 and TH2 cells, and mature DCs is higher than that of resting/nonstimulated cells (data not shown).
Summary and classification of sequenced tags in the combined leukocyte libraries
| Frequency . | Unique transcripts . | % . | Mass fraction mRNA (%) . | Unique genes . |
|---|---|---|---|---|
| More than 500 | 75 | 0.2 | 27.5 | 73 |
| 51-500 | 609 | 1.6 | 26.2 | 573 |
| 6-50 | 6 545 | 16.8 | 28.2 | 5 441 |
| 5 or fewer | 31 732 | 81.4 | 18.1 | 21 236 |
| Total | 38 961 | 100.0 | 100.0 | 27 323 |
| Frequency . | Unique transcripts . | % . | Mass fraction mRNA (%) . | Unique genes . |
|---|---|---|---|---|
| More than 500 | 75 | 0.2 | 27.5 | 73 |
| 51-500 | 609 | 1.6 | 26.2 | 573 |
| 6-50 | 6 545 | 16.8 | 28.2 | 5 441 |
| 5 or fewer | 31 732 | 81.4 | 18.1 | 21 236 |
| Total | 38 961 | 100.0 | 100.0 | 27 323 |
Frequency denotes the category of expression level analyzed in transcript copies per cell in the combined libraries. Unique genes represent a total number of unique genes matched to the Unigene cluster and to no reliable genes.
Genes selectively expressed in each type of leukocytes
At each stage of differentiation—from pluripotent hematopoietic stem cells to monocytes/macrophages, DCs, granulocytes, T cells, B cells, and NK cells—a different combination of restricted and ubiquitous regulatory factors may be important in inducing a distinct pattern of gene expression. Therefore, we examined the gene expression in each cell type. To assess a rigorous statistical significance of the observed differences, multiple statistical tests were conducted. We detected transcripts that were selectively expressed in each population of granulocytes, monocytes, macrophages, DCs, T cells, NK cells, and B cells (Supplemental Table S1 on Blood website; see the Supplemental Data Set link at the top of the online article). These data included well-known genes related to each cell type, such as the T-cell receptor gene for T cells, the perforin 1 gene for NK cells, and immunoglobulin-related genes for B cells.
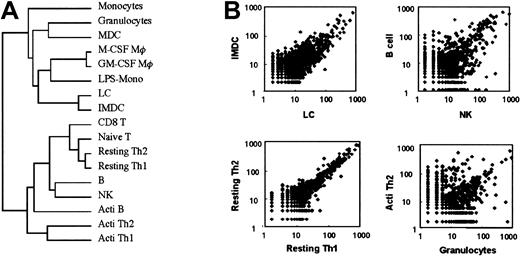
Next, to identify the overall similarities of the libraries derived from each leukocyte population, we used hierarchical cluster analysis. Resultant dendrograms were divided into myeloid and lymphoid cells (Figure 1A). Furthermore, to evaluate the resultant dendrograms, scatter plots for comparisons between arbitrarily selected cell types were also shown in Figure 1B.
Comparisons of cell types.
(A) Dendrogram of clustered libraries. (B) Scatter plot for comparisons between arbitrarily selected cell types. Frequency tables of the 17 libraries were first normalized to expression levels per 55 000 tags. Cluster and tree view programs were used for clustering of SAGE data. Briefly, the frequency tables of the 17 libraries were first normalized to expression levels per 55 000 tags. SAGE tags appearing at least 5 times in all 17 libraries were subjected to the clustering analysis described in “Materials and methods.” Acti indicates activated; Mφ, macrophages.
Comparisons of cell types.
(A) Dendrogram of clustered libraries. (B) Scatter plot for comparisons between arbitrarily selected cell types. Frequency tables of the 17 libraries were first normalized to expression levels per 55 000 tags. Cluster and tree view programs were used for clustering of SAGE data. Briefly, the frequency tables of the 17 libraries were first normalized to expression levels per 55 000 tags. SAGE tags appearing at least 5 times in all 17 libraries were subjected to the clustering analysis described in “Materials and methods.” Acti indicates activated; Mφ, macrophages.
Gene expression patterns of resting TH1 and TH2 cells were close, as shown in the mean of the tree view and scatter plots. In contrast, gene expression patterns in granulocytes and activated TH1 cells were entirely unrelated. Therefore, the tree view is shown as the similarity between the gene expression patterns. These data suggest that the genes are differentially expressed in each leukocyte population, depending on their differentiation stages.
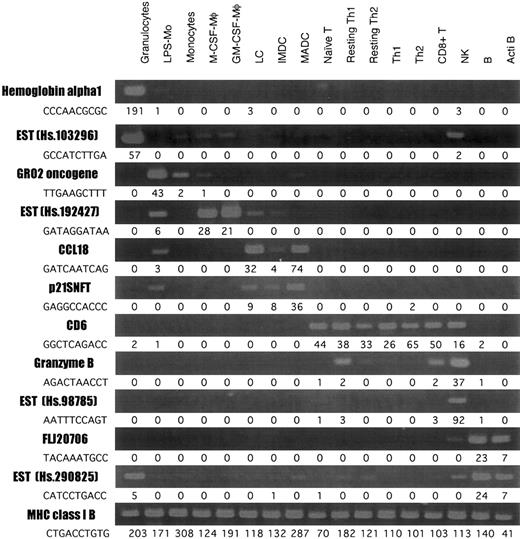
RT-PCR of genes selected in the SAGE analysis
To validate our SAGE data for leukocytes, we arbitrarily selected 12 differently expressed genes and evaluated them in cells obtained from more than 5 donor-derived samples by RT-PCR (Figure2), and the expression of each transcript was compared with SAGE data. MHC class IB was almost equally expressed in all cell types; and hemoglobin α 1 and EST (Hs. 103296) (granulocyte), Gro2 oncogene (monocyte), EST (Hs. 192427) (macrophage), CCL18 and p21SNFT (DCs), CD6 (T cell), granzyme B and EST (Hs. 98785) (NK cells), and FLJ20706 and EST (Hs. 290825) (B cells) were highly expressed in each group.
RT-PCR analysis of the genes differently expressed in leukocytes.
RT-PCR was performed on total RNA isolated from each cell type, as described in “Materials and methods.” The number next to each tag sequence is the number of tags per cell.
RT-PCR analysis of the genes differently expressed in leukocytes.
RT-PCR was performed on total RNA isolated from each cell type, as described in “Materials and methods.” The number next to each tag sequence is the number of tags per cell.
Comparison of expression patterns between myeloid and lymphoid cells
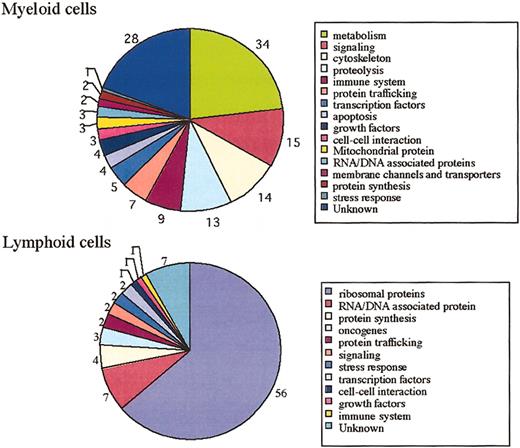
We next analyzed the genes differentially expressed in myeloid and lymphoid cells. Ubiquitously and highly expressed genes in myeloid and lymphoid cells are shown in Supplemental Table S2. The total number of genes significantly expressed in myeloid cells was 147 (P = .0 to 1 × 10−20). The total number of genes significantly expressed in lymphoid cells was 88 (P = .0 to 1 × 10−20). Functional breakdowns of genes selectively expressed in myeloid and lymphoid cells are depicted in Figure 3. Genes related to metabolism, signaling, cytoskeleton, proteolysis, membrane channels, and transporters were predominantly expressed in myeloid cells or at no or low levels in lymphoid cells. On the other hand, the transcripts for ribosomal proteins were highly expressed in lymphoid cells.
Functional classification of myeloid and lymphoid cells.
Putative functional breakdown of myeloid and lymphoid cell genes. The number next to each category indicates the total number of genes in that class. The total number of genes significantly and highly expressed in myeloid cells is 147. The total number of genes significantly and highly expressed in lymphoid cells is 88.
Functional classification of myeloid and lymphoid cells.
Putative functional breakdown of myeloid and lymphoid cell genes. The number next to each category indicates the total number of genes in that class. The total number of genes significantly and highly expressed in myeloid cells is 147. The total number of genes significantly and highly expressed in lymphoid cells is 88.
Gene expression in antigen-presenting cells
Genes commonly expressed in antigen-presenting cells (APCS), such as monocytes, macrophages, DCs, and B cells, are shown in Supplemental Table S3. As expected, genes related to major histocompatibility complex (MHC) class II were specifically expressed in APC libraries. APC-specific genes were composed of genes encoding proteins related to cytoskeleton, metabolism, proteolysis, capping (gelsolin-like protein), acid phosphatase 5, D component of complement,N-acetylglucosamine kinase, and solute carrier family 16.
Selectively and commonly expressed genes in leukocytes
To find out the leukocyte-specific genes, SAGE tags in leukocytes were compared with 4 280 231 tags in other tissues from the SAGE database (http://www.ncbi.nlm.nih.gov/SAGE/). L-plastin 1 (P = 7.8 × 10–636; UniGene no. 76506), proteoglycan 1 (P = 1.3 × 10–491; UniGene no. 1908), and dual-specificity phosphatase 2 (P = 3.1 × 10–387; UniGene no. 1883) were found to be more selectively and commonly expressed in leukocytes than in other tissues.
Discussion
Leukocytes play pivotal roles in inflammation and immunity. To molecularly define the type and function of human leukocytes, we performed SAGE in human leukocytes. Among 38 961 distinct tags that appeared more than once in all libraries combined, 27 323 tags were represented as unique genes. The number of genes assessed in leukocytes is reasonable for the current estimate of 30 000 to 40 000 genes predicted in the human genome.14 15
Hierarchical cluster analysis of expressed genes in leukocytes revealed 2 main branches as myeloid and lymphoid cells. In the lymphocyte branch, resting/circulating T cells were more alike as a group. Functionally, NK cells and CD8+ T cells played pivotal roles as cytotoxic lymphocytes in host defense; however, NK cells did not resemble CD8 T cells but did resemble B cells. In fact, several reports showed that NK and B cells were generated from the same precursor cells.16 17 Therefore, gene expression profiles in NK cells and CD8 T cells depended on origin rather than function.
Genes differentially expressed in each leukocyte type depended not only on the differentiation pathways but also on their functions. Metabolism, signaling, cytoskeleton, proteolysis, membrane channels, and transporter-related genes were expressed in myeloid cells but only at low or no levels in lymphoid cells. Myeloid cells responded to environmental stimuli as a major player in the primary host defense. Because myeloid cells infiltrated various tissues or made contact with other types of cells through immunologic synapses or adhesion proteins to process and present antigens, they may need specified gene expression to maintain these functions and higher amounts of energy than lymphoid cells. On the other hand, the transcripts of ribosomal proteins were highly expressed in lymphoid cells. Ribosomal proteins are known to play an important role in translational regulation, and they have been implicated in the control of differentiation, cellular transformation, tumor growth, and metastasis. However, the interrelation between individual cells and the massive transcription of ribosomal protein is still unclear. Transcription of a set of ribosomal proteins may be involved in enhancing the transcription of specific genes.
Through a comparison of leukocytes and other tissue, L-plastin 1, proteoglycan 1, and dual-specificity phosphatase 2 were observed as the specifically and commonly expressed genes in leukocytes. One of the specific genes, L-plastin, is capable of bundling actin filaments through its actin-binding domains and is related to cancer progression (eg, metastasis).18,19 L-plastin may regulate cell invasion. The function of proteoglycan 1, which is constitutively secreted by leukocytes, is less clear. Dual-specificity phosphatase 2 acts as a dual-specific protein phosphatase with stringent substrate specificity for MAP kinase.20 The genes may be important for the maintenance of the leukocyte function and can be good common markers for leukocytes.
This SAGE analysis showed the significant number of sequences that do not give reliable matches. However, at present, we cannot explain whether “no reliable matches” are real tags or not. Several possibilities may account for the appearance of these tags—sequences derived from hnRNA, PCR amplification error, unknown splicing variant of mRNA, random binding of oligo-dT to mRNA except for the poly A tail during construction of the cDNA library, and rear mRNA (highly specific expression in rear population). Recently, Saha et al21developed the Long SAGE methods that generate 21-bp tags derived from the 3′ ends of transcripts that can be rapidly analyzed and precisely matched to genomic sequence data. We will attempt the Long SAGE method to identify “not reliable matches” or “multiple matches” in leukocyte SAGE libraries, and we plan to improve this database.
In conclusion, identification of the genes specifically expressed in different types of leukocytes not only provides novel insight into the ontogeny and function of leukocytes, but also provides the diagnostic basis for blood and immune disorders such as rheumatism, diabetes, and atopic dermatitis.
Prepublished online as Blood First Edition Paper, January 9, 2003; DOI 10.1182/blood-2002-06-1866.
Supported by CREST/SORST and by a Grant-in-Aid for Scientific Research on Priority Areas (C) “Medical Genome Science” from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
S.N. and J.S. contributed equally to this work.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kouji Matsushima, Department of Molecular Preventive Medicine, School of Medicine, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan; e-mail:koujim@m.u-tokyo.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal