Tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL) and Fas ligand (FasL) have been implicated in antitumor immunity and therapy. In the present study, we investigated the sensitivity of Philadelphia chromosome (Ph1)–positive leukemia cell lines to TRAIL- or FasL-induced cell death to explore the possible contribution of these molecules to immunotherapy against Ph1-positive leukemias. TRAIL, but not FasL, effectively induced apoptotic cell death in most of 5 chronic myelogenous leukemia–derived and 7 acute leukemia–derived Ph1-positive cell lines. The sensitivity to TRAIL was correlated with cell-surface expression of death-inducing receptors DR4 and/or DR5. The TRAIL-induced cell death was caspase-dependent and enhanced by nuclear factor κB inhibitors. Moreover, primary leukemia cells from Ph1-positive acute lymphoblastic leukemia patients were also sensitive to TRAIL, but not to FasL, depending on DR4/DR5 expression. Fas-associated death domain protein (FADD) and caspase-8, components of death-inducing signaling complex (DISC), as well as FLIP (FLICE [Fas-associating protein with death domain–like interleukin-1–converting enzyme]/caspase-8 inhibitory protein), a negative regulator of caspase-8, were expressed ubiquitously in Ph1-positive leukemia cell lines irrespective of their differential sensitivities to TRAIL and FasL. Notably, TRAIL could induce cell death in the Ph1-positive leukemia cell lines that were refractory to a BCR-ABL–specific tyrosine kinase inhibitor imatinib mesylate (STI571; Novartis Pharma, Basel, Switzerland). These results suggested the potential utility of recombinant TRAIL as a novel therapeutic agent and the possible contribution of endogenously expressed TRAIL to immunotherapy against Ph1-positive leukemias.
Introduction
Tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL) is a proapoptotic member of the TNF superfamily that also includes TNF-α and Fas ligand (FasL).1 TRAIL induces apoptosis in a variety of tumor cells2,3 by interacting with its cell-surface receptors DR4 (TRAIL-R1)4 and DR5 (TRAIL-R2).5 DR4 and DR5 contain a cytoplasmic region designated as the “death domain,” which is responsible for transducing the death signal. Ligation by TRAIL results in recruitment of a cytoplasmic adaptor molecule, Fas-associated death domain protein (FADD), to the death domain of DR4 and DR5,6 leading to activation of caspase-8, which subsequently initiates proteolytic activation of downstream effector caspases such as caspase-3, -6 and -7, and finally induces apoptosis.7 In contrast to the DR4 and DR5 receptors, 2 other cell-surface TRAIL receptors, DcR1 (TRAIL-R3)5,8 and DcR2 (TRAIL-R4),9 lack a functional death domain and cannot transduce a proapoptotic signal. These “decoy receptors” compete with DR4 and DR5 for TRAIL binding. Thus, the preferential expression of decoy receptors on normal cells is regarded as one of the mechanisms for the preferential proapoptotic action of TRAIL against tumor cells but not normal cells. In addition to the perforin- and FasL-mediated pathways,10-12 it has been recently suggested that the TRAIL-mediated pathway also plays an important role in antitumor immunity. TRAIL is expressed on human CD4+T-cell clones13 and murine activated natural killer (NK) cells,14 and is involved in their cytotoxic activities against tumor target cells. Interferon-alpha (IFN-α) enhances the TRAIL expression on anti-CD3–stimulated human peripheral blood T cells, and this is involved in their cytotoxic activities against renal cell carcinomas.15 Although there are a few reports that TRAIL selectively induced apoptosis in multiple myeloma cells16,17 and some myeloid leukemia cells,18it remains to be determined whether TRAIL also participates in antitumor immunity against hematologic malignancies.
Chronic myelogeneous leukemia (CML) is a clonal myeloproliferative expansion of transformed hematopoietic progenitor cells characterized by Philadelphia chromosome (Ph1) created from t(9;22) (q34; q11).19-21 Approximately 20% of adult22 and 5% of childhood23 acute lymphoblastic leukemia (ALL) and 2% of acute myeloblastic leukemia (AML)20 are also Ph1 positive. The Ph1 results in the juxtaposition ofbcr and abl genes, and this generates a chimeric protein termed BCR-ABL with a marked tyrosine kinase activity.19,20 Virtually all cases of CML express a 210-kDa form of BCR-ABL (p210), while half of adult and most of childhood Ph1-positive ALLs express a shorter form of BCR-ABL termed p190. IFN-α is the first-line therapy for patients with CML in chronic phase who are not eligible for allogeneic hematopoietic stem cell transplantation (allo-SCT).24-26 IFN-α can prolong survival, but complete elimination of Ph1-positive leukemia cells has been attained in only 5% to 10% of the patients. Several reports have suggested a direct antileukemic effect of IFN-α on CML cells27 and indirect effects modulating the immune system.28-30 Allo-SCT is presently an only curative therapy not only for chronic phase of CML, but also for blastic crisis of CML (CML-BC) and Ph1-positive ALL.31 A critical role for the immune system, termed graft-versus-leukemia (GVL) effect, in achieving a cure in patients with CML and Ph1-positive ALL has been well documented.20 21
Although the clinical observations strongly suggest a pivotal role of cytotoxic T lymphocytes (CTLs) in suppressing Ph1-positive leukemias, the underlying effector mechanisms have not been well characterized. A role of FasL has been suggested, but its contribution to the elimination of Ph1-positive leukemia cells is still controversial. Selleri et al reported that Fas was expressed on CD34+cells from CML patients and up-regulated by IFN-α.28They also reported that CD34+ cells from CML patients who showed an optimal response to IFN-α therapy underwent apoptosis upon Fas triggering, whereas those derived from patients with a poor response were resistant to Fas-mediated killing.29 These results suggested a pivotal role of FasL in the elimination of CML clones by IFN-α therapy. However, Gora-Tybor et al reported that most of Ph1-positive leukemia cell lines did not express Fas and were resistant to Fas-mediated apoptosis.32 In addition, a large-scale clinical study has indicated that the higher Fas expression on CD34+ cells from CML patients was associated rather with poor cytogenetic response to IFN-α therapy.33 Moreover, using a retrovirally induced murine CML model, Shlomchik and Pear reported that GVL effect was not impaired in Fas-deficient mice, suggesting that the mechanism other than the Fas/FasL interaction may be sufficient for the immune-mediated eradication of CML clones.34 Given the IFN-α–inducible expression of TRAIL on human T cells,15 TRAIL may participate in the process of antileukemic effects against Ph1-positive leukemias. In the present study, by using Ph1-positive leukemia cell lines and primary leukemia cells, we showed that TRAIL, but not FasL, effectively induced apoptosis in Ph1-positive leukemia cells, expressing surface DR4 and/or DR5. Notably, TRAIL was also effective against leukemia cells that were refractory to the BCR-ABL kinase inhibitor imatinib mesylate (STI571; Novartis Pharma, Basel, Switzerland). These results suggest not only the potential utility of recombinant TRAIL for clinical treatment of Ph1-positive leukemias but also shed light on the possible involvement of the TRAIL/TRAIL receptor interaction in the IFN-α therapy for CML and/or the GVL effect after allo-SCT for CML and Ph1-positive ALL.
Materials and methods
Leukemia cells
A total of 12 Ph1-positive leukemia cell lines were used in this study and are listed in Table 1. K56235 and Nalm136 were established from patients with erythroid and lymphoid crisis of CML, respectively, while other cell lines have been established in our laboratory and described previously.37 KOPM28, KOPM53, and KOPN55bi were established from CML-BC cases rearranged in major bcr, while KOPM30, KOPN30bi, KOPN57bi, KOPN66bi, KOPN72bi, and YAMN91 were established from Ph1-positive acute leukemia (AL) cases rearranged in minor bcr. YAMN73 was established from a patient with Ph1-positive ALL expressing the Abl exon 2–spliced BCR-ABL (203-kDa) protein.38 KOPM30 and KOPN30bi were established from the same patient at different stages of the disease.39 These cell lines were classified into 2 groups based on cytochemistry and immunophenotype as follows: “myeloid” type, positive for myeloperoxidase (POX) and/or expressing myeloid antigens (K562, KOPM28, KOPM30, and KOPM53); and “lymphoid” type, negative for POX and expressing immature B-lymphoid antigens (Nalm1, KOPN30bi, KOPN55bi, KOPN57bi, KOPN66bi, KOPN72bi, YAMN73, and YAMN91). T-leukemic cell lines (Jurkat and MOLT4F) were used as controls. All cell lines were maintained in RPMI1640 medium supplemented with 10% fetal calf serum (FCS) in a humidified atmosphere of 5% CO2 at 37°C. For the use of patient samples, the informed consents were obtained from the patients and/or their parents. Mononuclear cells (blasts > 95%) isolated from bone marrow aspirates or peripheral blood samples by Ficoll-Hypaque density centrifugation were stored in liquid nitrogen with 15% dimethyl sulfoxide in FCS. For experiments, each sample was thawed, washed with serum-free RPMI1640 medium, and resuspended in RPMI1640 with 10% FCS.
Characteristics and sensitivity to FasL in Ph1-positive leukemia cell lines
| Cell line . | BCR-ABL . | Linage . | 3H-thymidine uptake, cpm* . | % inhibition by FasL† . | Expression of Fas, RFI‡ . | |
|---|---|---|---|---|---|---|
| Control . | FasL . | |||||
| CML-BC-derived | ||||||
| KOPM28 | p210 | Myeloid | 226 181 ± 2256 | 244 361 ± 1326 | −8 | 3.0 |
| KOPM53 | p210 | Myeloid | 178 218 ± 1645 | 174 291 ± 1364 | 2 | 2.4 |
| KOPN55bi | p210 | Lymphoid | 8 184 ± 340 | 7 969 ± 493 | 4 | 1.0 |
| Nalm1 | p210 | Lymphoid | 40 925 ± 269 | 36 365 ± 1361 | 12 | 1.9 |
| K562 | p210 | Myeloid | 9 611 ± 110 | 9 906 ± 295 | −3 | 1.0 |
| Ph1-positive AL-derived | ||||||
| KOPM30 | p190 | Myeloid | 10 849 ± 141 | 7 511 ± 375 | 30 | 2.2 |
| KOPN30bi | p190 | Lymphoid | 186 993 ± 4663 | 180 406 ± 3438 | 4 | 1.1 |
| KOPN57bi | p190 | Lymphoid | 90 605 ± 2044 | 86 368 ± 5687 | 4 | 1.5 |
| KOPN66bi | p190 | Lymphoid | 117 272 ± 2663 | 114 914 ± 2197 | 2 | 1.3 |
| KOPN72bi | p190 | Lymphoid | 145 957 ± 3415 | 140 332 ± 4242 | 4 | 1.2 |
| YAMN73 | p2031-153 | Lymphoid | 60 397 ± 991 | 59 396 ± 1840 | 2 | 1.0 |
| YAMN91 | p190 | Lymphoid | 13 884 ± 246 | 14 143 ± 167 | −2 | 1.3 |
| Cell line . | BCR-ABL . | Linage . | 3H-thymidine uptake, cpm* . | % inhibition by FasL† . | Expression of Fas, RFI‡ . | |
|---|---|---|---|---|---|---|
| Control . | FasL . | |||||
| CML-BC-derived | ||||||
| KOPM28 | p210 | Myeloid | 226 181 ± 2256 | 244 361 ± 1326 | −8 | 3.0 |
| KOPM53 | p210 | Myeloid | 178 218 ± 1645 | 174 291 ± 1364 | 2 | 2.4 |
| KOPN55bi | p210 | Lymphoid | 8 184 ± 340 | 7 969 ± 493 | 4 | 1.0 |
| Nalm1 | p210 | Lymphoid | 40 925 ± 269 | 36 365 ± 1361 | 12 | 1.9 |
| K562 | p210 | Myeloid | 9 611 ± 110 | 9 906 ± 295 | −3 | 1.0 |
| Ph1-positive AL-derived | ||||||
| KOPM30 | p190 | Myeloid | 10 849 ± 141 | 7 511 ± 375 | 30 | 2.2 |
| KOPN30bi | p190 | Lymphoid | 186 993 ± 4663 | 180 406 ± 3438 | 4 | 1.1 |
| KOPN57bi | p190 | Lymphoid | 90 605 ± 2044 | 86 368 ± 5687 | 4 | 1.5 |
| KOPN66bi | p190 | Lymphoid | 117 272 ± 2663 | 114 914 ± 2197 | 2 | 1.3 |
| KOPN72bi | p190 | Lymphoid | 145 957 ± 3415 | 140 332 ± 4242 | 4 | 1.2 |
| YAMN73 | p2031-153 | Lymphoid | 60 397 ± 991 | 59 396 ± 1840 | 2 | 1.0 |
| YAMN91 | p190 | Lymphoid | 13 884 ± 246 | 14 143 ± 167 | −2 | 1.3 |
Data are shown as means ± SE of triplicate cultures.
% inhibition by FasL was determined by the 3H-thymidine uptake assay in the presence or absence of 100 ng/mL rhsFasL as described in “Materials and methods.”
Relative fluorescence intensity (RFI) was determined by the ratio of mean fluorescence intensity for specific staining to that for control staining.
p203 corresponds to the Abl exon2-spliced BCR-ABL protein.
Reagents
Recombinant human soluble FasL (rhsFasL, SUPER FasL) and recombinant human soluble TRAIL (rhsTRAIL, Killer TRAIL), purchased from Alexis Biochemicals (San Diego, CA), are constituted from the extracellular domains of human FasL and human TRAIL, respectively; fused at the N-terminal to linker peptides and FRAG- and His-tags, respectively; and do not require a cross-linker for biologic activities. z-VAD-fmk, a caspase inhibitor with broad spectrum, was purchased from Enzyme Systems Products (Livermore, CA). A proteasome inhibitor N-acetyl-Leu-Leu-norLeu-al (LLnL), a cell-permeable nuclear factor κB (NF-κB) inhibitory peptide SN50,40 and its control peptide SN50M were purchased from Sigma-Aldrich (Tokyo, Japan) and BIOMOL (Plymouth Meeting, PA), respectively. Imatinib mesylate41 was kindly provided by Novartis Pharma (Basel, Switzerland).
3H-thymidine uptake assay
Leukemia cell lines (2-5 × 104 cells/well) were cultured in triplicate in 200 μL RPMI1640 medium supplemented with 10% FCS in a flat-bottomed 96-well plate (Costar, Cambridge, MA). The plates were incubated for the indicated periods, pulsed for the last 6 hours with 3H-thymidine (1 μCi/well [0.037 MBq/well]), and harvested onto glass-fiber filters. Radioactivity incorporated into DNA was measured by liquid scintillation counting.
The effects of rhsFasL and rhsTRAIL were determined by the last 6-hour pulse of the 42-hour culture in the absence or presence of 3-fold diluted concentrations (3.7, 11, 33, 100, and 300 ng/mL) of rhsFasL or rhsTRAIL. In some experiments, a neutralizing anti-TRAIL monoclonal antibody (mAb) (RIK-2; 10 μg/mL)13 or z-VAD-fmk (20 μM) was used to block the activity of rhsTRAIL and caspases, respectively. In other experiments, cell lines were preincubated for 3 hours with LLnL (2.5 μM), SN50, or SN50M (100 μg/mL) and then cultured in the absence or presence of rhsTRAIL. The effect of imatinib mesylate was also determined after 42-hour incubation in the absence or presence of imatinib mesylate (1.0 μM). The percent inhibition by TRAIL or imatinib mesylate was calculated as follows: {1 − [(cpm of treated well)/(cpm of untreated well)]} × 100. The percent3H-thymidine uptake was calculated as follows: [(cpm of treated well)/(cpm of untreated well)] × 100.
Viability and apoptosis assays
The cytotoxic effects of FasL and TRAIL were examined by the dye exclusion assay. Cell lines (1 × 105 cells/well) were cultured in the presence of rhsFasL or rhsTRAIL at 100 ng/mL, harvested at 12, 24, and 36 hours, and the viability was determined by staining with trypan blue. The early apoptotic event in leukemia cell lines was examined by binding of Annexinne-V to surface-exposed phosphatidylserine. Cell lines (4 × 105 cells/mL) were cultured in the absence or presence of rhsTRAIL (100 ng/mL), harvested at 12 hours, stained with fluorescein isothiocyanate (FITC)–conjugated Annexin-V (MBL, Nagoya, Japan), and analyzed by flow cytometry (EPICS PROFILE; Coulter, Miami, FL). In experiments with primary leukemias, cells (2 × 104 cells/well) were incubated in the absence or presence of rhsFasL or rhsTRAIL at 100 ng/mL with or without a neutralizing anti-TRAIL mAb RIK-2 (10 μg/mL) in triplicate in a 96-well plate for 24 hours, and the viability was determined by staining with trypan blue. In some patient samples, apoptosis was also examined by binding of FITC-conjugated Annexin-V after 12-hour culture.
Cell-surface expression of TRAIL receptors and Fas
mAbs specific for DR4 (DJR1, mouse immunoglobulin G 1 [IgG 1]), DR5 (DJR2, mouse IgG 1), DcR1 (DJR3, mouse IgG 1), and DcR2 (DJR4, mouse IgG 1) were raised against soluble human IgG1 Fc fusion proteins containing the extracellular domain of each TRAIL receptor and identified by their specific reactivity with the respective fusion protein in enzyme-linked immunosorbent assay (ELISA). Leukemia cell lines, primary leukemias, and baby hamster kidney (BHK) cell lines stably expressing DR4, DR5, DcR1, or DcR2 cDNA (1 × 106 cells) were incubated with 1 μg of biotinylated control mouse IgG 1 or mAb for 30 minutes on ice. After washing, the cells were incubated with phycoerythrin-conjugated streptavidin (Biomeda, Foster City, CA) for 30 minutes on ice, and then analyzed by flow cytometry. The relative fluorescence intensity (RFI) was determined by calculating the ratio of mean fluorescence intensity for specific staining to that for control staining. For the Fas expression, each cell line was incubated with mouse antihuman Fas (4A5; MBL) or irrelevant mouse IgG for 30 minutes on ice, and subsequently with FITC-conjugated anti–mouse IgG, and analyzed by flow cytometry.
Western blot analysis
The nonidet P-40 lysates of cell lines were separated on a sodium dodecyl sulfate–polyacrylamide gel under reducing conditions and then transferred to polyvinyl difluoride membranes as previously described.42 After blocking with 5% nonfat dry milk in 0.05% Tween-20 Tris (tris(hydroxymethyl)aminomethane)–buffered saline (TBS), membranes were incubated with mouse antihuman FADD (1:250 dilution; BD Transduction Laboratories, Lexington, KY), antihuman caspase-8 (1:1000 dilution; MBL), antihuman kinase domain of c-ABL (1:400 dilution; Pharmingen, San Diego, CA), or rat antihuman FLIP (FLICE [Fas-associating protein with death domain–like interleukin-1–converting enzyme]/caspase-8 inhibitory protein; 1:1000 dilution; Kamiya Biochemical, Seattle, WA) antibodies in 5% milk TBS at 4°C overnight. Membranes were incubated with horseradish peroxidase–conjugated goat antimouse or rat IgG (1:1000 dilution; MBL) at room temperature for 1 hour and were then developed using the enhanced chemiluminescence kit (Amersham Pharmacia Biotec, Buckinghamshire, United Kingdom).
Results
Cytotoxic effect of FasL against Ph1-positive leukemia cell lines
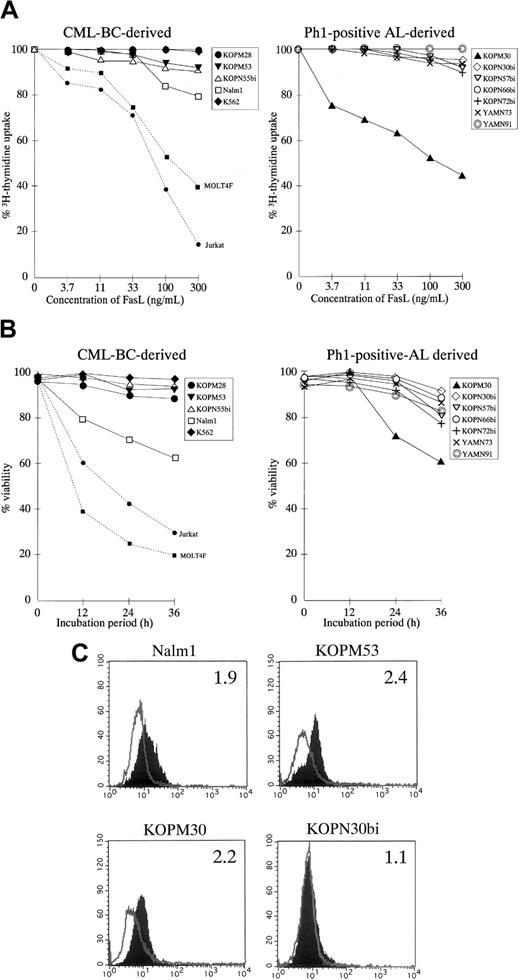
We first examined the susceptibility of 12 Ph1-positive leukemia cell lines to rhsFasL by the 3H-thymidine uptake for the last 6 hours of the 42-hour culture with various concentrations of rhsFasL. As shown Figure 1A, a marked growth inhibition was observed in a dose-dependent manner against well-characterized FasL-sensitive T-leukemia cell lines (Jurkat and MOLT4F).43 This growth inhibition was due to loss of cell viability, rather than cytostasis, as estimated by the trypan blue exclusion assay (Figure 1B). Although 1 of 5 CML-BC–derived (Nalm1) and 1 of 7 AL-derived (KOPM30) cell lines were moderately susceptible, the other 10 Ph1-positive cell lines were highly resistant to FasL as estimated by either the 3H-thymidine uptake (Figure 1A and Table 1) or the trypan blue exclusion assay (Figure1B).
Fas expression and cytotoxic effect of FasL against Ph1-positive leukemia cell lines.
(A) Dose response of growth inhibition. CML-BC–derived (left panel) or Ph1-positive AL-derived (right panel) cell lines were cultured for 42 hours in the absence or presence of indicated concentrations of rhsFasL, and 3H-thymidine uptake was evaluated for the last 6 hours. FasL-sensitive T-cell leukemia cell lines (Jurkat and MOLT4F) were also included as positive controls and are shown by the dotted lines. Data with myeloid cell lines are shown as the closed symbols. All data are represented as the mean of triplicate samples. Standard errors were always less than 10% (not shown). (B) Time course of cell viability. CML-BC–derived (left panel) or Ph1-positive AL-derived (right panel) cell lines were cultured with or without 100 ng/mL rhsFasL for 12, 24, or 36 hours, and then the viability was determined by the trypan blue exclusion assay. Data with Jurkat and MOLT4F are also shown by dotted lines. Data with myeloid cell lines are shown as closed symbols. All data are represented as the mean of triplicate samples. Standard errors were always less than 10% (not shown). (C) Cell-surface staining of Ph1-positive cell lines with anti-Fas mAb. CML-BC–derived FasL-sensitive (Nalm1) or -resistant (KOPM53) cell lines and Ph1-positive AL-derived FasL-sensitive (KOPM30) or -resistant (KOPN30bi) cell lines were stained with control mouse IgG or antihuman Fas mAb and analyzed by flow cytometry. Shaded and unshaded peaks correspond to specific and control stainings, respectively. RFI is indicated in each panel.
Fas expression and cytotoxic effect of FasL against Ph1-positive leukemia cell lines.
(A) Dose response of growth inhibition. CML-BC–derived (left panel) or Ph1-positive AL-derived (right panel) cell lines were cultured for 42 hours in the absence or presence of indicated concentrations of rhsFasL, and 3H-thymidine uptake was evaluated for the last 6 hours. FasL-sensitive T-cell leukemia cell lines (Jurkat and MOLT4F) were also included as positive controls and are shown by the dotted lines. Data with myeloid cell lines are shown as the closed symbols. All data are represented as the mean of triplicate samples. Standard errors were always less than 10% (not shown). (B) Time course of cell viability. CML-BC–derived (left panel) or Ph1-positive AL-derived (right panel) cell lines were cultured with or without 100 ng/mL rhsFasL for 12, 24, or 36 hours, and then the viability was determined by the trypan blue exclusion assay. Data with Jurkat and MOLT4F are also shown by dotted lines. Data with myeloid cell lines are shown as closed symbols. All data are represented as the mean of triplicate samples. Standard errors were always less than 10% (not shown). (C) Cell-surface staining of Ph1-positive cell lines with anti-Fas mAb. CML-BC–derived FasL-sensitive (Nalm1) or -resistant (KOPM53) cell lines and Ph1-positive AL-derived FasL-sensitive (KOPM30) or -resistant (KOPN30bi) cell lines were stained with control mouse IgG or antihuman Fas mAb and analyzed by flow cytometry. Shaded and unshaded peaks correspond to specific and control stainings, respectively. RFI is indicated in each panel.
These results suggested that Ph1-positive leukemia cell lines are generally resistant to the FasL-induced cell death. Next, we analyzed the cell-surface expression of Fas by flow cytometry. As indicated in Figure 1C and summarized in Table 1, Fas was detectable on myeloid cell lines except for K562 but rarely detectable on lymphoid cell lines except for Nalm1. Accordingly, the relatively low expression of Fas could explain the resistance to FasL in lymphoid cell lines, while some factor other than the Fas expression might contribute to the resistance to FasL in myeloid cell lines.
Cytotoxic effect of TRAIL against Ph1-positive leukemia cell lines
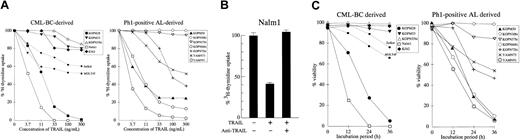
We next investigated the cytotoxic effect of rhsTRAIL against Ph1-positive leukemia cell lines. Among the CML-BC–derived cell lines (Figure 2A, left panel), 2 cell lines (Nalm1 and KOPM28) showed a marked growth inhibition in a dose-dependent manner, whereas the other 3 cell lines (KOPM53, KOPN55bi, and K562) were rather resistant. Among the Ph1-positive AL-derived cell lines (Figure 2A, right panel), 3 cell lines (YAMN91, KOPN66bi, and KOPM30) were highly sensitive, 2 cell lines (YAMN73 and KOPN72bi) were moderately sensitive, but 2 cell lines (KOPN57bi and KOPN30bi) were resistant. To demonstrate that these antileukemic effects were really mediated by TRAIL, we performed the blocking experiment using a neutralizing anti-TRAIL mAb (RIK-2).13 As shown in Figure 2B, the growth inhibition in Nalm1 was totally abolished by RIK-2, substantiating the specific activity of rhsTRAIL. As summarized in Table2, the sensitivity of Ph1-positive leukemia cell lines to TRAIL was not correlated with the type of disease (CML-BC or AL), the type of BCR-ABL fusion protein (p210, p203, or p190), or the type of lineage (myeloid or lymphoid).
Cytotoxic effect of TRAIL against Ph1-positive leukemia cell lines.
(A) Dose response of growth inhibition. CML-BC–derived (left panel) or Ph1-positive AL-derived (right panel) cell lines were cultured for 42 hours in the absence or presence of the indicated concentrations of rhsTRAIL, and the 3H-thymidine uptake was evaluated for the last 6 hours. T-cell leukemia cell lines (Jurkat and MOLT4F) were also included as controls and are shown by the dotted lines. Data with myeloid cell lines are shown as the closed symbols. All data are represented as the mean of triplicate samples. Standard errors were always less than 10% (not shown). (B) Neutralization by anti-TRAIL mAb. Nalm1 cells were cultured for 42 hours with rhsTRAIL (10 ng/mL) in the presence or absence of neutralizing anti-TRAIL mAb (10 μg/mL), and the 3H-thymidine uptake was evaluated for the last 6 hours. Data are represented as the mean ± SD of triplicate samples. (C) Time course of cell viability. CML-BC–derived (left panel) or Ph1-positive AL-derived (right panel) cell lines were cultured with 100 ng/mL of rhsTRAIL for 12, 24, or 36 hours, and then the viability was determined by the trypan blue exclusion assay. Data with Jurkat and MOLT4F are also shown by the dotted lines. Data with myeloid cell lines are shown as the closed symbols. All data are represented as the mean of triplicate samples. Standard errors were always less than 10% (not shown).
Cytotoxic effect of TRAIL against Ph1-positive leukemia cell lines.
(A) Dose response of growth inhibition. CML-BC–derived (left panel) or Ph1-positive AL-derived (right panel) cell lines were cultured for 42 hours in the absence or presence of the indicated concentrations of rhsTRAIL, and the 3H-thymidine uptake was evaluated for the last 6 hours. T-cell leukemia cell lines (Jurkat and MOLT4F) were also included as controls and are shown by the dotted lines. Data with myeloid cell lines are shown as the closed symbols. All data are represented as the mean of triplicate samples. Standard errors were always less than 10% (not shown). (B) Neutralization by anti-TRAIL mAb. Nalm1 cells were cultured for 42 hours with rhsTRAIL (10 ng/mL) in the presence or absence of neutralizing anti-TRAIL mAb (10 μg/mL), and the 3H-thymidine uptake was evaluated for the last 6 hours. Data are represented as the mean ± SD of triplicate samples. (C) Time course of cell viability. CML-BC–derived (left panel) or Ph1-positive AL-derived (right panel) cell lines were cultured with 100 ng/mL of rhsTRAIL for 12, 24, or 36 hours, and then the viability was determined by the trypan blue exclusion assay. Data with Jurkat and MOLT4F are also shown by the dotted lines. Data with myeloid cell lines are shown as the closed symbols. All data are represented as the mean of triplicate samples. Standard errors were always less than 10% (not shown).
TRAIL-sensitivity and cell-surface expression of TRAIL receptors in Ph1-positive leukemia cell lines
| Cell line . | 3H-thymidine uptake, cpm* . | % inhibition by TRAIL† . | Expression of TRAIL receptors, RFI‡ . | ||||
|---|---|---|---|---|---|---|---|
| Control . | TRAIL . | DR4 . | DR5 . | DcR1 . | DcR2 . | ||
| CML-BC-derived | |||||||
| KOPM28 | 103 712 ± 1849 | 6 000 ± 54 | 95 | 1.0 | 3.4 | 1.1 | 1.0 |
| KOPM53 | 200 509 ± 9527 | 165 725 ± 8340 | 18 | 1.1 | 1.0 | 1.0 | 1.0 |
| KOPN55bi | 11 705 ± 1730 | 9 954 ± 398 | 10 | 1.0 | 1.3 | 1.1 | 1.1 |
| Nalm1 | 28 920 ± 621 | 167 ± 14 | 99 | 1.7 | 2.1 | 1.0 | 1.0 |
| K562 | 12 099 ± 213 | 10 034 ± 61 | 20 | 1.5 | 5.0 | 1.0 | 1.0 |
| Ph1-positive AL-derived | |||||||
| KOPM30 | 29 182 ± 1796 | 7 055 ± 368 | 75 | 1.6 | 3.2 | 1.0 | 1.0 |
| KOPN30bi | 114 528 ± 4592 | 116 241 ± 2666 | −1 | 1.0 | 1.7 | 1.1 | 1.1 |
| KOPN57bi | 40 975 ± 2021 | 35 681 ± 2001 | 5 | 1.0 | 1.3 | 1.0 | 1.1 |
| KOPN66bi | 75 603 ± 1175 | 9 031 ± 157 | 85 | 1.5 | 2.6 | 1.0 | 1.0 |
| KOPN72bi | 112 855 ± 5916 | 68 357 ± 4469 | 52 | 1.7 | 1.1 | 1.0 | 1.0 |
| YAMN73 | 36 437 ± 716 | 20 553 ± 498 | 45 | 1.2 | 2.9 | 1.2 | 1.0 |
| YAMN91 | 27 788 ± 1529 | 1 062 ± 79 | 96 | 1.0 | 3.4 | 1.1 | 1.0 |
| Cell line . | 3H-thymidine uptake, cpm* . | % inhibition by TRAIL† . | Expression of TRAIL receptors, RFI‡ . | ||||
|---|---|---|---|---|---|---|---|
| Control . | TRAIL . | DR4 . | DR5 . | DcR1 . | DcR2 . | ||
| CML-BC-derived | |||||||
| KOPM28 | 103 712 ± 1849 | 6 000 ± 54 | 95 | 1.0 | 3.4 | 1.1 | 1.0 |
| KOPM53 | 200 509 ± 9527 | 165 725 ± 8340 | 18 | 1.1 | 1.0 | 1.0 | 1.0 |
| KOPN55bi | 11 705 ± 1730 | 9 954 ± 398 | 10 | 1.0 | 1.3 | 1.1 | 1.1 |
| Nalm1 | 28 920 ± 621 | 167 ± 14 | 99 | 1.7 | 2.1 | 1.0 | 1.0 |
| K562 | 12 099 ± 213 | 10 034 ± 61 | 20 | 1.5 | 5.0 | 1.0 | 1.0 |
| Ph1-positive AL-derived | |||||||
| KOPM30 | 29 182 ± 1796 | 7 055 ± 368 | 75 | 1.6 | 3.2 | 1.0 | 1.0 |
| KOPN30bi | 114 528 ± 4592 | 116 241 ± 2666 | −1 | 1.0 | 1.7 | 1.1 | 1.1 |
| KOPN57bi | 40 975 ± 2021 | 35 681 ± 2001 | 5 | 1.0 | 1.3 | 1.0 | 1.1 |
| KOPN66bi | 75 603 ± 1175 | 9 031 ± 157 | 85 | 1.5 | 2.6 | 1.0 | 1.0 |
| KOPN72bi | 112 855 ± 5916 | 68 357 ± 4469 | 52 | 1.7 | 1.1 | 1.0 | 1.0 |
| YAMN73 | 36 437 ± 716 | 20 553 ± 498 | 45 | 1.2 | 2.9 | 1.2 | 1.0 |
| YAMN91 | 27 788 ± 1529 | 1 062 ± 79 | 96 | 1.0 | 3.4 | 1.1 | 1.0 |
Data are shown as means ± SE.
% inhibition by TRAIL was determined by the 3H-thymidine uptake assay in the presence or absence of 100 ng/mL rhsTRAIL as described in “Materials and methods.”
RFI was determined by the ratio of mean fluorescence intensity for specific staining to that for control staining.
To more directly evaluate the cytotoxic activity of TRAIL, we performed the dye exclusion assay (Figure 2C).
Consistent with the growth inhibition as estimated by the3H-thymidine uptake assay, 7 of 12 Ph1-positive leukemia cell lines were moderately or highly sensitive to the TRAIL-induced cell death.
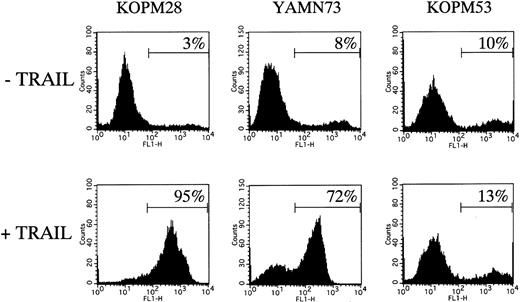
We also examined the binding of Annexin-V by flow cytometry after 12-hour treatment with rhsTRAIL (100 ng/mL). As shown in Figure 3, the Annexin-V–positive population was increased to 95% in the highly sensitive cell line (KOPM28), 72% in the moderately sensitive cell line (YAMN73), but only 13% in the resistant cell line (KOPM53), indicating that the TRAIL-induced cell death in Ph1-positive leukemia cell lines was caused by apoptosis.
Flow cytometric analysis of TRAIL-induced apoptosis.
TRAIL-sensitive cell lines (KOPM28 and YAMN73) and TRAIL-resistant cell line (KOPM53) were cultured for 12 hours with or without rhsTRAIL (100 ng/mL) and then stained with FITC-conjugated Annexin-V. The percentages of positive cells are indicated.
Flow cytometric analysis of TRAIL-induced apoptosis.
TRAIL-sensitive cell lines (KOPM28 and YAMN73) and TRAIL-resistant cell line (KOPM53) were cultured for 12 hours with or without rhsTRAIL (100 ng/mL) and then stained with FITC-conjugated Annexin-V. The percentages of positive cells are indicated.
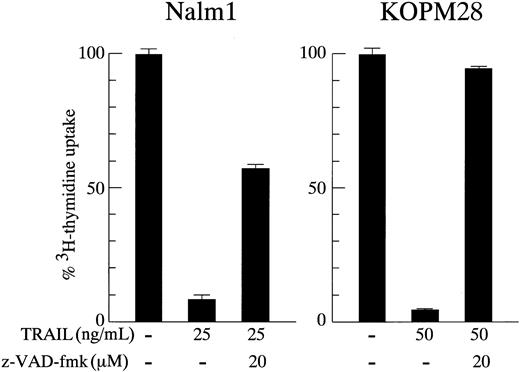
To confirm that the apoptotic cell death induced by TRAIL was dependent on the activation of caspases, we performed the 3H-thymidine uptake assay in the presence of z-VAD-fmk, a broad caspase inhibitor. As shown in Figure4, the TRAIL-induced growth inhibition was abrogated by z-VAD-fmk partially in Nalm1 and almost completely in KOPM28, indicating that the activation of caspases was required for the TRAIL-induced cell death in Ph1-positive leukemia cells.
Effect of a caspase inhibitor on growth inhibition by TRAIL.
Ph1-positive cell lines sensitive to TRAIL (Nalm1 and KOPM28) were cultured for 30 hours in the absence or presence of rhsTRAIL (25 and 50 ng/mL, respectively) with or without z-VAD-fmk (20 μM) pretreatment, and the 3H-thymidine uptake was determined for the last 6 hours. Data are represented as the mean ± SD of triplicate samples.
Effect of a caspase inhibitor on growth inhibition by TRAIL.
Ph1-positive cell lines sensitive to TRAIL (Nalm1 and KOPM28) were cultured for 30 hours in the absence or presence of rhsTRAIL (25 and 50 ng/mL, respectively) with or without z-VAD-fmk (20 μM) pretreatment, and the 3H-thymidine uptake was determined for the last 6 hours. Data are represented as the mean ± SD of triplicate samples.
Expression of TRAIL receptors on Ph1-positive leukemia cell lines
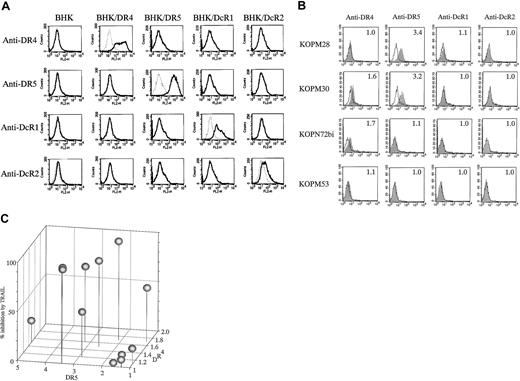
To investigate whether the sensitivity of Ph1-positive leukemia cells to TRAIL depends on the expression of TRAIL receptors, we analyzed the cell-surface expression of DR4, DR5, DcR1, and DcR2 by flow cytometry. The specificity of each mAb is shown against BHK cell lines transfected with respective TRAIL receptor cDNAs (Figure5A).
Cell-surface expression of TRAIL receptors on Ph1-positive leukemia cell lines.
(A) Specificity of mAbs against human TRAIL receptors. The parental BHK and the transfectants stably expressing human DR4, DR5, DcR1, or DcR2 were stained with control mouse IgG or murine antihuman DR4, DR5, DcR1, or DcR2 mAbs and analyzed by flow cytometry. Dotted and solid lines correspond to control and specific stainings, respectively. (B) Cell-surface staining of Ph1-positive cell lines with anti-TRAIL receptor mAbs. CML-BC–derived TRAIL-sensitive (KOPM28) or -resistant (KOPM53) cell lines and Ph1-positive AL-derived TRAIL-sensitive cell lines (KOPM30 and KOPN72bi) were stained with control mouse IgG or antihuman DR4, DR5, DcR1, and DcR2 mAbs, and analyzed by flow cytometry. Shaded and unshaded peaks correspond to specific and control stainings, respectively. RFI is indicated in each panel. (C) Correlation between the cell-surface DR4/DR5 expression and the growth inhibition by TRAIL. The horizontal axes represent the RFI of DR4 and DR5. The vertical axes represent the percent inhibition by TRAIL (100 ng/mL).
Cell-surface expression of TRAIL receptors on Ph1-positive leukemia cell lines.
(A) Specificity of mAbs against human TRAIL receptors. The parental BHK and the transfectants stably expressing human DR4, DR5, DcR1, or DcR2 were stained with control mouse IgG or murine antihuman DR4, DR5, DcR1, or DcR2 mAbs and analyzed by flow cytometry. Dotted and solid lines correspond to control and specific stainings, respectively. (B) Cell-surface staining of Ph1-positive cell lines with anti-TRAIL receptor mAbs. CML-BC–derived TRAIL-sensitive (KOPM28) or -resistant (KOPM53) cell lines and Ph1-positive AL-derived TRAIL-sensitive cell lines (KOPM30 and KOPN72bi) were stained with control mouse IgG or antihuman DR4, DR5, DcR1, and DcR2 mAbs, and analyzed by flow cytometry. Shaded and unshaded peaks correspond to specific and control stainings, respectively. RFI is indicated in each panel. (C) Correlation between the cell-surface DR4/DR5 expression and the growth inhibition by TRAIL. The horizontal axes represent the RFI of DR4 and DR5. The vertical axes represent the percent inhibition by TRAIL (100 ng/mL).
Representative cytofluorographic data on highly sensitive (KOPM28, KOPM30), moderately sensitive (KOPN72bi), and resistant (KOPM53) cell lines are shown in Figure 5B, and the RFI in each cell line is summarized in Table 2. Among 4 TRAIL receptors, DR4 and DR5 were detectable (RFI ≤ 1.5) on 5 and 8, respectively, of 12 Ph1-positive leukemic cell lines. DcR1 and DcR2 were not detectable on any tested cell lines.
Correlation between the cell-surface DR4/DR5 expression and the percent inhibition by TRAIL (Figure 5C) revealed that all TRAIL-sensitive cell lines expressed DR4 and/or DR5 at significant levels. In contrast, the TRAIL-resistant cell lines except for K562 showed undetectable or low levels of DR4 and DR5. These results suggested that the sensitivity of Ph1-positive leukemia cells to TRAIL is mostly correlated with the cell-surface expression levels of DR4 and DR5.
Cytotoxic effects of TRAIL and FasL against primary Ph1-positive leukemia cells
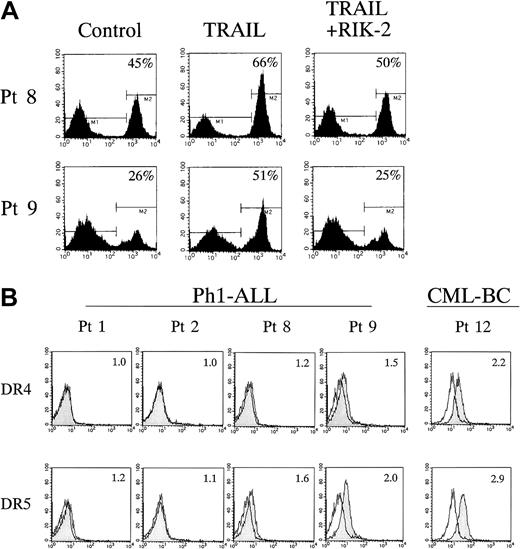
To verify the antileukemic effects of TRAIL and FasL against primary leukemia cells, leukemic blasts from 10 Ph1-positive ALL cases and 2 CML-BC cases were tested. Each sample was cultured for 24 hours in the absence or presence of 100 ng/mL of rhsFasL or rhsTRAIL in combination with a neutralizing anti-TRAIL mAb, RIK-2, and the viability was determined by the trypan blue exclusion assay. As summarized in Table 3, viability of the cells was significantly reduced by the addition of TRAIL in 6 of 10 Ph1-positive ALL cases, while only 1 case was moderately sensitive to FasL. The specific activity of TRAIL was demonstrated by the significant recovery of viability with the RIK-2 treatment. Induction of apoptosis by TRAIL was also confirmed in leukemic blasts from patients 8 and 9 by Annexin-V binding on flow cytometry (Figure 6A). Importantly, primary leukemia cells from patient 3 were sensitive to TRAIL, while the cell line (KOPN57bi) established from this patient was resistant to TRAIL (Figure 2A and Table 2). Leukemic blasts from patients 4 and 8, the origin of TRAIL-sensitive YAMN91 and YAMN73, respectively, were sensitive to TRAIL, while those from patient 2, the origin of TRAIL-resistant KOPN30bi, were resistant. Regarding TRAIL sensitivity in leukemic blasts from CML-BC, only one case (patient 11) was evaluated and showed resistance.
Sensitivity to FasL/TRAIL and TRIL receptors expression in primary Ph1-positive leukemia cells
| Patient no. . | Sex . | Age . | Derived cell line . | Onset/Relapse . | Sample . | Type of BCR-ABL . | Viability after 24-h culture3-150 . | Expression of TRAIL receptors, RFI3-152 . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control, % . | FasL, % . | TRAIL, % . | TRAIL + RIK2, % . | DR4 . | DR5 . | |||||||
| Ph1-ALL | ||||||||||||
| 1 | M | 7 | — | Onset | BM | p190 | 89 ± 2 | 91 ± 1 | 90 ± 3 | 90 ± 1 | 1.0 | 1.2 |
| 2 | M | 8 | KOPN30bi | Onset | PB | p190 | 75 ± 4 | 71 ± 6 | 77 ± 4 | 78 ± 2 | 1.0 | 1.1 |
| 3 | M | 11 | KOPN57bi | Onset | PB | p190 | 84 ± 1 | 77 ± 2 | 34 ± 53-151 | 72 ± 33-151,3-153 | ND | ND |
| 4 | M | 4 | YAMN91 | Onset | PB | p190 | 80 ± 4 | 76 ± 1 | 52 ± 33-151 | 80 ± 23-153 | 1.0 | 1.5 |
| 5 | F | 12 | — | Onset | BM | p190 | 85 ± 1 | 93 ± 2 | 90 ± 4 | 95 ± 2 | 1.0 | 1.1 |
| 6 | M | 16 | — | Relapse | PB | p210 | 85 ± 1 | 95 ± 3 | 90 ± 3 | 95 ± 2 | ND | ND |
| 7 | M | 9 | — | Relapse | BM | p190 | 89 ± 1 | 90 ± 1 | 62 ± 53-151 | 86 ± 23-153 | ND | ND |
| 8 | F | 3 | YAMN73 | Relapse | PB | p2033-155 | 87 ± 1 | 69 ± 23-151 | 46 ± 33-151 | 78 ± 33-151,3-153 | 1.2 | 1.6 |
| 9 | F | 6 | — | Relapse | PB | p190 | 92 ± 2 | 87 ± 3 | 46 ± 33-151 | 88 ± 13-153 | 1.5 | 2.0 |
| 10 | F | 15 | — | Relapse | BM | ND | 93 ± 3 | 83 ± 3 | 66 ± 33-151 | 77 ± 23-151,3-153 | ND | ND |
| CML-BC | ||||||||||||
| 11 | M | 3 | — | — | PB | p210 | 91 ± 4 | 91 ± 3 | 88 ± 4 | 92 ± 2 | ND | ND |
| 12 | F | 11 | — | — | PB | p210 | NA# | NA | NA | NA | 2.2 | 2.9 |
| Patient no. . | Sex . | Age . | Derived cell line . | Onset/Relapse . | Sample . | Type of BCR-ABL . | Viability after 24-h culture3-150 . | Expression of TRAIL receptors, RFI3-152 . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control, % . | FasL, % . | TRAIL, % . | TRAIL + RIK2, % . | DR4 . | DR5 . | |||||||
| Ph1-ALL | ||||||||||||
| 1 | M | 7 | — | Onset | BM | p190 | 89 ± 2 | 91 ± 1 | 90 ± 3 | 90 ± 1 | 1.0 | 1.2 |
| 2 | M | 8 | KOPN30bi | Onset | PB | p190 | 75 ± 4 | 71 ± 6 | 77 ± 4 | 78 ± 2 | 1.0 | 1.1 |
| 3 | M | 11 | KOPN57bi | Onset | PB | p190 | 84 ± 1 | 77 ± 2 | 34 ± 53-151 | 72 ± 33-151,3-153 | ND | ND |
| 4 | M | 4 | YAMN91 | Onset | PB | p190 | 80 ± 4 | 76 ± 1 | 52 ± 33-151 | 80 ± 23-153 | 1.0 | 1.5 |
| 5 | F | 12 | — | Onset | BM | p190 | 85 ± 1 | 93 ± 2 | 90 ± 4 | 95 ± 2 | 1.0 | 1.1 |
| 6 | M | 16 | — | Relapse | PB | p210 | 85 ± 1 | 95 ± 3 | 90 ± 3 | 95 ± 2 | ND | ND |
| 7 | M | 9 | — | Relapse | BM | p190 | 89 ± 1 | 90 ± 1 | 62 ± 53-151 | 86 ± 23-153 | ND | ND |
| 8 | F | 3 | YAMN73 | Relapse | PB | p2033-155 | 87 ± 1 | 69 ± 23-151 | 46 ± 33-151 | 78 ± 33-151,3-153 | 1.2 | 1.6 |
| 9 | F | 6 | — | Relapse | PB | p190 | 92 ± 2 | 87 ± 3 | 46 ± 33-151 | 88 ± 13-153 | 1.5 | 2.0 |
| 10 | F | 15 | — | Relapse | BM | ND | 93 ± 3 | 83 ± 3 | 66 ± 33-151 | 77 ± 23-151,3-153 | ND | ND |
| CML-BC | ||||||||||||
| 11 | M | 3 | — | — | PB | p210 | 91 ± 4 | 91 ± 3 | 88 ± 4 | 92 ± 2 | ND | ND |
| 12 | F | 11 | — | — | PB | p210 | NA# | NA | NA | NA | 2.2 | 2.9 |
M indicates male; F, female; BM, bone marrow; PB, peripheral blood; ND, not determined; NA, not available; and —, not applicable.
Data are shown as mean ± SE.
Significant reduction in viability (P < .01,t test).
RFI was determined by the ratio of mean fluorescence intensity for specific staining to that for control staining.
Significant increase in viability (P < .01,t test) in comparison with TRAIL treatment alone.
p203 corresponds to the Abl exon 2-spliced BCR-ABL protein.
#Sensitivity could not be evaluated because of complete spontaneous cell death in control culture.
Flow cytometric analysis of TRAIL-induced apoptosis and cell-surface expression of DR4 and DR5 in primary Ph1-positive ALL cells.
(A) Leukemic blasts from patients 8 and 9 listed in Table 3 were cultured for 12 hours with or without rhsTRAIL (100 ng/mL) in the presence or absence of neutralizing anti-TRAIL mAb RIK-2 (10 μg/mL), and then stained with FITC-conjugated Annexin-V. The percentages of positive cells are indicated in each panel. (B) Cell-surface staining of Ph1-positive primary leukemia cells with anti-DR4 and DR5 mAbs. Leukemic blasts from Ph1-positive ALL cases (patients 1, 2, 8, and 9 listed in Table 3) and a CML-BC case (patient 12) were stained with control mouse IgG or antihuman DR4 and DR5 mAbs, and analyzed by flow cytometry. Shaded and unshaded peaks correspond to specific and control stainings, respectively. Relative fluorescence intensity (RFI) is indicated in each panel.
Flow cytometric analysis of TRAIL-induced apoptosis and cell-surface expression of DR4 and DR5 in primary Ph1-positive ALL cells.
(A) Leukemic blasts from patients 8 and 9 listed in Table 3 were cultured for 12 hours with or without rhsTRAIL (100 ng/mL) in the presence or absence of neutralizing anti-TRAIL mAb RIK-2 (10 μg/mL), and then stained with FITC-conjugated Annexin-V. The percentages of positive cells are indicated in each panel. (B) Cell-surface staining of Ph1-positive primary leukemia cells with anti-DR4 and DR5 mAbs. Leukemic blasts from Ph1-positive ALL cases (patients 1, 2, 8, and 9 listed in Table 3) and a CML-BC case (patient 12) were stained with control mouse IgG or antihuman DR4 and DR5 mAbs, and analyzed by flow cytometry. Shaded and unshaded peaks correspond to specific and control stainings, respectively. Relative fluorescence intensity (RFI) is indicated in each panel.
Next, the cell-surface expression of DR4 and DR5 was analyzed by flow cytometry as indicated in Figure 6B and Table 3. In Ph1-positive ALL cases, the expression of DR4/DR5 was detectable on TRAIL-sensitive leukemia cells from patients 4, 8, and 9, but almost undetectable on TRAIL-resistant leukemia cells from patients 1, 2, and 5. In the CML-BC case (patient 12), both DR4 and DR5 were clearly detectable, although its TRAIL-sensitivity could not be evaluated because of excessive spontaneous cell death in control culture.
Expression of molecules consisting of death-inducing signaling complex (DISC)
Ligation of death receptors by FasL44 and TRAIL45,46 triggers a series of protein-protein interactions that leads to assembly of a DISC. Thus, expression levels of the molecules consisting of DISC are critical determinants for sensitivity. It is known that Fas and DR4/DR5 recruit FADD47,48 and caspase-849,50 into DISC, and that FLIP51 acts as a negative regulator of caspase-8.
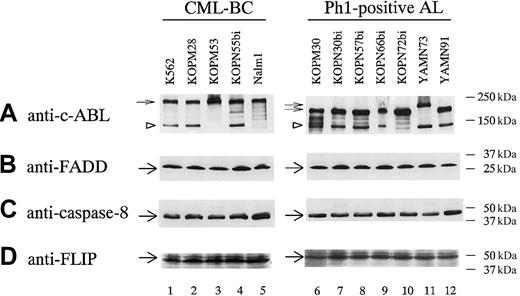
We therefore performed Western blot analysis of these molecules in Ph1-positive leukemia cell lines (Figure7). FADD, caspase-8, and FLIP were almost ubiquitously expressed in Ph1-positive leukemia cell lines irrespective of their differential sensitivities to FasL and TRAIL. These observations indicated that an absence of FADD and/or caspase-8 or an excessive expression of FLIP could not be a mechanism for resistance to TRAIL and FasL.
Western blot analysis of molecules consisting of DISC in Ph1-positive leukemia cell lines.
Lysates from 12 Ph1-positive leukemia cell lines were analyzed by Western blot using antibodies against c-ABL kinase domain, FADD, caspase-8, and FLIP as described in “Materials and methods.” Small arrows and arrowheads indicate BCR-ABL proteins (p210, p203, and p190) and c-ABL protein, respectively, in panel A. Large arrows indicate FADD in panel B, caspase-8 in panel C, and FLIP in panel D.
Western blot analysis of molecules consisting of DISC in Ph1-positive leukemia cell lines.
Lysates from 12 Ph1-positive leukemia cell lines were analyzed by Western blot using antibodies against c-ABL kinase domain, FADD, caspase-8, and FLIP as described in “Materials and methods.” Small arrows and arrowheads indicate BCR-ABL proteins (p210, p203, and p190) and c-ABL protein, respectively, in panel A. Large arrows indicate FADD in panel B, caspase-8 in panel C, and FLIP in panel D.
Modulation of TRAIL sensitivity by NF-κB inhibitors
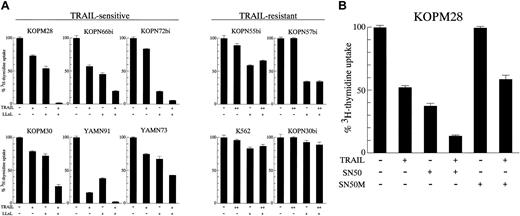
It has been reported that NF-κB is constitutively activated by BCR-ABL in Ph1-positive leukemias52 and that NF-κB could modulate TRAIL-induced cell death.6,9,53 We attempted to examine whether the NF-κB inhibitors could modulate the TRAIL sensitivity of Ph1-positive leukemia cells. For this purpose, 10 leukemia cell lines (6 sensitive and 4 resistant) were preincubated for 3 hours with 2.5 μM proteasome inhibitor LLnL,54 which is known to inhibit the activation of NF-κB by blocking the degradation of the IκB inhibitory protein, followed by 42-hour exposure to lower concentrations of rhsTRAIL (10 ng/mL for sensitive and 50 ng/mL for resistant cell lines), and then the3H-thymidine uptake was assessed for the last 6 hours (Figure 8A). As expected from a considerable role of NF-κB in the BCR-ABL–mediated transformation, the treatment with LLnL alone moderately repressed the3H-thymidine uptake in most of Ph1-positive leukemia cell lines. In the TRAIL-sensitive cell line KOPM28, for instance, the treatment with either TRAIL or LLnL reduced the3H-thymidine uptake to approximately 70% or 50%, respectively, but the treatment with both TRAIL and LLnL almost completely abrogated the 3H-thymidine uptake. Similar results were obtained in all TRAIL-sensitive cell lines examined. In contrast, LLnL did not enhance the proapoptotic activity of TRAIL in all TRAIL-resistant cell lines, including K562, which considerably expressed DR5. These results suggested that the inhibition of NF-κB activation by LLnL either augmented the TRAIL sensitivity or synergistically acted with TRAIL in the process of apoptosis induction, but could not convert the TRAIL-resistant cells to be TRAIL sensitive.
Modulation of TRAIL sensitivity by NF-κB inhibitors.
(A) Effect of LLnL. TRAIL-sensitive or resistant cell lines were cultured for 42 hours in the absence or presence of rhsTRAIL (10 ng/mL for sensitive cell lines, 50 ng/mL for resistant cell lines) with or without LLnL (2.5 μM) pretreatment, and the 3H-thymidine uptake was determined for the last 6 hours. Data are represented as the mean ± SD of triplicate sample. (B) Effect of SN50. The TRAIL-sensitive cell line KOPM28 was cultured for 42 hours in the absence or presence of rhsTRAIL (10 ng/mL) with or without SN50 or SN50M (100 μg/mL) pretreatment, and the 3H-thymidine uptake was determined for the last 6 hours. Data are represented as the mean ± SD of triplicate sample.
Modulation of TRAIL sensitivity by NF-κB inhibitors.
(A) Effect of LLnL. TRAIL-sensitive or resistant cell lines were cultured for 42 hours in the absence or presence of rhsTRAIL (10 ng/mL for sensitive cell lines, 50 ng/mL for resistant cell lines) with or without LLnL (2.5 μM) pretreatment, and the 3H-thymidine uptake was determined for the last 6 hours. Data are represented as the mean ± SD of triplicate sample. (B) Effect of SN50. The TRAIL-sensitive cell line KOPM28 was cultured for 42 hours in the absence or presence of rhsTRAIL (10 ng/mL) with or without SN50 or SN50M (100 μg/mL) pretreatment, and the 3H-thymidine uptake was determined for the last 6 hours. Data are represented as the mean ± SD of triplicate sample.
A similar experiment was also performed in KOPM28 using another NF-κB–specific inhibitor, SN50, which blocks the nuclear translocation of NF-κB.40 As shown in Figure 8B, SN50 (100 μg/mL), but not its mutant control peptide SN50M, reduced the 3H-thymidine uptake to nearly 60% by itself and enhanced sensitivity to TRAIL, further substantiating the modulation of TRAIL sensitivity by NF-κB.
Comparison of antileukemic effects of TRAIL and imatinib mesylate
Imatinib mesylate is a specific inhibitor of BCR-ABL tyrosine kinase activity41 and shows a potent cytotoxic effect on Ph1-positive leukemias.55,56 In a clinical trial, imatinib mesylate was reported to be very effective in CML patients who were resistant to the IFN-α therapy.55
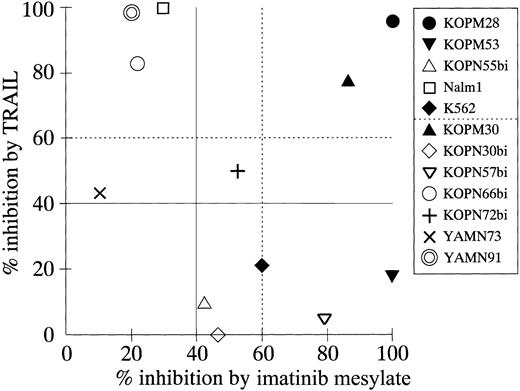
Thus, we compared the cytotoxic effects of TRAIL and imatinib mesylate against 12 Ph1-positive cell lines (Figure9). All these cell lines were established from the patients who had not been treated with imatinib mesylate and were not selected by imatinib mesylate treatment in vitro. All myeloid cell lines (KOPM28, KOPM30, KOPM53, and K562; closed symbols) showed high sensitivity to imatinib mesylate (percent inhibition, > 60%). In contrast, only 1 of 8 lymphoid cell lines (KOPN57bi) was highly sensitive, and 4 lymphoid cell lines (Nalm1, KOPN66bi, YAMN73, and YAMN91) were naturally resistant to imatinib mesylate (percent inhibition, < 40%). Two myeloid cell lines (KOPM28 and KOPM30) showed high sensitivity to both TRAIL and imatinib mesylate. The 4 lymphoid cell lines resistant to imatinib mesylate were sensitive to TRAIL, while 5 cell lines (myeloid 2; lymphoid 3) were resistant to TRAIL but sensitive to imatinib mesylate. Most importantly, none of 12 cell lines were resistant to both TRAIL and imatinib mesylate. These results suggested that TRAIL and imatinib mesylate could be used complementarily to eliminate Ph1-positive leukemia cells.
Correlation between percent inhibition by TRAIL and percent inhibition by imatinib mesylate.
The indicated cell lines were cultured for 42 hours in the presence or absence of rhsTRAIL (100 ng/mL) or imatinib mesylate (1.0 μM), and the 3H-thymidine uptake was assessed for the last 6 hours. Data on myeloid cell lines are shown by closed symbols.
Correlation between percent inhibition by TRAIL and percent inhibition by imatinib mesylate.
The indicated cell lines were cultured for 42 hours in the presence or absence of rhsTRAIL (100 ng/mL) or imatinib mesylate (1.0 μM), and the 3H-thymidine uptake was assessed for the last 6 hours. Data on myeloid cell lines are shown by closed symbols.
Discussion
In the present study, to explore the possible contribution of FasL and TRAIL to the immune-mediated antileukemic effects against Ph1-positive leukemia cells, we first investigated whether FasL or TRAIL could induce cell death in 12 Ph1-positive leukemia cell lines. Consistent with a previous report,32 most of the Ph1-positive cell lines were resistant to FasL. In contrast, 2 of 5 CML-BC–derived and 5 of 7 Ph1-AL–derived cell lines were highly or moderately sensitive to TRAIL-induced apoptotic cell death. Similar results were obtained with primary Ph1-positive ALL cells. Consistent with our data, Plasilova et al recently reported that the growth of CML progenitors in chronic phase as well as in accelerated or blastic phases was significantly suppressed by recombinant TRAIL.57
We next analyzed the cell-surface expression of TRAIL receptors to understand the differences in TRAIL sensitivity among the Ph1-positive leukemia cell lines. All TRAIL-sensitive cell lines and primary cells expressed the death-inducing receptors DR4 and/or DR5 on their surface, whereas the TRAIL-resistant cell lines, except for K562, and primary cells expressed neither DR4 nor DR5. In addition, none of these cell lines expressed DcR1 and DcR2, which are thought to act as decoy receptors. These results suggested that the sensitivity of Ph1-positive leukemia cells to TRAIL is primarily determined by the cell-surface expression of DR4 and/or DR5.
In addition to the receptor expression, regulators in the death-signaling pathway would be critical for the determination of sensitivity to FasL and TRAIL. We demonstrated that caspase-8, FADD, and FLIP were ubiquitously expressed in Ph1-positive leukemia cell lines irrespective of their differential sensitivities to FasL and TRAIL, suggesting that these molecules are not critically involved in the determination of sensitivity to FasL and TRAIL at least in their expression levels. We observed a marked discrepancy between sensitivity to TRAIL and FasL in several Ph1-positive cell lines despite expression of both death receptors. In particular, KOPM28 expressing both DR5 and Fas at highest levels showed a high sensitivity to TRAIL but not to FasL. Recently, a similar discrepancy has also been documented in some solid tumors.58,59 In addition, distinct intracellular signaling pathways in TRAIL- and FasL-mediated apoptosis have been reported.60 Thus, KOPM28 would serve as a useful subject to explore distinct intracellular signaling via TRAIL and FasL in further studies.
It has been demonstrated that the binding of TRAIL to DR4 and DR5 as well as DcR2 induced the NF-κB activation,6,9,53 and that the increased NF-κB activity protects tumor cells from various proapoptotic stimuli.62 Therefore, we examined whether the NF-κB activity could modulate the sensitivity to TRAIL in Ph1-positive leukemia cells. The pretreatment with NF-κB inhibitors, LLnL and SN50, enhanced sensitivity to TRAIL in the TRAIL-sensitive cell lines, suggesting that the DR4/DR5-mediated or BCR-ABL–mediated NF-κB activation plays a substantial role in protecting Ph1-positive leukemia cells from TRAIL-induced cell death. Of importance, in TRAIL-resistant cell lines not expressing DR4 or DR5, NF-κB inhibitors could not overcome their TRAIL resistance. Since a proteasome inhibitor that inhibits NF-κB activation (PS-341) has already been in phase 3 clinical trials against multiple myelomas,62 63 the combination with PS-341 would be a promising way to enhance the TRAIL-mediated elimination of Ph1-positive leukemia cells in a clinical setting.
We previously demonstrated that IFN-α up-regulated the cell-surface expression of TRAIL, on TCR/CD3-stimulated T cells in vitro, which mediated cytotoxicity against TRAIL-sensitive renal cell carcinomas.15 In addition, Wen et al reported that antileukemic agents including Ara-C increased the expression levels of DR5 on leukemia cell lines.64 Moreover, it has been reported that a combination of Ara-C and IFN-α increased the rate of remission and prolonged survival in CML patients.65Therefore, it may be feasible to expect that Ara-C enhances the DR5 expression in CML progenitors and IFN-α induces the TRAIL expression on CTLs in CML patients, leading to elimination of Ph1-positive leukemias through the TRAIL/DR5 interaction. To address this possibility, the correlation between the in vitro susceptibility of CML progenitors to TRAIL and the clinical responses to AraC and/or IFN-α therapy have to be determined.
Allo-SCT is a potentially curative therapy against CML and Ph1-positive ALL, and its effectiveness is thought to be achieved mainly by the immune-mediated GVL effect. We herein showed that TRAIL could efficiently induce apoptosis in Ph1-positive AL-derived cell lines as well as CML-BC–derived cell lines. In this regard, endogenously expressed TRAIL on CTLs or NK cells may be involved in the GVL effect. To address this hypothesis, the correlation between the in vitro TRAIL sensitivity of leukemia cells and the leukemia-free survival after allo-SCT in CML and Ph1-positive ALL patients has to be determined in future study.
A significant number of CML patients cannot tolerate the IFN-α therapy because of lethargy, malaise, anorexia, depression, and autoimmune-like syndromes. The GVL effect after allo-SCT is frequently associated with life-threatening graft-versus-host disease. In contrast, at least in mice and nonhuman primates, administration of rhsTRAIL did not exhibit a serious toxicity.66 67 Thus, rhsTRAIL could be a novel therapeuetic agent for CML and Ph1-positive ALL patients as a safer alternative.
Finally, we investigated the correlation between TRAIL sensitivity and imatinib mesylate sensitivity. imatinib mesylate exerted a potent antileukemic effect against Ph1-positive cells not only in patients with CML in chronic phase who failed to the IFN-α therapy,56 but also in patients with CML in advanced phase or Ph1-positive ALL.56 61 Importantly, we demonstrated that imatinib mesylate efficiently repressed most of the TRAIL-resistant cell lines, while TRAIL repressed most of the imatinib mesylate-resistant cell lines. This suggests a potential clinical utility of TRAIL particularly for patients with imatinib mesylate-resistant CML and Ph1-positive ALL.
Prepublished online as Blood First Edition Paper, December 27, 2002; DOI 10.1182/blood-2002-06-1770.
Supported in part by research grants from the Ministry of Education, Science, and Culture, Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Takeshi Inukai, Department of Pediatrics, School of Medicine, University of Yamanashi, Tamaho, Nakakoma, Yamanashi 409-3898, Japan; e-mail:tinukai@res.yamanashi-med.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal