We developed a method to generate dendritic cells (DCs) from mouse embryonic stem (ES) cells. We cultured ES cells for 10 days on feeder cell layers of OP9, in the presence of granulocyte-macrophage colony-stimulating factor in the latter 5 days. The resultant ES cell–derived cells were transferred to bacteriologic Petri dishes without feeder cells and further cultured. In about 7 days, irregularly shaped floating cells with protrusions appeared and these expressed major histocompatibility complex class II, CD11c, CD80, and CD86, with the capacity to stimulate primary mixed lymphocyte reaction (MLR) and to process and present protein antigen to T cells. We designated them ES-DCs (ES cell–derived dendritic cells), and the functions of ES-DCs were comparable with those of DCs generated from bone marrow cells. Upon transfer to new dishes and stimulation with interleukin-4 plus tumor necrosis factor α, combined with anti-CD40 monoclonal antibody or lipopolysaccharide, ES-DCs completely became mature DCs, characterized by a typical morphology and higher capacity to stimulate MLR. Using an expression vector containing the internal ribosomal entry site–puromycinN-acetyltransferase gene or a Cre-lox–mediated exchangeable gene-trap system, we could efficiently generate ES cell transfectants expressing the products of introduced genes after their differentiation to DCs. ES-DCs expressing invariant chain fused to a pigeon cytochrome C epitope presented the epitope efficiently in the context of Ek. We primed ovalbumin (OVA)–specific cytotoxic T lymphocytes in vivo by injecting mice with ES-DCs expressing OVA, thus demonstrating immunization with ES-DCs genetically engineered to express antigenic protein. The methods may be applicable to immunomodulation therapy and gene-trap investigations of DCs.
Introduction
Dendritic cells (DCs) are the most potent antigen-presenting cells (APCs) responsible for priming of naive T cells in the immune response. DCs are involved in the maintenance of immunologic self-tolerance in the periphery, inducing regulatory T cells or anergy of autoreactive T cells.1-3 It has been reported that distinct subpopulations of DCs preferentially induce differentiation of either T helper 1 (Th1) or Th2 cells.4-6 Therefore, DCs physiologically play a central role in immunoregulation. Manipulation of functions of DCs by genetic modification and in vivo transfer of DCs with modified property is considered a promising means to control immune responses in an antigen-specific manner.7 As for the methods for gene transfer to DCs, electroporation, lipofection, and virus vector–mediated transfection have been developed. However, there are several problems related to presently used means (ie, efficiency of gene transfer, stability of gene expression, potential risk accompanying the use of virus vectors, and the immunogenicity of virus vectors). Although improvements have been made in these methods, development of more efficient and safer means is desirable.8 9
Embryonic stem (ES) cells are characterized by pluripotency and infinite propagation capacity. Non–virus-mediated methods for gene transfer, including targeted gene integration and procedures for isolation of appropriate recombinant cell clones, have been established for ES cells. Recently, a novel Cre-lox–mediated exchangeable gene-trap system has been developed using TT2 ES cells.10The method enables efficient gene-trap, plasmid rescue for the analysis of trapped genes, and targeted integration of replacement vectors to the gene-trapped sites. Genetic modification of ES cells and their subsequent in vitro differentiation to DCs would be an attractive strategy for genetic manipulation of DCs and for analysis of gene functions in DCs.
For hematopoietic differentiation of ES cells in vitro, embryoid body–mediated methods and the OP9-coculture method have been established.11,12 OP9 is a bone marrow stromal cell line that originated from macrophage colony-stimulating factor (M-CSF)–defective op/op mouse,13 and generation of various hematopoietic cells from ES cells using OP9 cells as feeder cells has been reported, including granulocytes, erythrocytes, B lymphocytes, and osteoclasts.11-15 The method has been applied to several molecular and cellular analyses for investigations of hematopoiesis.16-19
In the current study, we attempted to establish a method to develop DCs from ES cells in vitro. We adapted the OP9-coculture method for hematopoietic differentiation of ES cells. For induction of differentiation to DCs, we used granulocyte-macrophage colony-stimulating factor (GM-CSF), the cytokine essential for in vitro generation of DCs from hematopoietic cells.20-22 The generated ES cell–derived DCs were morphologically and functionally comparable with those differentiated in vitro from bone marrow cells. For gene transfer to ES cells to generate genetically modified DCs, we adopted 2 means: an expression vector containing β-actin promoter and internal ribosomal entry site (IRES)–puromycinN-acetyltransferase gene and Cre-lox–mediated targeted gene integration into gene-trapped ES cell clones.
Materials and methods
Mice
Balb/c, CBA, and C57BL/6 mice were obtained from Clea Animal (Tokyo, Japan) or Charles River (Hamamatsu, Japan) and kept in specific pathogen-free conditions. Male CBA and female C57BL/6 mice were mated to generate (CBA × C57BL/6) F1 mice.
Peptides, cell lines, and cytokines
The Ek-binding peptide pigeon cytochrome C (PCC)88-104, KAERADLIAYLKQATAK, and Kb-binding peptide ovalbumin (OVA)257-264, SIINFEKL, were synthesized with 9-fluorenylmethyloxycarbonyl (F-MOC) method on an automatic peptide synthesizer (PSSM8; Shimadzu, Kyoto, Japan) and purified using high-performance liquid chromatography. The ES cell line TT2, derived from (CBA × C57BL/6) F1 blastocysts,23 was maintained as described.24 The T-cell hybridomas, RF33.70, recognizing OVA257-264 in the context of Kb, and 2B4, recognizing PCC88-104 in the context of Ek, have been described elsewhere.25,26 The M-CSF–defective bone marrow–derived stromal cell line, OP9,12 was maintained in α–minimum essential medium (α-MEM) supplemented with 20% fetal calf serum (FCS); and to form feeder cell layers, OP9 cells were seeded onto culture plates precoated with gelatin. Recombinant mouse GM-CSF was provided by Kirin Brewery (Tokyo, Japan). Recombinant mouse interleukin-4 (IL-4), tumor necrosis factor α (TNF-α), and IL-1β were purchased from Peprotec (London, United Kingdom).
Hematopoietic differentiation of TT2 ES cells
The induction of hematopoietic differentiation of TT2 ES cells was done as described.11 After 15 days of culture on feeder cell layers of OP9 without exogenous cytokines, ES cell–derived cells were harvested by pipetting and were then subjected to cytospin preparation and May-Giemsa staining.
Induction of differentiation of ES cells into DCs
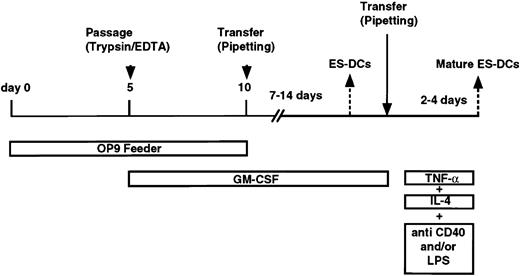
The procedure for induction of differentiation of ES cells into DCs is shown in Figure 1. ES cells were suspended in α-MEM supplemented with 20% FCS and seeded (1.5 × 104/2 mL medium/well) onto OP9 cell layers in 6-well plates. On day 3, half of the medium was removed and 2 mL fresh medium was added to each well. On day 5, cells were harvested using phosphate-buffered saline (PBS)/0.25% trypsin/1 mM EDTA (ethylenediaminetetraacetic acid), reseeded onto fresh OP9 cell layers, and cultured in α-MEM supplemented with 20% FCS and GM-CSF (1000 U/mL). At this step, cells recovered from 3 wells of 6-well culture plates were suspended in 20 mL medium and seeded into one 150-mm dish. On day 10 (5 days after the transfer), floating cells were recovered by pipetting. On average, 4 to 8 × 106cells were recovered from one 150-mm dish, thus indicating 100 to 200 times increase in cell number from undifferentiated ES cells. The recovered cells were transferred to bacteriologic Petri dishes (2.5 × 105 cells/90-mm dish) without feeder cells, and cultured in RPMI-1640 medium supplemented with 10% FCS, GM-CSF (500 U/mL), and 2-mercaptoethanol. After days 17 to 19, 1.5 to 2 × 105 floating or loosely adherent cells were recovered per dish (ES cell–derived dendritic cells [ES-DCs]), the number of cells increasing about 100 times over the number of undifferentiated ES cells. When over half the number of cells became adherent after day 12, the transfer of floating cells to fresh dishes on around day 15 improved the purity and yield of ES-DCs. To induce a complete maturation of ES-DCs, cells cultured for longer than 10 days in Petri dishes were transferred to fresh Petri dishes and cultured in RPMI/10% FCS without GM-CSF. The next day, IL-4 (10 ng/mL), TNF-α (5 ng/mL), plus anti-CD40 mAb (10 μg/mL, clone 3/23), or IL-4, TNF-α, plus lipopolysaccharide (LPS; 1 μg/mL) were added. After 2 or 3 days, cells were harvested by pipetting and used for functional and flow cytometric analysis. In most experiments, some cells harvested on days 5 and 10 were freeze-stocked for future use.
Generation of DCs from mouse bone marrow cells
Generation of dendritic cells from mouse bone marrow cells was done according to the reported procedures20 27 with some modifications. In brief, bone marrow cells were isolated from (C57BL/6 × CBA) F1 mice and cultured in bacteriologic Petri dishes (1.5 × 106/90-mm dish) in RPMI-1640 medium supplemented with 10% FCS, GM-CSF (500 U/mL), and 2-ME (50 μM). Culture medium was changed by half on days 5 and 10, and floating cells harvested by pipetting between 9 to 12 days of the culture were used as bone marrow–derived DCs (BM-DCs) in functional experiments. For the purpose of maturation, TNF-α (5 ng/mL) was added on the day before analysis.
Flow cytometric analysis
Staining of cells and analysis on a flow cytometer (FACScan, Becton Dickinson, San Jose, CA) was done as described previously.28 The procedure for intracellular staining with anti-human CD74 mAb was also described previously.29Antibodies used for staining were as follows: fluorescein isothiocyanate (FITC)–conjugated anti–H-2Kb(clone CTKb, mouse IgG2a; Caltag, Burlingame, CA), anti–I-Ab (clone 3JP, mouse IgG2a), anti–I-Ek(clone 8705-A, mouse IgG2a; Cedarlane, Hornby, Canada), anti-mouse CD11c (clone N148, hamster IgG; Chemicon, Temecula, CA), R-PE–conjugated anti-mouse CD80 (clone RMMP-1, rat IgG2a; Caltag), R-PE–conjugated anti-mouse CD86 (clone RMMP-2, rat IgG2a; Caltag), anti-mouse CD40 (clone 3/23, rat IgG2a; Serotec, Oxford, United Kingdom), R-PE–conjugated anti-F4/80 (A3-1, rat IgG2b; Serotec), anti-mouse CD205 (clone NLDC-145, rat IgG2a; Serotec), FITC-conjugated anti-mouse CD8α (clone 53-6.7, rat IgG2a; Pharmingen, San Diego, CA), FITC-conjugated anti-human CD74 (clone M-B741, mouse IgG2a; Pharmingen), FITC-conjugated goat anti–mouse Ig (Pharmingen), FITC-conjugated goat anti–hamster IgG (Caltag), FITC-conjugated goat anti–rat Ig (Pharmingen), mouse IgG2a control (clone G155-178; Pharmingen), FITC-conjugated mouse IgG2a control (clone G155-178; Pharmingen), R-PE–conjugated rat IgG2a control (clone LO-DNP-16; Caltag), FITC-conjugated rat IgG2a control (clone LODNP-57; Beckman-Coulter, Tokyo, Japan), hamster IgG control (clone 530-6; Caltag), and rat IgG2a control (clone LO-DNP-16; Caltag).
Mixed lymphocyte reaction (MLR)
Splenic mononuclear cells were prepared from unprimed female Balb/c mice, and T cells were isolated from the splenic mononuclear cells by magnetic cell sorting using anti-CD90 (Thy1.2) supermagnetic MicroBeads (Miltenyi Biotec, Bergisch-Gladbach, Germany), and then used as responders. Graded numbers of stimulator cells were x-ray irradiated (35 Gy) and cocultured with responder cells (1.5 × 105) in wells of 96-well round-bottomed culture plates and cultured for 4 days. [3H]-thymidine (6.7 Ci/mmol [247.9 GBq/mmol]) was added to the culture (1 μCi/well [0.037 MBq/well]) in the last 16 hours. At the end of the culture, cells were harvested onto glass fiber filters (Wallac, Turku, Finland), and the incorporation of [3H]-thymidine was measured by scintillation counting.
Antigen presentation assay
DCs were seeded onto 96-well flat-bottomed culture plates with or without PCC protein (50 μg/mL; Sigma, St Louis, MO) or peptide (10 μM). After 6 hours, 2B4 hybridoma cells were added (5 × 104/well). After 24 hours of culture, the supernatant (50 μL/well) was collected and added to cultures of the IL-2–dependent cell line, CTLL-20 (5 × 103/100 μL/well), in 96-well flat-bottomed culture plates. After 16 hours, [3H]-thymidine was added and cells were incubated for a further 8 hours. The incorporation of [3H]-thymidine by CTLL-20 was measured by scintillation counting. In some experiments, ES-DCs were fixed as follows. Cells were washed with PBS and suspended in PBS at 106 cells/mL, and diluted glutaraldehyde was added to the final concentration of 0.002%. After incubation for 30 seconds at room temperature, equal volume of 0.2 ML-lysine/PBS was added and mixed gently, and cells were incubated for 1 minute at room temperature. Cells were sequentially washed with PBS and with culture medium and used in the experiments. In experiments using BM-DCs pulsed with peptide, OVA or PCC peptide was added to the final concentration of 1 or 10 μM, incubated for 4 hours, washed twice, and used as stimulator cells.
Plasmid construction
To obtain pCI-PCC, the expression vector presenting PCC epitope on major histocompatibility complex (MHC) class II molecules and driven by the SRα promoter, double-stranded oligo DNA encoding the PCC epitope, 5′-AAGGCAGAAAGGGCAGACCTAATAGCTTATCTTAAACAAGCTACTGCCAAG-3′, was inserted into the previously reported human invariant chain–based epitope-presenting vector, pCI.30 The coding region of this construct was transferred to pCAG-IP,31 a mammalian expression vector containing the chicken β-actin promoter and IRES-puromycin N-acetyltransferase gene cassette, to generate pCAG-PCC-IP. The replacement vector, p6SEFPPF, and the Cre expression vector, pCAGGS-Cre, have been described elsewhere.10 32-35 A cDNA fragment coding for OVA protein was inserted into a mammalian expression vector pCAGGS to make pCAGGS-OV. The DNA fragment containing β-actin promoter–cDNA for OVA-rabbit β-globin poly (A) signal was excised from this construct and inserted into p6SEFPPF replacing the enhanced green fluorescent protein–coding sequence to obtain p6AOVP.
Quantitative analysis of β-galactosidase activity of ES-DCs
Generation and characterization of gene-trapped TT2 ES cell clones, in which lox-β-geo(β-galactosidase/neomycin resistance fusion gene)–lox cassette was introduced as a reporter gene, have been reported elsewhere.10 ES cell clones were differentiated to DCs according to the procedure described above. After differentiation, ES-DCs were suspended in lysis buffer (150 mM NaCl/20 mM tris(hydroxymethyl)aminomethane [Tris-HCl], pH 8.0), and cell lysates were prepared by 2 cycles of freezing and thawing. For each cell lysate sample, the protein concentration was measured using a protein assay kit (MIRCO BCA assay kit; Pierce, Rockford, IL) and the β-galactosidase (β-gal) activity was quantified using a β-gal assay kit (Gene Therapy Systems, San Diego, CA) using chlorophenol red-β-D-galactopyranoside as substrate. Relative β-gal activity (β-gal activity/protein concentration) was calculated for each sample, and the gene-trapped ES cell clone showing the highest β-gal activity after DC differentiation was selected for transfection with the replacement vector.
Transfection of ES cells
TT2 ES cells maintained on layers of primary embryonic fibroblasts (PEFs) were harvested and suspended in Dulbecco modified Eagle medium at a concentration of 2.5 × 107/mL, and 1 × 107 cells were electroporated in a 4-mm gap cuvette under the condition of 200 V and 950 μF. For transfection with pCI-PCC or pCAG-PCC-IP, 40 μg linearized plasmid DNA was used. For Cre-mediated targeted integration of the replacement vector into a gene-trap ES clone, 20 μg each of p6AOVP and pCAGGS-Cre in circular form were used for transfection. Site-specific integration into ES cells using a pair of mutant lox, lox71 and lox66, has been described previously.33Transfected ES cells were cultured on PEF feeder layers in 90-mm culture dishes and selected with puromycin (2 μg/mL) on days 3, 5, and 7 for 24 hours each, and drug-resistant colonies were picked up on day 9 into 24-well culture plates with PEFs.
Transfer of ES-DCs into mice and cytotoxity assay
ES-DCs were stimulated with IL-4, TNF-α, and anti-CD40 mAb and injected intraperitoneally into mice (5 × 105cells/mouse). Injections were given twice at a 7-day interval, and 7 days after the second injection, mice were killed and spleen cells isolated. Spleen cells were treated with hemolysis buffer (140 mM NH4Cl, 10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.4) for one minute, washed, and cultured in 24-well culture plates (2.5 × 106/well) in 45% RPMI/45% AIMV/10% horse serum supplemented with recombinant human IL-2 (100 U/mL) and OVA257-264 peptide (0.1 μM). After 5 days, cells were recovered, and viable cells isolated with lympholyte-M (Cedarlane) were used as effector cells. As target cells, EL-4, a thymoma cell line which originated from a C57BL/6 mouse, was labeled with sodium [51Cr]-chromate for one hour at 37°C and washed twice with RPMI/10% FCS. Subsequently, target cells were incubated in 24-well culture plates (1 × 106cells/well) with or without 10 μM OVA peptide for 3 hours, harvested, washed with RPMI/10% FCS, and seeded onto 96-well round-bottomed culture plates (5 × 103 cells/well). Effector cells were added (5 × 104cells/well) to the target cells and incubated for 4 hours at 37°C. At the end of the incubation, the plates were centrifuged, and supernatants (50 μL/well) were harvested and counted on a gamma counter. The percentage of specific lysis was calculated as: 100 × [(experimental release − spontaneous release)/(maximal release − spontaneous release)]. Spontaneous release and maximal release were determined in the presence of either medium or 1% Triton X-100, respectively.
Results
Application of OP9-coculture method to generate hematopoietic cells from TT2 ES cells
For induction of hematopoietic differentiation of ES cells, we used OP9 cells, a bone marrow stromal cell line defective in the M-CSF gene,13 as feeder cells. Successful hematopoietic differentiation by the OP9-coculture method of ES cell lines derived from 129 strain of mice, such as D3 and E14, has been reported.11,12,14,17 In the current study, we used another line of ES cell, TT2, established from (CBA × C57BL/6) F1 blastocysts,23 which has been used in gene targeting to generate many lines of mutant mice and also for a large-scale gene-trap project.10 24
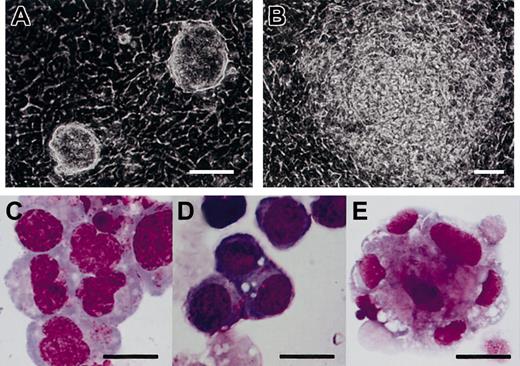
To determine if the OP9-coculture method can be applied to TT2, we cultured TT2 ES cells following the reported culture procedure.11 TT2 ES cells, maintained on PEFs in the presence of leukemia inhibitory factor (LIF), were transferred onto the OP9 cell layer and cultured without exogenous cytokines (Figure 2A). Most of the ES cell colonies showed a differentiated morphology in 4 to 5 days (Figure 2B). After 5 days of culture on OP9 feeder layers, ES cell–derived cells were transferred onto freshly prepared OP9 feeder layers and cultured for another 10 days. Cells of various morphologies floating or loosely adherent to the feeder cells appeared. We harvested the differentiated cells and examined them after May-Giemsa staining. As shown in Figure 2C-E, we observed hematopoietic cells of at least erythroid, myeloid, and megakaryocytic lineage. Therefore, this method is applicable also to TT2 ES cells.
Hematopoietic differentiation of TT2 ES cells on feeder cell layers of OP9.
(A-B) Phase-contrast micrographs of TT2 ES cell colonies on OP9 feeder cell layers on day 3 (A) and day 5 (B) are shown. (C-E) May-Giemsa staining of cytospin specimens of hematopoietic cells derived from TT2 ES cells. TT2 cells were cultured on OP9 feeder cell layer for 15 days in total, without addition of exogenous cytokines. Floating cells were applied to cytospin preparations and stained with May-Giemsa. Cells of myeloid (C), erythroid (D), and megakaryocytic (E) lineages are shown. Scale bars represent 50 μm (A-B); and 20 μm, (C-E).
Hematopoietic differentiation of TT2 ES cells on feeder cell layers of OP9.
(A-B) Phase-contrast micrographs of TT2 ES cell colonies on OP9 feeder cell layers on day 3 (A) and day 5 (B) are shown. (C-E) May-Giemsa staining of cytospin specimens of hematopoietic cells derived from TT2 ES cells. TT2 cells were cultured on OP9 feeder cell layer for 15 days in total, without addition of exogenous cytokines. Floating cells were applied to cytospin preparations and stained with May-Giemsa. Cells of myeloid (C), erythroid (D), and megakaryocytic (E) lineages are shown. Scale bars represent 50 μm (A-B); and 20 μm, (C-E).
Development of DCs from ES cells
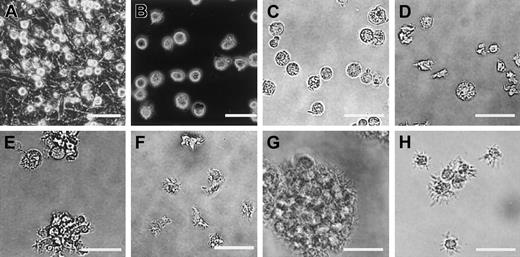
To induce differentiation to DCs, the above-described mesodermally differentiated ES cell–derived cells harvested from a 5-day culture on OP9 feeder layers were cultured on fresh OP9 cell layers in the presence of exogenous GM-CSF (Figure 1). In comparison with the culture without exogenous GM-CSF, addition of this cytokine resulted in appearance of a larger number of floating cells. On day 8 (3 days after the transfer), we observed many round and relatively homogenous floating cells (Figure 3A) and most expressed CD11b, suggesting their commitment to myeloid cell lineage. On day 10, cells floating or loosely adherent to feeder cells were recovered by pipetting and transferred to bacteriologic Petri dishes without feeder cells (Figure 3B-C). After this passage, some (< 30%) of the transferred cells adhered to the dish surface and resembled macrophages. On days 15 to 18, floating cells could be divided roughly into 2 types, 1 with a round shape and of a larger size, the other smaller and irregularly shaped with protrusions (Figure3D). In addition, clusters of floating cells (Figure 3E) of the latter type were observed after days 17 to 19, and the cell clusters gradually increased. Floating cells were positive for MHC class I, MHC class II, CD80, CD86, DEC205, and CD11c (Figure 4A-B). Based on the morphology, surface phenotype, and function (described below), we referred to cells with protrusions as ES-DCs. ES-DCs were positive for F4/80 and CD11b and negative for CD8α, suggesting that they were of myeloid lineage. However, further analysis may be necessary to definitely determine the lineage of ES-DCs. Further culture in Petri dishes in the presence of GM-CSF resulted in appearance of cells with longer protrusions showing morphology of mature DCs (Figure3F).
Morphology of ES cell–derived DCs.
ES cell–derived cells on day 8 (A), day 12 (B-C), day 17 (D-E), and day 27 (F) of differentiation culture are shown. Cells on day 24 were recovered and stimulated for 2 days with IL-4, TNF-α, plus agonistic anti-CD40 mAb (G), or with IL-4, TNF-α, plus LPS (H). Panels A and B are phase-contrast micrographs. Scale bars represent 20 μm.
Morphology of ES cell–derived DCs.
ES cell–derived cells on day 8 (A), day 12 (B-C), day 17 (D-E), and day 27 (F) of differentiation culture are shown. Cells on day 24 were recovered and stimulated for 2 days with IL-4, TNF-α, plus agonistic anti-CD40 mAb (G), or with IL-4, TNF-α, plus LPS (H). Panels A and B are phase-contrast micrographs. Scale bars represent 20 μm.
Induction of maturation of ES-DCs
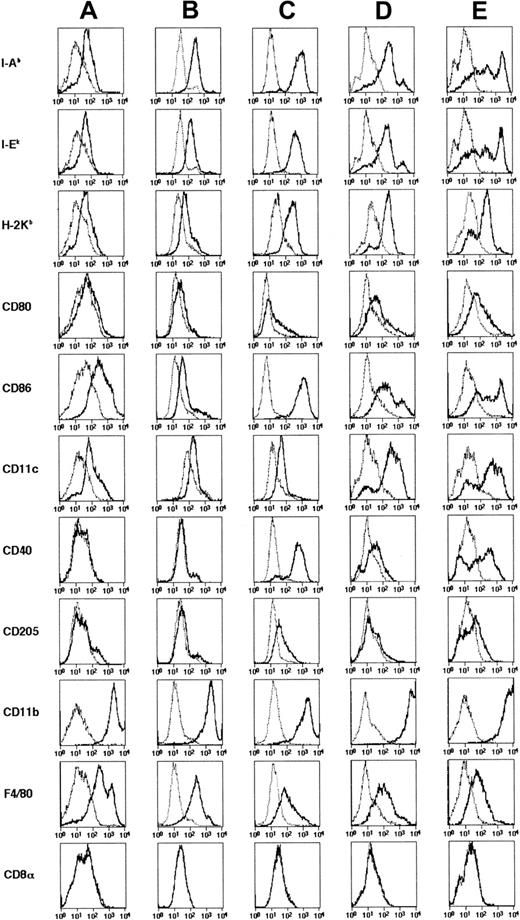
The induction of maturation by separating floating cells from adherent cells was noted in culture of bone marrow– and skin-derived DCs.20,27,36 37 To induce further maturation of ES-DCs, we recovered floating cells from 14- to 16-day cultures in Petri dishes (on days 24-26 in Figure 1) and transferred these to new Petri dishes. Some transferred cells became typical mature DCs in 2 to 3 days, bearing large veils or long dendritic protrusions, along with dead cells and adherent cells. We tested several factors reported to have effects on maturation of DCs (IL-4, TNF-α, IL-1β, anti-CD40 mAb, and LPS) in single use or in combinations. As a result, we found that the combination of IL-4, TNF-α, plus anti-CD40, or IL-4, TNF-α, plus LPS drastically increased the number of typical mature DCs. Most of the mature ES-DCs formed clusters if stimulated with IL-4, TNF-α, plus anti-CD40 (Figure 3G). On the other hand, most floating cells did not cluster if stimulated with IL-4, TNF-α, plus LPS (Figure 3H). The stimulated ES-DCs expressed higher levels of surface MHC class I, MHC class II, CD80, and CD86 than did cells before the treatment (Figure 4A-C). In addition, cells stimulated with IL-4, TNF-α, plus LPS expressed CD40, for which ES-DCs before stimulation were negative. On the other hand, expression of CD11b and F4/80 decreased slightly and that of CD11c did not change significantly. We compared the surface phenotype of ES-DCs with that of BM-DCs generated by culture in the presence of GM-CSF for 10 days (Figure 4D) and those further stimulated with TNF-α (Figure 4E). Levels of expression of MHC class II, CD40, CD80, and CD86 were almost comparable between mature ES-DCs and BM-DCs.
Surface phenotypes of DCs differentiated from ES cells and bone marrow cells.
ES-DCs on day 17 (A), day 26 (B), and when given maturation stimuli (C) were analyzed using flow cytometry on the surface expression of indicated molecules. (C) For analysis on expression of CD11c, CD40, and CD205, cells were stimulated with IL-4, TNF-α, plus LPS, and for the analysis on expression of other molecules, cells stimulated with IL-4, TNF-α, plus anti-CD40 mAb were used. For comparison, DCs generated from bone marrow cells by 10-day culture in the presence of GM-CSF (D) and those stimulated with TNF-α for 20 hours (E) were also analyzed. Staining patterns with specific antibodies (thick lines) and isotype-matched controls (thin dotted lines) are shown.
Surface phenotypes of DCs differentiated from ES cells and bone marrow cells.
ES-DCs on day 17 (A), day 26 (B), and when given maturation stimuli (C) were analyzed using flow cytometry on the surface expression of indicated molecules. (C) For analysis on expression of CD11c, CD40, and CD205, cells were stimulated with IL-4, TNF-α, plus LPS, and for the analysis on expression of other molecules, cells stimulated with IL-4, TNF-α, plus anti-CD40 mAb were used. For comparison, DCs generated from bone marrow cells by 10-day culture in the presence of GM-CSF (D) and those stimulated with TNF-α for 20 hours (E) were also analyzed. Staining patterns with specific antibodies (thick lines) and isotype-matched controls (thin dotted lines) are shown.
Stimulation of primary MLR by ES-DCs
To test ES-DCs for the capacity to activate naive T cells, we did primary MLR assays using ES-DCs as stimulators. Because H-2 of TT2 cells is of the k/b haplotype, allogenic splenic T cells purified from unprimed Balb/c mice (H-2d) were used as responders. ES-DCs prepared from 7-day culture in Petri dishes (day 17 in Figure 1) had a capacity to stimulate MLR comparable with that of BM-DCs cultured for 10 days27 (Figure5A), although it is possible that BM-DCs generated by different culture protocols have stronger stimulation capacity. In contrast, ES cell–derived cells harvested from a 10-day culture on OP9 feeder cell layers (day 10 in Figure 1) did not stimulate T cells. We also tested DCs given maturation stimuli (Figure5B). We tested the T-cell stimulatory capacity of ES-DCs treated with 3 different stimulation cocktails: IL-4/TNF-α/anti-CD40 mAb, IL-4/TNF-α/LPS, or IL-4/TNF-α/anti-CD40 mAb/LPS. Consistent with the elevation of surface MHC class II and costimulatory molecules, the maturation stimuli enhanced the capacity to stimulate T cells.
Allogeneic MLR-stimulating capacity of ES-DCs.
(A) Graded numbers of ES-DCs harvested on day 17 (⋄), ES-derived cells on day 10 (■), and (CBA × C57BL/6) F1 BM-DCs cultured for 11 days (○) were irradiated and cocultured with splenic T cells (1.5 × 105) isolated from Balb/c mice for 4 days, and the proliferative response of T cells in the last 16 hours of the culture was measured by [3H]-thymidine uptake. (B) ES-DCs harvested on day 24 and stimulated with IL-4/TNF-α/anti-CD40 (⋄), IL-4/TNF-α/LPS (▵), or IL-4/TNF-α/anti-CD40/LPS (▪) were used as stimulators. BM-DCs cultured for 10 days and stimulated with TNF-α (○) for 20 hours were also used as stimulators. For control, ES-derived cells cultured on OP9 feeder cell layers for 10 days were also used as stimulators (■). Mean values in kilo cpm (kcpm) ± standard deviation for triplicate cultures are shown.
Allogeneic MLR-stimulating capacity of ES-DCs.
(A) Graded numbers of ES-DCs harvested on day 17 (⋄), ES-derived cells on day 10 (■), and (CBA × C57BL/6) F1 BM-DCs cultured for 11 days (○) were irradiated and cocultured with splenic T cells (1.5 × 105) isolated from Balb/c mice for 4 days, and the proliferative response of T cells in the last 16 hours of the culture was measured by [3H]-thymidine uptake. (B) ES-DCs harvested on day 24 and stimulated with IL-4/TNF-α/anti-CD40 (⋄), IL-4/TNF-α/LPS (▵), or IL-4/TNF-α/anti-CD40/LPS (▪) were used as stimulators. BM-DCs cultured for 10 days and stimulated with TNF-α (○) for 20 hours were also used as stimulators. For control, ES-derived cells cultured on OP9 feeder cell layers for 10 days were also used as stimulators (■). Mean values in kilo cpm (kcpm) ± standard deviation for triplicate cultures are shown.
Antigen processing and presentation by ES-DCs
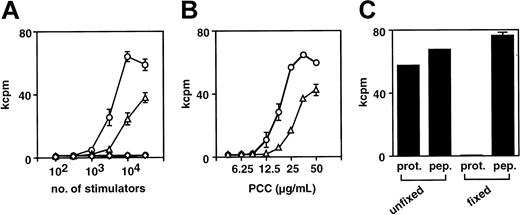
To examine the APC function of ES-DCs, presentation of PCC epitope to a T-cell hybridoma, 2B4, recognizing PCC88-104 in the context of I-Ek, was tested. ES-DCs cultured for 17 days in Petri dishes (day 27 in Figure 1) and bone marrow–derived DCs cultured for 10 days were harvested and incubated with PCC protein, to allow for capture of the antigen. After 6 hours, 2B4 cells were added and presentation of the PCC epitope by DCs was measured by quantifying the IL-2 produced by 2B4. We compared functions of these 2 kinds of DCs under the conditions of different numbers of APCs and concentrations of antigenic protein. As shown in Figure 6A-B, APC activity of ES-DCs was even stronger than BM-DCs. To rule out the possibility that the response of T cells was induced by direct binding of peptide fragments contaminated in the PCC protein to I-E molecules, we fixed ES-DCs with glutaraldehyde. As shown in Figure 6C, fixation of ES-DCs before addition of the protein abrogated the response of T cells, whereas ES-DCs fixed under the same condition and added with PCC88-104 synthetic peptide induced the response of 2B4. These results indicate that ES-DCs can capture and process soluble protein antigens and present the resultant peptides in the context of MHC class II molecules.
Antigen-presenting capacity of ES-DCs.
(A) Graded numbers of ES-DCs (○) and BM-DCs (▵) were incubated with PCC protein (50 μg/mL) for 6 hours, and subsequently 2B4 hybridoma cells were added (5 × 104/well). ES-DCs (●) and BM-DCs (⋄) were also tested in the absence of the protein. After 24 hours, culture supernatant was collected and IL-2 produced by the hybridoma was measured by proliferation of CTLL-20 cells. (B) ES-DCs (○) and BM-DCs (▵) were plated (3 × 104/well) and incubated with graded doses of PCC protein for 6 hours and subsequently added with 2B4 cells. IL-2 production by 2B4 was measured as in panel A. (C) ES-DCs fixed with glutaraldehyde or left unfixed were plated (3 × 104/well) and incubated with PCC protein (50 μg/mL, prot.) or PCC88-104 peptide (10 μM, pep.) for 6 hours, and subsequently 2B4 hybridoma cells were added. Results were expressed as mean cpm of triplicate (A-B) or duplicate (C) cultures ± standard deviation.
Antigen-presenting capacity of ES-DCs.
(A) Graded numbers of ES-DCs (○) and BM-DCs (▵) were incubated with PCC protein (50 μg/mL) for 6 hours, and subsequently 2B4 hybridoma cells were added (5 × 104/well). ES-DCs (●) and BM-DCs (⋄) were also tested in the absence of the protein. After 24 hours, culture supernatant was collected and IL-2 produced by the hybridoma was measured by proliferation of CTLL-20 cells. (B) ES-DCs (○) and BM-DCs (▵) were plated (3 × 104/well) and incubated with graded doses of PCC protein for 6 hours and subsequently added with 2B4 cells. IL-2 production by 2B4 was measured as in panel A. (C) ES-DCs fixed with glutaraldehyde or left unfixed were plated (3 × 104/well) and incubated with PCC protein (50 μg/mL, prot.) or PCC88-104 peptide (10 μM, pep.) for 6 hours, and subsequently 2B4 hybridoma cells were added. Results were expressed as mean cpm of triplicate (A-B) or duplicate (C) cultures ± standard deviation.
Genetic modification of ES-DCs using an expression vector containing IRES-puromycin N-acetyltransferase cassette
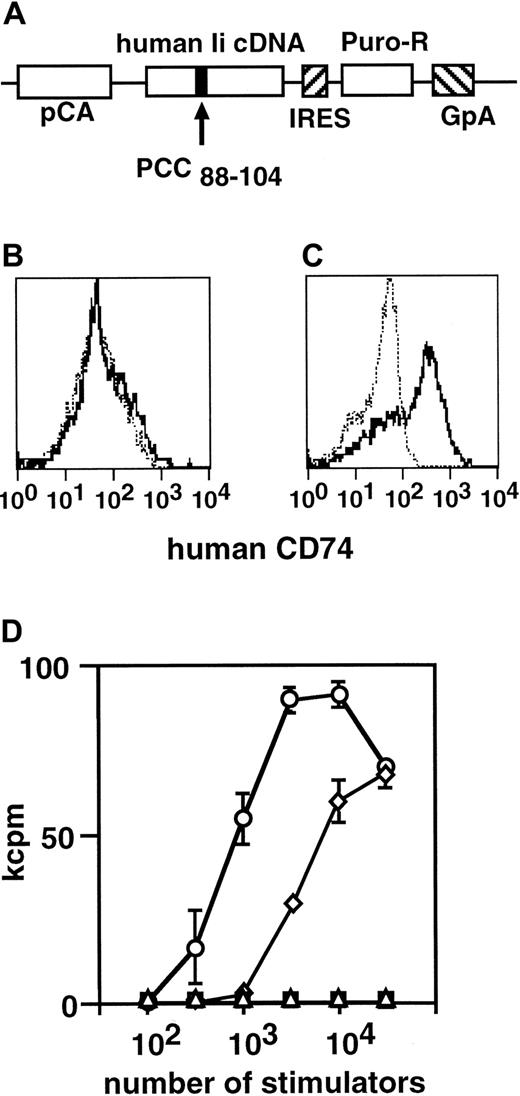
For the genetic modification of ES-DCs, we planned to introduce expression vectors into ES cells and develop DCs from the ES cell transfectants. For this purpose, we first used the MHC class II–restricted epitope presentation vector, pCI,29 30 in which the class II–associated invariant chain peptide (CLIP) region of the human invariant chain (Ii) is substitutive with antigenic peptides and the expression in mammalian cells is driven by the SRα promoter. We inserted a DNA fragment coding PCC88-104 into pCI to make pCI-PCC and transfected ES cells with this vector. We differentiated 48 transfected ES cell clones to DCs, and examined their expression of human Ii (CD74), using flow cytometry. To our disappointment, only 1 of 48 clones expressed human Ii and the level of expression of the clone was very low.
To improve the efficiency of expression of the introduced genes, we used a vector containing a β-actin promoter and an IRES-puromycin N-acetyltransferase cassette, pCAG-IP.31 We transferred the DNA fragment coding for Ii-PCC88-104 fusion protein from pCI-PCC to pCAG-IP to obtain pCAG-PCC-IP (Figure 7A) and transfected ES cells with this construct. We picked up puromycin-resistant ES cell clones and differentiated them to DCs. We observed that 24 (85%) of 28 clones analyzed expressed human Ii at the level readily detectable by flow cytometric analysis (Figure 7B-C). We examined ES-DCs presenting the PCC epitope generated by this procedure for the potential to stimulate the PCC-specific T-cell hybridoma, 2B4. As shown in Figure 7D, the ES-DCs introduced with the Ii-PCC88-104 expression vector stimulated the T-cell hybridoma more efficiently than that of BM-DCs prepulsed with 10 μM synthetic PCC peptide for 4 hours.
Introduction of an epitope-presenting vector to ES-DCs.
(A) The structure of PCC epitope presentation vector, pCAG-PCC-PI, is shown. CLIP region of human Ii cDNA was replaced with an oligo DNA encoding PCC88-104. The expression of this gene is driven by the chicken β-actin promoter (pCA). The mutant Ii coding sequence is followed by IRES-puromycin N-acetyltransferase gene (Puro-R) and polyadenylation signal sequence of the human growth hormone (GpA).(B-C) Flow cytometric analysis of ES-DCs on the expression of human Ii. ES-DCs without (B) or with (C) expression construct were stained intracellularly with antihuman Ii (CD74) (thick lines) or isotype-matched control mAb (thin dotted lines). (D) Stimulation of PCC-specific T-cell hybridoma, 2B4, by ES-DCs with (○) or without (▪) PCC epitope-presenting vector. As a control, BM-DCs pulsed with PCC peptide (10 μM) for 4 hours (⋄) or left unpulsed (▵) were also used as stimulators. Stimulator cells and hybridomas were cocultured for 24 hours, and IL-2 produced by 2B4 was measured by proliferation of CTLL-20 cells. Results were expressed as mean cpm of triplicate cultures ± standard deviation.
Introduction of an epitope-presenting vector to ES-DCs.
(A) The structure of PCC epitope presentation vector, pCAG-PCC-PI, is shown. CLIP region of human Ii cDNA was replaced with an oligo DNA encoding PCC88-104. The expression of this gene is driven by the chicken β-actin promoter (pCA). The mutant Ii coding sequence is followed by IRES-puromycin N-acetyltransferase gene (Puro-R) and polyadenylation signal sequence of the human growth hormone (GpA).(B-C) Flow cytometric analysis of ES-DCs on the expression of human Ii. ES-DCs without (B) or with (C) expression construct were stained intracellularly with antihuman Ii (CD74) (thick lines) or isotype-matched control mAb (thin dotted lines). (D) Stimulation of PCC-specific T-cell hybridoma, 2B4, by ES-DCs with (○) or without (▪) PCC epitope-presenting vector. As a control, BM-DCs pulsed with PCC peptide (10 μM) for 4 hours (⋄) or left unpulsed (▵) were also used as stimulators. Stimulator cells and hybridomas were cocultured for 24 hours, and IL-2 produced by 2B4 was measured by proliferation of CTLL-20 cells. Results were expressed as mean cpm of triplicate cultures ± standard deviation.
Cre-lox–mediated targeted gene introduction into gene-trapped ES cell clones
As an alternative strategy to efficiently obtain ES cell transfectants, which expressed the introduced genes after differentiation to DCs, we used targeted gene integration into gene-trapped ES cell clones. Cell lineage–specific gene-expression patterns are regulated not only by transcription factors that bind specific nucleotide sequences but also by epigenetic mechanisms; that is, activation and inactivation of specific chromatin domains are controlled by the status of histone acetylation and DNA methylation. We supposed that, by integrating expression constructs into certain chromosomal region replacing some gene, which is actively transcribed in DCs, we could efficiently obtain ES cell transfectant clones expressing the introduced genes after differentiation to DCs.
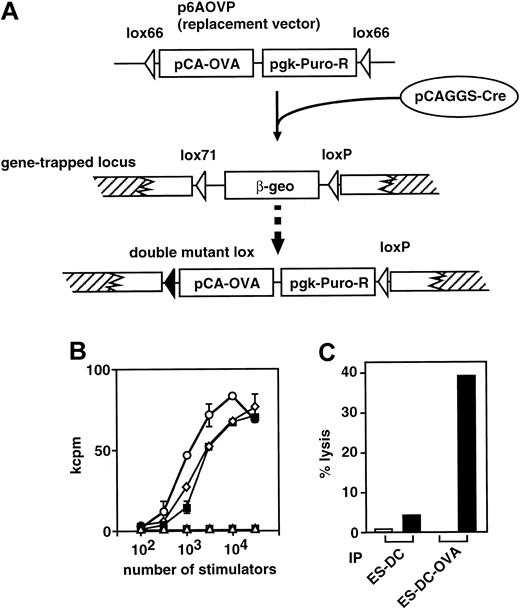
A library of gene-trapped TT2 ES cells has been recently generated, in which a lox-β-geo-lox cassette as reporter gene was inserted into various chromosomal regions, trapping various genes.10 From the gene-trapped ES cell library, we first selected 20 ES cell clones that showed strong β-gal activity when differentiated into mesodermal cell lineage. These ES cell clones were subjected to the DC-differentiation culture, and resultant ES-DCs were analyzed for β-gal activity. From the 20 gene-trapped ES cell clones, we chose 1 clone that showed the highest β-gal activity after differentiation to DCs and that had a DC-differentiation capacity comparable with that of parental TT2 ES cells. We transfected the selected clone with a replacement vector, p6AOVP, to introduce OVA expression cassette (OVA cDNA driven by the rabbit β-actin promoter ligated with the puromycin N-acetyltransferase gene driven by the pgk promoter) replacing the β-geo sequence (Figure8A). After cotransfection with p6AOVP and pCAGGS-Cre, which was for transient expression of Cre recombinase, and selection with puromycin, we picked up puromycin-resistant clones and induced their differentiation to DCs. We analyzed the expression of OVA protein after differentiation by detecting the capacity of each cell clone to stimulate RF33.70, a T-cell hybridoma specific to OVA257-264. Of 10 clones examined, 9 (90%) strongly stimulated the hybridoma. The result of analysis using ES-DCs generated from one of the ES cell clones introduced with the replacement vector is shown in Figure 8B. The ES-DCs were superior to BM-DCs prepulsed with 1 or 10 μM OVA257-264 peptide in stimulating the hybridoma. Taken together, this exchangeable gene-trap system allows one to efficiently generate ES cell transfectants, which highly express the introduced genes after differentiation to DCs.
Generation of ES-DCs expressing OVA antigen and in vivo priming of OVA-specific cytotoxic lymphocytes.
(A) Cre/mutant lox–mediated targeted integration of the expression cassette to a gene-trapped locus is schematically depicted. The expression cassette, the OVA cDNA driven by the chicken β-actin promoter (pCA-OVA) ligated with the puromycinN-acetyltransferase gene driven by the pgk promoter (pgk-Puro-R), was inserted to the gene-trapped site, replacing the β-geo sequence by the effect of Cre recombinase transiently expressed from pCAGGS-Cre. The combination of a lox66 of the replacement vector and a lox71 of a trapped locus resulted in a double-mutant lox, which inhibited further recombination of the replaced allele and increased the frequency of proper recombinant ES cell clones. (B) Stimulation of OVA-specific T-cell hybridoma, RF33.70, with ES-DCs with (○) or without (■) expression of OVA was analyzed. As a control, bone marrow–derived DCs pulsed with 10 μM (⋄) or 1 μM (▪) of OVA257-264 for 4 hours or left unpulsed (▵) are also used as stimulators. Stimulators and hybridomas were cocultured for 24 hours, and IL-2 produced by 2B4 was measured by proliferation of CTLL-20 cells. Results were expressed as mean cpm of triplicate cultures ± standard deviation. (C) ES-DCs with (ES-DC-OVA) or without (ES-DC) expression of OVA protein were injected intraperitoneally on days 0 and 7 into syngenic F1 mice. Splenocytes from the injected mice were harvested on day 14 and cultured in the presence of OVA257-264 (0.1 μM) for 5 days. The resultant cells were assayed for the capacity to lyse EL-4 tumor cells either pulsed with 10 μM OVA peptide (▪) or left unpulsed (■). Results were expressed as mean specific lysis of triplicate assays and the standard deviation of triplicates was less than 2.5%.
Generation of ES-DCs expressing OVA antigen and in vivo priming of OVA-specific cytotoxic lymphocytes.
(A) Cre/mutant lox–mediated targeted integration of the expression cassette to a gene-trapped locus is schematically depicted. The expression cassette, the OVA cDNA driven by the chicken β-actin promoter (pCA-OVA) ligated with the puromycinN-acetyltransferase gene driven by the pgk promoter (pgk-Puro-R), was inserted to the gene-trapped site, replacing the β-geo sequence by the effect of Cre recombinase transiently expressed from pCAGGS-Cre. The combination of a lox66 of the replacement vector and a lox71 of a trapped locus resulted in a double-mutant lox, which inhibited further recombination of the replaced allele and increased the frequency of proper recombinant ES cell clones. (B) Stimulation of OVA-specific T-cell hybridoma, RF33.70, with ES-DCs with (○) or without (■) expression of OVA was analyzed. As a control, bone marrow–derived DCs pulsed with 10 μM (⋄) or 1 μM (▪) of OVA257-264 for 4 hours or left unpulsed (▵) are also used as stimulators. Stimulators and hybridomas were cocultured for 24 hours, and IL-2 produced by 2B4 was measured by proliferation of CTLL-20 cells. Results were expressed as mean cpm of triplicate cultures ± standard deviation. (C) ES-DCs with (ES-DC-OVA) or without (ES-DC) expression of OVA protein were injected intraperitoneally on days 0 and 7 into syngenic F1 mice. Splenocytes from the injected mice were harvested on day 14 and cultured in the presence of OVA257-264 (0.1 μM) for 5 days. The resultant cells were assayed for the capacity to lyse EL-4 tumor cells either pulsed with 10 μM OVA peptide (▪) or left unpulsed (■). Results were expressed as mean specific lysis of triplicate assays and the standard deviation of triplicates was less than 2.5%.
Priming of antigen-specific cytotoxic T cells with genetically modified ES-DCs
The capacity of ES-DCs introduced with an OVA-expression vector as described above to prime OVA-specific T cells in vivo was analyzed. ES-DCs (5 × 105) with or without OVA expression vector were injected intraperitoneally into syngenic (CBA × C57BL/6) F1 mice twice with a 7-day interval. Splenocytes were isolated 7 days after the second injection and cultured in vitro in the presence of a suboptimal concentration (0.1 μM) of OVA257-264 peptide, the major Kb-binding epitope of OVA protein. After 5 days, viable cells were recovered and assayed for their capacity to kill EL-4 thymoma cells (H-2b) prepulsed with the OVA peptide. The results shown in Figure 8C indicated that cytotoxic T cells specific to the OVA epitope were primed in vivo with ES-DCs expressing OVA protein but not with ES-DC without OVA. These results demonstrate that ES-DCs genetically engineered to express an antigenic protein have the capacity to prime antigen-specific cytotoxic T cells in vivo.
Discussion
We developed a method to generate DCs from mouse ES cells in vitro. The ES-DCs were comparable with bone marrow cell–derived DCs in morphology, surface phenotypes, and function. Several improvements have been made in the procedure for differentiation culture. For feeder cells used in days 5 to 10 of culture, we compared OP9 with other bone marrow stromal cell lines, PA6 and ST2, both producing M-CSF. Although DCs could differentiate when PA6 or ST2 cells were used as feeder cells, the number of generated DCs was fewer and the phenotype of the generated DCs differed. With PA6 or ST2, generated DCs did not express CD80 and CD205, and their activity to stimulate MLR was weaker than that of DCs produced with OP9. The use of tissue culture–grade dishes in the culture after transfer from the OP9 feeder cell layer (after day 10 in Figure 1) gave rise to a fewer number and a lower purity of ES-DCs than did the use of bacterial-quality Petri dishes. If we used dishes of tissue-culture grade, many cells firmly adhered to the dish surface, resembling macrophages or fibroblasts, and inhibited the generation of ES-DCs. The beneficial effect of bacterial-quality Petri dishes to DC development has been noted also for culture of BM-DCs.27 GM-CSF has been reported to be essential for in vitro generation of DCs from hematopoietic cells.20-22GM-CSF is also necessary for generation of ES-DCs. We applied the current culture protocol to ES cell lines other than TT2. We tested 3 lines of ES cells, D3, R1, and CCE, and observed that all of these lines also differentiated to DCs, thereby suggesting that the method is applicable to most lines of mouse ES cells.
Recently, another method to generate DCs in vitro from mouse ES cells has been reported.38 In the procedure, embryoid bodies (EBs) made from ES cells were cultured for 14 days in the presence of GM-CSF and IL-3. Resultant DCs, referred to as esDCs, lack expression of CD8α and Dec-205 (CD205), suggesting myeloid origin. Upon stimulation with LPS, they were matured and became highly competent to stimulate T cells. Characteristics of the EB-derived esDCs seemed to be similar to those of our ES-DCs. Compared with our method, the EB-mediated method is simple in that feeder cells are not necessary. On the other hand, our method allows one to microscopically observe individual cells following differentiation from ES cells to mature DCs.
For investigation of the physiologic function of genes, generation of mutant mice by gene targeting in ES cells is a potent and widely used means. However, it takes a relatively long time to develop mutant mice by gene targeting, and if the disrupted gene is essential for embryogenesis, homozygous mutant mice cannot be obtained because of embryonic lethality. ES cell clones homozygous for mutated allele can be obtained from single-allele mutant cell clones by selection with a high dose of a selection drug39 or sequential gene targeting with 2 targeting vectors bearing different selection markers.28 The method for in vitro generation of DCs from ES cells may prove useful for analyzing genes essential for both embryogenesis and function of DCs. Using the TT2 ES cell line, the generation of a large-scale library of gene-trap ES clones is now in progress. Using the method established in the current study, it is possible to efficiently screen large numbers of gene-trap ES cell clones to search for genes expressed in DCs. After selection of ES cell clones in which genes expressed in DCs were trapped, one can generate homozygous mutant ES cell clones by high-dose drug selection, differentiate them to DCs, and evaluate the functional significance of the identified genes in DCs.
For genetic modification of ES-DCs, we found 2 means for gene transfer to ES cells useful: an expression vector containing IRES-puromycin N-acetyltransferase gene and Cre-lox–mediated targeted gene integration into gene-trapped ES cell clones. Both worked very efficiently to generate ES cell transfectant clones expressing the gene products after DC differentiation; the frequency of appropriate transfectant clones in picked-up drug-resistant clones was 85% and 90% when we used the vector with IRES-puromycin N-acetyltransferase gene and targeted gene integration, respectively. We are now preparing ES-DCs expressing both antigenic protein and immunomodulating molecules, such as immunostimulatory or inhibitory molecules, cytokines, or chemokines. One of our goals is to manipulate immune responses in an antigen-specific manner by in vivo transfer of such engineered DCs. We have already succeeded in introducing 2 different plasmid vectors with different selection markers, the puromycin- and neomycin-resistance gene, by sequential transfection, and confirmed that generation of DCs from double-transfected ES cells is also feasible. Possible future applications of this method may be treatment of autoimmune and allergic diseases, inhibition of graft rejection in transplantation medicine, antitumor immunotherapy, and vaccination against intractable infectious diseases. Recently, human ES cells have been generated.40,41 In the mouse system, a method was devised to generate ES cell lines with an appropriate genetic background by nuclear transfer from somatic cells to already established ES cells.42 43 With the advances in ES cell–related technology, immunomodulation therapy using DCs generated from genetically engineered ES cells may be considered.
We thank Drs T. Nakano, K. Inaba, K. Matsushima, K. Iwabuchi, and K. Matsuno for valuable suggestions; Dr S. Aizawa for TT2; Drs N. Takakura and T. Suda for OP9 and PA6; Dr K. Lock for RF33.70; Drs T. Koda and M. Bevan for a cDNA clone for OVA; Dr H. Niwa for pCAG-IP; and Kirin Brewery for rGM-CSF. M. Ohara provided helpful comments on the manuscript.
Prepublished online as Blood First Edition Paper, October 24, 2002; DOI 10.1182/blood-2002-07-2254.
Supported by Grants-in-Aid 11557027, 12213111, 14370115, 14657082, and 14570421 from the Ministry of Education, Science, Technology, Sports, and Culture of Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Yasuharu Nishimura, Division of Immunogenetics, Department of Neuroscience and Immunology, Kumamoto University Graduate School of Medical Sciences, 2-2-1 Honjo, Kumamoto 860-0811, Japan; e-mail:mxnishim@gpo.kumamoto-u.ac.jp.

![Fig. 5. Allogeneic MLR-stimulating capacity of ES-DCs. / (A) Graded numbers of ES-DCs harvested on day 17 (⋄), ES-derived cells on day 10 (■), and (CBA × C57BL/6) F1 BM-DCs cultured for 11 days (○) were irradiated and cocultured with splenic T cells (1.5 × 105) isolated from Balb/c mice for 4 days, and the proliferative response of T cells in the last 16 hours of the culture was measured by [3H]-thymidine uptake. (B) ES-DCs harvested on day 24 and stimulated with IL-4/TNF-α/anti-CD40 (⋄), IL-4/TNF-α/LPS (▵), or IL-4/TNF-α/anti-CD40/LPS (▪) were used as stimulators. BM-DCs cultured for 10 days and stimulated with TNF-α (○) for 20 hours were also used as stimulators. For control, ES-derived cells cultured on OP9 feeder cell layers for 10 days were also used as stimulators (■). Mean values in kilo cpm (kcpm) ± standard deviation for triplicate cultures are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/9/10.1182_blood-2002-07-2254/4/m_h80934224005.jpeg?Expires=1769232241&Signature=BumZ7bKGzH~VCrqcS1nMtfH9pfZGQq0CTDANDdfvprSM~PM543PRRdpTUztQVXIQMV8qupBefKe8kwrrh9m1IQaEU5zfcqSVV2PE77R8jMgC8sa-cVl3H0uDNYGjRGkNaPKvLaKs4cJ5pFGMc4AiA8VUVp-3GIneF17wt8pJGPrzFlKATomHz2D4yzhXQrHO8Y2Gxzx0fiRAynFlL6eYNDdZx6Ypgrx~Np~o3FZvvUqSIaLMvz40WZQfxTorzuxh4FoZVBmK7NX6xTHy5JLKD7juVrgh30-a3SECAWKh98wxrupCAyQ7F2MG~vJufBf9SKq4oniMkcCA3PRLmWi-rQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal