Structural aberrations of the short arm of chromosome 2, mostly resulting in gains of 2p13∼16, have recently been described as being highly recurrent in Hodgkin and Reed-Sternberg (HRS) cells of classical Hodgkin lymphoma (cHL). As these gains consistently lead to increased copy numbers of the REL oncogene locus, we investigated the expression of the c-Rel protein in a series of 30 cHL cases with known genomic REL status as determined by comparative genomic hybridization and interphase cytogenetics. Expression of the c-Rel protein was investigated in 26 biopsies by immunohistochemistry. Distinct patterns were observed in HRS cells with no staining, cytoplasmic, and/or nuclear staining for c-Rel. All 13 samples with additional copies of the REL locus displayed nuclear staining for c-Rel, while 13 cHL samples lacking chromosome 2 (2p) gains displayed a significantly lower proportion or complete absence of HRS cells with nuclear c-Rel expression. Detailed analysis using combined immunophenotyping and interphase cytogenetics of individual HRS cells demonstrated that REL gains correlated with the presence of nuclear c-Rel staining. Additionally, in 2 cHL samples with translocation breakpoints in 2p13∼16, nuclear staining of c-Rel was observed; in one of them the staining pattern was indicative of a truncated c-Rel protein. The correlation between structural aberrations involving the REL locus and nuclear c-Rel accumulation in HRS cells qualifies REL as a target gene of the frequent gains in 2p in cHL. The data suggest thatREL aberrations are a genetic mechanism contributing to constitutive nuclear factor (NF)–κB/Rel activation in cHL.
Introduction
Classical Hodgkin lymphoma (cHL) accounts for approximately one third of all malignant lymphomas. It is widely accepted that in most instances the neoplastic Hodgkin and Reed-Sternberg (HRS) cells represent clonal populations of transformed germinal center B cells.1,2 The HRS cells are embedded in a background of reactive cells that are attracted by cytokines produced by HRS cells.3,4 The reasons for this particular feature of cHL are largely unknown; however, recent advances in the field have enabled new mechanisms to be identified, such as a constitutive activation of transcription factor nuclear factor (NF)–κB, which may account for the up-regulated expression of genes typically associated with HRS cells.5 6
Very recently, using comparative genomic hybridization (CGH) from microdissected HRS cells as well as simultaneous fluorescence immunophenotyping and interphase cytogenetics (FICTION;fluorescence immunophenotyping and interphasecytogenetics as a tool forinvestigation of neoplasms), we were able to identify structural aberrations affecting the short arm of chromosome 2 (2p) as the most frequent chromosomal imbalance in cHL. Changes in 2p, mostly leading to gains, were observed in up to 55% of all cHL cases, with a predominance in the nodular-sclerosis (cHL-NS) subtype.7-9
The high frequency of gains of 2p in cHL might indicate that this chromosomal region harbors one or more genes potentially involved in the pathogenesis of this disease. The critical region of gain in cHL was narrowed to 2p13∼16 by CGH. The 2 candidate oncogenesBCL11A and REL have been mapped within this critical region.9 Using interphase cytogenetics, the HRS cells of 3 cHL cases with changes in 2p were found to display aberrant signal patterns only for a fluorescence in situ hybridization (FISH) probe spanning the REL locus but not for a probe spanning the BCL11A locus.9 These data suggest thatREL, rather than BCL11A, is the target of the 2p alterations in cHL.
REL encodes a member of the family of NF-κB transcription factors. The NF-κB family is involved in the control of cell proliferation, inflammatory response, and apoptosis. Moreover, deregulation of members of this family has been shown to drive tumor development.10-14 In HRS cells, NF-κB is constitutively activated, but the reason for this activation is largely unknown. Mutations in the NFKBIA gene coding for the potent NF-κB inhibitor IκBα have been described in HRS cells of a small subset of cHL and might account for NF-κB activation in these tumors. However, most cHL cases lack NFKBIAmutations.15-17 Alternatively, genomic changes of theREL locus in 2p might be a further mechanism resulting in NF-κB activation in cHL by increased expression of the c-Rel protein in HRS cells.
We herein show by immunohistochemistry and by simultaneous interphase cytogenetics and fluorescence immunocytology that structural chromosomal aberrations in 2p correlate with nuclear c-Rel protein accumulation in HRS cells of cHL.
Materials and methods
Tissue samples
Twenty-six biopsies from 25 cHL patients, which had been previously analyzed by CGH from microdissected HRS cells for genomic imbalances (for case numbers, see Table 1),8 were selected for the present study based on the availability of paraffin-embedded tissue suitable for immunohistochemistry. Lymphomas were classified according to the guidelines of the World Health Organization (WHO) classification on the basis of routinely stained paraffin.18 For immunophenotyping of HRS cells, monoclonal antibodies against CD3, CD15, CD20, CD30, the joining (J-) chain, and the epithelial membrane antigen (EMA) were used. The series comprised 9 cHL cases of mixed cellularity (MC), 9 of nodular sclerosis (NS), 6 of lymphocyte-rich (LR) subtype, and 1 of the lymphocyte-depleted (LD) subtype. Samples cHL16a and b were subsequent biopsies taken at diagnosis and 6 months later. In addition, 5 cHL were included from an independent molecular cytogenetic study (case nos. 1, 2, 4, 17, and 46 in Martin-Subero et al, 2002).9Furthermore, to demonstrate c-Rel expression in reactive lymphoid tissue, paraffin sections of 3 hyperplastic tonsils were used.
Correlation between c-Rel expression and RELgenomic status as detected by immunohistology, CGH, and FICTION
| Case no.* . | Subtype of cHL . | Immunohistology of HRS cells . | Partial CGH karyotype (2p) . | FICTION data . | |||
|---|---|---|---|---|---|---|---|
| Nucleus . | Cytoplasm . | c-Rel expression . | REL/D2Z1ratio, median (range)† . | REL genomic status . | |||
| 1 | LR | − | − | Balanced | Slightly positive | 1.0 (1.0-1.0) | Intact |
| 2 | LR | − | − | Balanced | Negative | 1.0 (1.0-1.0) | Intact |
| 3 | LR | + | +++ | Balanced | NE | 1.0 (1.0-1.2)‡ | Intact |
| 4 | LR | ++ | − | Balanced | NE | NE | — |
| 5 | LR | ++ | + | rev ish enh(2p13-p21) | Positive | 2.0 (2.0-2.0) | Duplicated§ |
| 6 | LR | ++ | +++ | Balanced | NE | NE | — |
| 7 | MC | +++ | +++ | rev ish enh(2p13-p25) | ND | ND | — |
| 8 | MC | + | +++ | Balanced | NE | NE | — |
| 9 | MC | + | − | Balanced | Negative | 1.0 (1.0-1.5) | Intact |
| 10 | MC | + | ++ | Balanced | NE | NE | — |
| 11 | MC | NE | NE | Balanced | Negative | 1.0 (1.0-1.0) | Intact |
| 12 | MC | ++ | + | rev ish enh(2p11-p25) | NE | NE | — |
| 13 | MC | NE | NE | Balanced | Negative | 1.0 (1.0-2.0) | Intact |
| 14 | MC | ++ | +++ | Balanced | NE | 5.0 (1.0-6.5)‡ | Amplified |
| 15 | MC | +++ | − | rev ish amp(2p16) | Positive | 6.0 (2.0-6.5) | Amplified |
| 16a | NS | +++ | ++ | rev ish enh(2p12-p24) | ND | ND | — |
| 16b | NS | ++ | ++ | rev ish enh(2p13-p24) | Positive | 1.3 (1.0-1.7) | Duplicated |
| 17 | NS | ++ | ++ | rev ish enh(2p11-p24) | NE | NE | — |
| 18 | NS | +++ | +++ | rev ish enh(2p11-p25) | Positive | 1.5 (1.2-2.0) | Duplicated |
| 19 | NS | +++ | − | rev ish enh(2p12-p21) | Positive | 1.4 (1.2-1.8) | Duplicated |
| 20 | NS | ++ | +++ | Balanced | Positive | 1.0 (1.0-1.0) | Intact |
| 21 | NS | NE | NE | Balanced | Negative | 1.0 (1.0-1.0) | Intact |
| 22 | NS | +++ | +++ | rev ish enh(2p11-p25) | ND | ND | — |
| 23 | NS | ++ | + | rev ish enh(2p11-p25) | Positive | 1.5 (1.4-1.7) | Duplicated§ |
| 24 | NS | +++ | +++ | rev ish amp (2p13-p16) | Positive | 7.5 (5.0-10.0) | Amplified |
| 25 | LD | + | − | Balanced | Slightly positive | 1.0 (1.0-1.0) | Intact |
| Case no.* . | Subtype of cHL . | Immunohistology of HRS cells . | Partial CGH karyotype (2p) . | FICTION data . | |||
|---|---|---|---|---|---|---|---|
| Nucleus . | Cytoplasm . | c-Rel expression . | REL/D2Z1ratio, median (range)† . | REL genomic status . | |||
| 1 | LR | − | − | Balanced | Slightly positive | 1.0 (1.0-1.0) | Intact |
| 2 | LR | − | − | Balanced | Negative | 1.0 (1.0-1.0) | Intact |
| 3 | LR | + | +++ | Balanced | NE | 1.0 (1.0-1.2)‡ | Intact |
| 4 | LR | ++ | − | Balanced | NE | NE | — |
| 5 | LR | ++ | + | rev ish enh(2p13-p21) | Positive | 2.0 (2.0-2.0) | Duplicated§ |
| 6 | LR | ++ | +++ | Balanced | NE | NE | — |
| 7 | MC | +++ | +++ | rev ish enh(2p13-p25) | ND | ND | — |
| 8 | MC | + | +++ | Balanced | NE | NE | — |
| 9 | MC | + | − | Balanced | Negative | 1.0 (1.0-1.5) | Intact |
| 10 | MC | + | ++ | Balanced | NE | NE | — |
| 11 | MC | NE | NE | Balanced | Negative | 1.0 (1.0-1.0) | Intact |
| 12 | MC | ++ | + | rev ish enh(2p11-p25) | NE | NE | — |
| 13 | MC | NE | NE | Balanced | Negative | 1.0 (1.0-2.0) | Intact |
| 14 | MC | ++ | +++ | Balanced | NE | 5.0 (1.0-6.5)‡ | Amplified |
| 15 | MC | +++ | − | rev ish amp(2p16) | Positive | 6.0 (2.0-6.5) | Amplified |
| 16a | NS | +++ | ++ | rev ish enh(2p12-p24) | ND | ND | — |
| 16b | NS | ++ | ++ | rev ish enh(2p13-p24) | Positive | 1.3 (1.0-1.7) | Duplicated |
| 17 | NS | ++ | ++ | rev ish enh(2p11-p24) | NE | NE | — |
| 18 | NS | +++ | +++ | rev ish enh(2p11-p25) | Positive | 1.5 (1.2-2.0) | Duplicated |
| 19 | NS | +++ | − | rev ish enh(2p12-p21) | Positive | 1.4 (1.2-1.8) | Duplicated |
| 20 | NS | ++ | +++ | Balanced | Positive | 1.0 (1.0-1.0) | Intact |
| 21 | NS | NE | NE | Balanced | Negative | 1.0 (1.0-1.0) | Intact |
| 22 | NS | +++ | +++ | rev ish enh(2p11-p25) | ND | ND | — |
| 23 | NS | ++ | + | rev ish enh(2p11-p25) | Positive | 1.5 (1.4-1.7) | Duplicated§ |
| 24 | NS | +++ | +++ | rev ish amp (2p13-p16) | Positive | 7.5 (5.0-10.0) | Amplified |
| 25 | LD | + | − | Balanced | Slightly positive | 1.0 (1.0-1.0) | Intact |
LR indicates lymphocyte-rich; MC, mixed cellularity; NS, nodular sclerosis; LD, lymphocyte-depleted, NE: not evaluable, ND: not done; −, negative staining; +, staining in up to 30% of HRS cells; ++, staining in more than 30% and up to 70% of HRS cells; +++; staining in more than 70% of HRS cells; —, not applicable.
CGH data for cases 1 through 23 and 25 have been previously published. For comparison of the complete CGH karyotype, case numbers 1 through 23 and 25 correspond to case numbers in Joos et al8as follows: 2, 3, 4, 5, 6, 8, 13, 14, 15, 17, 19, 20, 21, 23, 24, 29a, 29b, 30, 31, 33, 34, 36, 37, 38, 40. Case cHL24 had the following CGH karyotype: rev ish enh(1q21-q44,4q32-q35,9,12q23-q24,17,20,21,22,Xq12-q28) dim(5q11-q35,11,13,14,16q21-q23) amp(2p13-p16).
The number of signals for the REL probe divided by the number of signals for the centromere of chromosome 2. Hybridization signals were scored in c- Rel+ HRS cells or in large atypical aneuploid cells when c-Rel expression was not detected.
Done by FISH.
Immunohistochemistry
Based on pilot studies, immunohistochemistry was performed on paraffin-embedded tissue sections, except in the case of cHL7, where cryostat sections were used. Staining was performed according to standardized protocols using the commercially available Envision kit for detection of bound antibody (Dako, Carpinteria, CA). Briefly, paraffin sections were dewaxed with xylene and passed through decreasing concentrations of acetone (100%, 70%, 40%; 10 minutes each). For antigen retrieval, sections were treated by microwave irradiation (750 W; 20 minutes) in citrate buffer (pH 6.0) for 20 minutes and then incubated with the primary antibody.
For detection of c-Rel protein, the polyclonal rabbit serum 265 raised against a synthetic peptide of the C-terminus of the human c-Rel protein was applied.19,20 Furthermore, in one sample we also applied the serum 1136 raised against amino acid sequence 340-353, which corresponds to a more N-terminally located epitope of human c-Rel.21
Evaluation of immunostaining was carried out in a blinded fashion. In tissue sections, strongly stained immunoblasts or lymph follicles were regarded as positive intrinsic controls. Several categories were created according to the proportion of HRS cells displaying positive staining for c-Rel: − stands for no staining detected, + indicates staining in up to 30%, ++ indicates staining in more than 30% and up to 70%, and +++ indicates staining in more than 70% of the total number of HRS cells analyzed (Table 1).
Transfection and blocking experiments
To test the specificity of the anti-Rel antisera, transfection experiments using human embryonic kidney cells 293 were carried out. These cells were transiently transfected with the Rc/CMV2 vector and the Rc/CMV2 plasmid with the full-length REL cDNA according to the manufacturer's protocol (Fugene; Roche Biochemicals, Mannheim, Germany). Cells were pelleted and fixed in formalin 48 hours after transfection and embedded in paraffin. Subsequently 2- to 4-μm-thick paraffin sections of the cell pellets were subjected to immunocytological staining (see “Immunohistochemistry”).
For blocking experiments, specific competing synthetic peptides, originally applied for immunization of rabbits, were used (1 μg peptide/μL serum). Prior to staining, 50 μL of the serum and 50 μL competitive peptide were incubated together on the slides and after incubation for 30 minutes, whereupon the subsequent staining procedure with secondary antibodies was carried out. In parallel, an irrelevant antirabbit serum was used as control.
Fluorescence in situ hybridization (FISH) and combined immunophenotyping and interphase cytogenetics (FICTION)
Probes for the REL and BCL11A loci used for interphase cytogenetic studies have been described recently.9,22 A chromosome 2 centromeric probe (CEP2) was obtained from Vysis (Downers Grove, IL). The FISH experiments were performed as described elsewhere.23
In order to study both c-Rel expression pattern andREL genomic status simultaneously, FICTION was carried out. Touch or cytospin preparations from lymphoma tissues were fixed in acetone for 10 minutes and air dried. Slides were then incubated with the rabbit c-Rel polyclonal antibody 265, followed by sequential application of Alexa 350–conjugated goat antirabbit and Alexa 350–conjugated donkey antigoat secondary antibodies (Molecular Probes, Leiden, the Netherlands). Incubation steps were performed at room temperature with a 30-minute incubation period and subsequent washings. Subsequent to fluorescence immunostaining, slides were incubated for 10 minutes in Carnoy fixative (methanol–acetic acid, 3:1), washed in dH2O and postfixed in 1% paraformaldehyde for 1 minute. Slides were then dehydrated by an ethanol series (70%, 85%, and absolute) and air dried in the dark. The appropriate FISH probe (1.5 μL) was applied to the cell-containing area of the slide and covered with a round 10-mm cover slip. Probe and target DNA were simultaneously denatured at 70°C for 7 minutes and incubated overnight at 37°C. Posthybridization washes were performed in 0.1 × standard saline citrate (SSC) 3 times for 5 minutes each at 60°C. Finally, slides were mounted in antifade solution.24 No DNA counterstaining was performed. Slides were analyzed using a Zeiss Axioskop2 fluorescence microscope (Göttingen, Germany) equipped with the appropriate filter sets (AHF, Tübingen, Germany) and documented using the ISIS imaging system (MetaSystems, Altlussheim, Germany). HRS cells were identified by virtue of their atypical cytology, hyperploidy, and, in positive cases, c-Rel expression. A median of 12 HRS cells (range, 2-31 cells) were evaluated per cHL case.
Statistical analysis
Correlations between c-Rel expression and genomic RELaberrations in various subgroups were calculated using the Fisher exact test application in SPSS Version 11.0 (SPSS, Erkrath, Germany);P values less than .05 were considered significant. For statistical analyses, only samples taken at diagnosis were considered (ie, case 16b was excluded).
Results
Determination of the genomic REL status
Twenty-six biopsies from 25 cHL patients, for which CGH data from microdissected HRS cells were available, were included in the study. Gains of chromosomal material on the short arm of chromosome 2, including the critical region in 2p13∼16 containing theREL gene, were detected in 12 cHL biopsies (Table1). Two of these lymphomas displayed a distinct high-level DNA amplification of the REL locus. In order to assess the REL copy number in these cases, we performed interphase cytogenetics (FISH and FICTION) using probes for the REL locus and the centromeric region of chromosome 2. Seventeen of the 26 biopsies were successfully analyzed in this way. In 9 lymphomas, the balanced status for 2p detected by CGH was also found by interphase cytogenetics (median REL/D2Z1 ratio = 1.0). The 2 cases with high-level amplification by CGH showed high medianREL/D2Z1 ratios of 6.0 and 7.5, confirming the presence of an amplification of the REL locus. In 5 cases with gain of 2p by CGH, the median REL/D2Z1 ratio was between 1.3 and 2.0, thus exceeding the balanced status. Overall, there was complete agreement between the findings obtained by the 2 techniques, except in one case: in cHL14, CGH revealed a balanced status while interphase FISH indicated amplification of the REL locus, with aREL/D2Z1 ratio of 5.0.
Immunohistochemical detection of the c-Rel protein
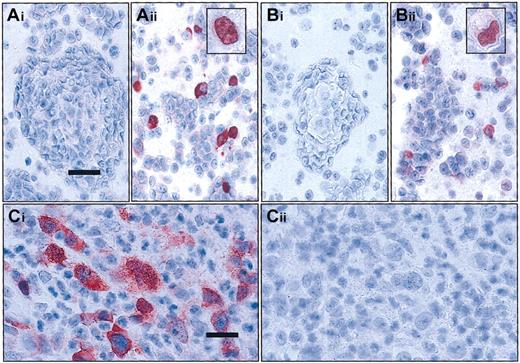
Specificity of the c-Rel antisera 265 and 1136 applied for immunohistochemistry was proven by transfection experiments in 293 human embryonic kidney cells as well as by blocking experiments with specific peptides in lymphoma tissue (see Figure1).
Evaluation of the specificity of the c-Rel antisera used by transfection and competition experiments.
(A-B) Transfection experiments. HEC 293 cells were transiently transfected with the Rc/CMV2 plasmid containing full-lengthREL cDNA (Aii and Bii) or with the Rc/CMV2 vector alone (Ai and Bi). In immunocytology with antisera 265 (A) and 1136 (B) on the paraffin sections of the pelleted cells, immunoreactivity is restricted to cells transfected with the vector containing full-lengthREL cDNA. Note that staining in some cells is exclusively localized in the nucleus (see inserts Aii,Bii); bar equals 37.5 μm; inset magnification, bar equals 18 μm. (C) Competitive experiments. cHL14 revealed a strong cytoplasmic and nuclear staining for c-Rel with c-Rel antisera 265 (Ci). Specificity of the staining was confirmed by complete absence of staining after simultaneous incubation with a specific peptide (Cii); bar = 25 μm. Stains described in “Materials and methods.”
Evaluation of the specificity of the c-Rel antisera used by transfection and competition experiments.
(A-B) Transfection experiments. HEC 293 cells were transiently transfected with the Rc/CMV2 plasmid containing full-lengthREL cDNA (Aii and Bii) or with the Rc/CMV2 vector alone (Ai and Bi). In immunocytology with antisera 265 (A) and 1136 (B) on the paraffin sections of the pelleted cells, immunoreactivity is restricted to cells transfected with the vector containing full-lengthREL cDNA. Note that staining in some cells is exclusively localized in the nucleus (see inserts Aii,Bii); bar equals 37.5 μm; inset magnification, bar equals 18 μm. (C) Competitive experiments. cHL14 revealed a strong cytoplasmic and nuclear staining for c-Rel with c-Rel antisera 265 (Ci). Specificity of the staining was confirmed by complete absence of staining after simultaneous incubation with a specific peptide (Cii); bar = 25 μm. Stains described in “Materials and methods.”
Expression of the c-Rel protein in tonsillar tissue and classical Hodgkin lymphoma
In hyperplastic tonsillar tissue, a strong, predominantly cytoplasmic staining confined to germinal center cells was obtained with both antisera. Inconsistently, there was a faint nuclear staining, equally confined to germinal center cells. In the extrafollicular areas a small number of intermingled immunoblasts revealed a strong cytoplasmic staining (Figure 2Ai-ii). Because of its superior staining quality, serum 265 was used for all subsequent immunohistochemical and FICTION studies.
Staining of the HRS cells revealed 4 different patterns: negative (Figure 2B), positive in the cytoplasm (Figure 2C), positive in the nucleus (Figure 2F), and positive in the cytoplasm and the nucleus (Figure 2Dii,2E). In some lymphomas, different staining patterns coexisted within the tumor cell population (Figure 2Hi).
Expression of c-Rel protein and detection of changes of the
REL locus in tonsillar tissue and in classical Hodgkin lymphoma. (A) In lymphofollicular tonsillar hyperplasia, the germinal center B cells stain c-Rel+ predominantly in the cytoplasm; follicular dentric cells and macrophages are c-Rel− (Ai). In the extrafollicular region (Aii), intermingled immunoblasts are c-Rel+; bars = 37.5μm. (B) In cHL1, with balanced genomic status for REL by CGH and FICTION, the HRS cells are c-Rel protein−; bar equals 25 μm. (C) Strong c-Rel staining is limited to the cytoplasm in the vast majority of HRS in cHL3, with balanced genomic status forREL; bar equals 25 μm. (Di) Partial CGH karyotype of cHL24 showing a high-level DNA amplification on the short arm of chromosome 2 including chromosomal bands 2p13-p16, as shown by the massive deviation of the ratio profile to the right. In this case, REL was shown to be amplified by FICTION analysis. (Dii) Immunohistochemically, the HRS cells of the case shown in Di show accumulation of c-Rel in the nucleus and in the cytoplasm; bar equals 25 μm. (E) cHL with a t(2;22)(p16;q12) showing a strong cytoplasmic and nuclear staining of c-Rel in HRS cells; bar equals 15 μm. (F) Immunhistochemistry of cHL15 reveals a consistent c-Rel nuclear staining of HRS cells; bar equals 37.5 μm. The inset figure shows a multicolor FICTION assay of this cHL sample combining FISH with probes for the REL(green) and BCL11A (pink) loci as well as for the centromere of chromosome 2 (CEP2; red) and simultaneous detection of the c-Rel protein using antiserum 265 (blue). The depicted Hodgkin cell displays a nuclear c-Rel expression and an amplification of the RELand BCL11A oncogene loci indicated by a number of green and pink signals clearly exceeding that of the red centromeric probe. (G) Application of the same multicolor FICTION assay as in panel F to cHL18. The Hodgkin cell with 5 copies of the centromere of chromosome 2 and 7 copies of the REL and BCL11A loci has strong nuclear c-Rel positivity; 2 bystander cells with regular signal patterns for the REL and BCL11A loci display only background staining of c-Rel. (Hi) Example of heterogeneous c-Rel expression in HRS cells (cHL5). Two HRS cells show strong nuclear positivity for c-Rel, while the HRS cell on the upper right side has staining limited to the cytoplasm; bar equals 15 μm. (Hii) FICTION assay of the same cHL sample as in Hi, combining the c-Rel antiserum 265 (blue) with probes for REL (green) and the centromere of chromosome 2 (CEP2; red). The binucleated HRS cell containing supernumerary copies of the REL locus as compared with the number of signals for CEP2 shows strong nuclear and slight cytoplasmic c-Rel staining; as internal control, surrounding bystander cells feature a regular hybridization pattern. Note that in the HRS cell the size of several signals derived from the REL probe is larger than that located in the nuclei of bystander cells. This is caused by the presence of small duplications involving theREL locus.
Expression of c-Rel protein and detection of changes of the
REL locus in tonsillar tissue and in classical Hodgkin lymphoma. (A) In lymphofollicular tonsillar hyperplasia, the germinal center B cells stain c-Rel+ predominantly in the cytoplasm; follicular dentric cells and macrophages are c-Rel− (Ai). In the extrafollicular region (Aii), intermingled immunoblasts are c-Rel+; bars = 37.5μm. (B) In cHL1, with balanced genomic status for REL by CGH and FICTION, the HRS cells are c-Rel protein−; bar equals 25 μm. (C) Strong c-Rel staining is limited to the cytoplasm in the vast majority of HRS in cHL3, with balanced genomic status forREL; bar equals 25 μm. (Di) Partial CGH karyotype of cHL24 showing a high-level DNA amplification on the short arm of chromosome 2 including chromosomal bands 2p13-p16, as shown by the massive deviation of the ratio profile to the right. In this case, REL was shown to be amplified by FICTION analysis. (Dii) Immunohistochemically, the HRS cells of the case shown in Di show accumulation of c-Rel in the nucleus and in the cytoplasm; bar equals 25 μm. (E) cHL with a t(2;22)(p16;q12) showing a strong cytoplasmic and nuclear staining of c-Rel in HRS cells; bar equals 15 μm. (F) Immunhistochemistry of cHL15 reveals a consistent c-Rel nuclear staining of HRS cells; bar equals 37.5 μm. The inset figure shows a multicolor FICTION assay of this cHL sample combining FISH with probes for the REL(green) and BCL11A (pink) loci as well as for the centromere of chromosome 2 (CEP2; red) and simultaneous detection of the c-Rel protein using antiserum 265 (blue). The depicted Hodgkin cell displays a nuclear c-Rel expression and an amplification of the RELand BCL11A oncogene loci indicated by a number of green and pink signals clearly exceeding that of the red centromeric probe. (G) Application of the same multicolor FICTION assay as in panel F to cHL18. The Hodgkin cell with 5 copies of the centromere of chromosome 2 and 7 copies of the REL and BCL11A loci has strong nuclear c-Rel positivity; 2 bystander cells with regular signal patterns for the REL and BCL11A loci display only background staining of c-Rel. (Hi) Example of heterogeneous c-Rel expression in HRS cells (cHL5). Two HRS cells show strong nuclear positivity for c-Rel, while the HRS cell on the upper right side has staining limited to the cytoplasm; bar equals 15 μm. (Hii) FICTION assay of the same cHL sample as in Hi, combining the c-Rel antiserum 265 (blue) with probes for REL (green) and the centromere of chromosome 2 (CEP2; red). The binucleated HRS cell containing supernumerary copies of the REL locus as compared with the number of signals for CEP2 shows strong nuclear and slight cytoplasmic c-Rel staining; as internal control, surrounding bystander cells feature a regular hybridization pattern. Note that in the HRS cell the size of several signals derived from the REL probe is larger than that located in the nuclei of bystander cells. This is caused by the presence of small duplications involving theREL locus.
Of the 26 cHL samples with available CGH data, 23 samples from 22 patients could be immunohistochemically evaluated for c-Rel protein expression. All 12 biopsies with gains or amplifications of 2p13∼16 by CGH displayed a nuclear staining for c-Rel. The same held true for cHL14, in which amplification of the REL locus was evidenced by interphase FISH. Eleven of these 13 cHL samples showed an additional cytoplasmic staining, while 2 samples with gains on 2p (including 1 sample with a high-level DNA amplification of the REL locus) showed nuclear staining only (nos. 15 and 19). In cHL19, withREL staining restricted to the nucleus, the gain on the short arm of chromosome 2 was the sole imbalance detected by CGH. In order to analyze the correlation between REL copy number and c-Rel protein expression on the single-cell level, FICTION analysis was successfully performed on 7 of the 13 cHL samples with gains in 2p. FICTION confirmed nuclear expression of c-Rel protein in HRS cells with supernumerary copies of the REL locus (Table 1; Figure 2F-G,Hii). FICTION analysis of 3 additional cHL samples with high-level amplification of the REL locus (previous case nos. 1, 2, and 4 in Martin-Subero et al, 2002)9 also confirmed abnormally strong nuclear c-Rel staining (data not shown).
Among the 13 cHL samples without gain on 2p according to CGH and interphase cytogenetics, 6 completely lacked c-Rel staining by immunohistochemistry or FICTION. In 2 of these, there was a slight discrepancy between the 2 techniques, most likely due to a very faint c-Rel expression as detected by fluorescence immunophenotyping (cHL1) or a low percentage of c-Rel+ HRS cells (cHL9). Nevertheless, over the complete study there was good overall accordance between immunohistochemistry and FICTION. Eight cHL samples without gains in 2p displayed nuclear staining by immunohistochemistry, including 5 with simultaneous cytoplasmic staining. Remarkably, the percentage of HRS cells with nuclear positivity for c-Rel on immunohistochemistry never exceeded 70% and was below 30% in 7 of 10 cHL samples without gain in 2p. In contrast, the percentage of nuclear c-Rel+ HRS cells in cHL samples with 2p gain was always more than 30% (P = .001) and exceeded 70% in 7 of 12 samples (P = .005; cHL16b excluded). The nuclear c-Rel accumulation (9 of 10 biopsies), like the genomic gain in 2p (8 of 10 biopsies), was particularly prominent in the nodular sclerosis subtype of cHL.
In addition to the cases studied by CGH, 2 cHL cases were analyzed in which previous interphase cytogenetics revealed evidence of a breakpoint within the region spanned by the probe for theREL locus.9 Immunohistochemistry of one of these cases, which had been previously shown to contain a t(2;22)(p16;q12),25 featured strong nuclear and cytoplasmic expression of c-Rel as revealed by the antiserum 265 (Figure 2E). The second case with a split of the probe spanning theREL locus completely lacked staining with serum 265. Since a fusion protein lacking the C-terminus of c-Rel has recently been described in the human diffuse large B-cell lymphoma (DLBCL) cell line RC-K8, we also applied the serum 1136, recognizing a more N-terminally located epitope of c-Rel in this latter case.26 Indeed, the HRS cells of this case strongly stained with this serum in the nucleus and cytoplasm, suggesting that the c-Rel protein expressed in these HRS cells lacks the wild-type C-terminus.
Discussion
In the present study, we show for the first time that HRS cells of most cHL samples express the c-Rel protein. This held particularly true for samples containing chromosomal aberrations in 2p13∼16, which in the present series consistently accumulated nuclear c-Rel protein in high percentage of HRS cells. Within the subset of cHL samples with alterations involving the REL locus, the percentage of nuclear c-Rel+ HRS cells was significantly higher than in cHL samples with apparently normal REL loci. These findings suggest that, at least in a subset of cHL, nuclear accumulation of c-Rel expression is related to chromosomal changes affecting the corresponding genomic region.
In nonneoplastic lymphoid tissue we found a high c-Rel expression in germinal center cells. These data are in line with mouse models indicating an important role of c-Rel in the development and differentiation of germinal center B cells.27,28Nevertheless, expression of c-Rel was not restricted to the germinal center but was also observed in a few intermingled immunoblasts in the extrafollicular area. This is in accordance with recent gene-expression profiling data showing strong REL transcript expression in activated B cells.29 Most germinal center cells and immunoblasts expressed c-Rel predominantly in the cytoplasm. Only rarely did we also observe the protein in the nucleus, suggesting a transitory nuclear translocation of c-Rel protein during germinal center B-cell reaction.
The most striking finding of the present study was the nuclear accumulation of c-Rel in a high percentage of HRS cells in cHL samples with genomic changes involving the REL oncogene locus. In the clear majority of cHL cases hitherto characterized, chromosomal alterations in the short arm of chromosome 2 led to gain of theREL gene. In these lymphomas, nuclear c-Rel accumulation might be caused by a gene dosage effect analogous to the mechanisms described for numerous other oncogenes, such as HER2/NEU,BCL2, and CCNE.30-32 In 2 cHL samples with nuclear c-Rel accumulation, we detected chromosomal breakpoints, using a FISH probe spanning the REL locus. One of those cases carried a translocation t(2;22), cytogenetically suggesting deregulation of c-Rel through juxtaposition of theREL gene and regulatory sequences of the immunoglobulin light chain locus λ (IGL) in 22q.9,25 Thus, it seems highly likely that in this case c-Rel expression is deregulated by juxtaposition of the REL gene toIG regulatory sequences, which is a well-known mechanism in B-cell lymphomas.33 34
In the second cHL sample with a break event detected by theREL-spanning FISH probe, the precise chromosomal rearrangement affecting the REL region is not yet clear.9 Strikingly, this case displayed strong nuclear staining using an antiserum directed to the REL homology domain recognizing amino acids 340 through 353. In contrast, no staining was observed using the serum 265 targeting amino acids 573 through 587 of the human c-Rel protein. These findings can be explained by the presence of a truncation of the C-terminus of the wild-type c-Rel protein caused by the chromosomal change. Such a mechanism of c-Rel activation has recently been demonstrated in the cell line RC-K8, derived from a human large B-cell lymphoma. In this cell line, the 5′ end of the REL gene is fused with the 3′ end of another gene called NRG (non-REL gene), resulting in a chimeric protein devoid of the wild-type C-terminus of c-Rel.26
Apart from the cell line RC-K8, genomic changes affecting theREL locus have been described in numerous primary lymphomas. In particular, gains including the REL locus have previously been reported in other B-cell lymphomas, such as diffuse large B-cell lymphoma,35,36 mediastinal B-cell lymphoma,37and follicular lymphomas.38 Nevertheless, the frequency of 2p gains in these non-Hodgkin lymphomas did not exceed 30% in most series and is thus considerably lower than in cHL. Moreover, genomic amplification of REL was recently shown to be restricted to the germinal center B cell–like subtype of DLBCLs defined by lymphochip technology.39 Finally, a recent abstract reports that in DLBCL, nuclear expression of c-Rel correlated with REL gene amplification.40
A well-known common characteristic of HRS cells is a high level of constitutive nuclear NF-κB/Rel activity, which stimulates proliferation and confers resistance to apoptosis.5 This constitutive NF-κB/Rel activity has been shown to maintain high expression of a characteristic gene network, including CD40, CD86, and a set of antiapoptotic genes in HRS cells.6 Various mechanisms have been proposed to confer NF-κB/Rel activation in HRS.4 These include alterations in signaling pathways mediated through receptors such as CD30, CD40, and RANK, or through infection by the LMP-1 protein of Epstein-Barr virus (EBV). Furthermore, mutations of the NFKBIA gene resulting in impairment of the IκBα-mediated control of NF-kappaB/Rel activity have been detected in HRS cells of a subset of cHL.15,16However, there are further transcription factors, such as activator protein-1 (AP-1)41 or signal transducer and activator of transcription (STAT), that may contribute to the activated status of HRS cells in a parallel manner to the NF-κB/Rel pathway.42 These alternative mechanisms may be active in the subset of cHL lacking nuclear c-Rel accumulation in our study.
We conclude that, based on the close correlation between structural chromosomal aberrations in 2p13∼16 and nuclear accumulation of c-Rel described in the present study, it seems likely that genomic changes of the REL locus provide an alternative genetic mechanism of constitutive nuclear NF-κB/Rel activation in HRS cells of cHL.
We wish to thank Dr N. Rice and Dr M. Ernst, National Cancer Institute – Frederick Cancer Research and Development Center (NCI-FCRDC), Frederick, MD, for helpful discussion and for providing the c-Rel antisera, the competing peptides, and the c-Rel vector. We thank Iwona Nerbas, Claudia Becher, and Dorit Schuster for excellent technical assistance and Caroline Higginson for editorial help.
Prepublished online as Blood First Edition Paper, January 2, 2003; DOI 10.1182/blood-2002-08-2577.
Supported by the Tumorzentrum Heidelberg/Mannheim (I.I.2.), the Deutsche Krebshilfe (70-2859-Ba2), the Landesforschungschwerpunkt Baden Württemberg (D.1526.1.4), and the Interdisziplinäre Zentrum für klinische Krebsforschung (IZKF) Kiel.
T.F.E.B. and J.I.M.-S. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Peter Möller, Abteilung für Pathologie, Universität Ulm, Albert-Einstein-Allee 11, D-89081 Ulm, Germany; e-mail:peter.moeller@medizin.uni-ulm.de; or Reiner Siebert, Institut für Humangenetik, Universitätsklinikum Schleswig-Holstein, Campus Kiel, Schwanenweg 24, D-24105 Kiel, Germany; e-mail: rsiebert@medgen.uni-kiel.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal