Abstract
Donor-derived T lymphocytes directed against minor histocompatibility antigens (mHags) exclusively expressed on cells of the hematopoietic lineages can eliminate hematologic malignancies. Transfer of T-cell receptors (TCRs) directed against these mHags into T lymphocytes may provide a strategy to generate antileukemic T cells. To investigate the feasibility of this strategy the TCR usage of mHag HA-2-specific T-cell clones was characterized. Thirteen different types of HA-2-specific T-cell clones were detected, expressing TCRs with diversity in TCR α- and β-chain usage, however, containing in the TCR α chain a single conserved gene segment Jα42, indicating that Jα42 is involved in HA-2-specific recognition. We transferred various HA-2 TCRs into T lymphocytes from HLA-A2-positive HA-2-negative individuals resulting in T cells with redirected cytolytic activity against HA-2-expressing target cells. Transfer of chimeric TCRs demonstrated that the HA-2 specificity is not only determined by the Jα42 region but also by the N-region of the α chain and the CDR3 region of the β chain. Finally, when HA-2 TCRs were transferred into T cells from HLA-A2-negative donors, the HA-2 TCR-modified T cells exerted potent antileukemic reactivity without signs of anti-HLA-A2 alloreactivity. These results indicate that HA-2 TCR transfer may be used as an alternative strategy to generate HA-2-specific T cells to treat hematologic malignancies of HLA-A2-positive, HA-2-expressing patients that received transplants from HLA-A2-matched or -mismatched donors. (Blood. 2003;102:3530-3540)
Introduction
Donor lymphocyte infusion (DLI) into patients with a relapse of their hematologic malignancy after allogeneic stem cell transplantation (allo-SCT) has been proven to be a successful treatment strategy.1-3 The beneficial graft-versus-leukemia (GVL) effect of donor lymphocytes following HLA-matched allo-SCT can be caused by T cells that are capable of recognizing minor histocompatibility antigens (mHags) on the malignant cell population from the patient.4-6 The tissue distribution of mHags probably determines the clinical relevance of T-cell responses against these antigens. T cells recognizing broadly expressed mHags may cause not only GVL but also graft-versus-host disease (GVHD).7,8 T cells recognizing mHags that are exclusively expressed on cells of the hematopoietic lineages may eliminate hematopoietic cells from the patient, including the malignant cells, without affecting donor-derived hematopoiesis or directly affecting nonhematopoietic tissues of the patient, causing only limited GVHD.9,10
The mHag HA-2 derived from a diallelic gene encoding a novel human class I myosin protein is one of the mHags that is exclusively expressed on cells of hematopoietic origin.11,12 Based on the limited tissue distribution of this particular mHag, T cells directed against HA-2 can be used to treat hematologic malignancies relapsing after allo-SCT. Recently, we demonstrated the emergence of HA-2-specific cytotoxic T cells in the peripheral blood of a patient with leukemia who relapsed after allo-SCT and was subsequently given a DLI. The appearance of these T cells coincided with a clinical antileukemic immune response resulting in durable remission.9 Since a high percentage of the human population expresses the HA-2,13 this mHag cannot be used for induction of HA-2-specific T cells in most cases of transplantation between HLA-identical individuals.
Due to the improved manipulation of the graft and the posttransplantation immune suppression, an increasing number of stem cell transplantations using partially HLA-mismatched related or unrelated donors is being performed.14,15 This approach opens new possibilities to use the mismatched HLA allele to specifically target an immune response against hematopoietic tissues from the patient, leading to eradication of the hematologic malignancy.16 In HLA-A2-mismatch transplantations in which cells derived from the patient are HLA-A2-positive and express the mHag HA-2 and the donor cells do not express the HLA-A2 restriction molecule, donor T-cell responses may be generated against mHag HA-2 peptide presented in HLA-A2 molecules on hematopoietic cells from the patient. The recent generation of mHag HA-1-specific cytotoxic T lymphocytes (CTLs) restricted by non-self-HLA molecules supports this approach.17 Several studies have shown the in vitro generation of antigen-specific T cells that are restricted by non-self-HLA molecules.18-21 Stimulation with non-self-HLA molecules, however, also induced undesired allo-HLA-specific T cells, which can cause damage of all HLA-A2-positive hematopoietic and nonhematopoietic tissues.17 A strategy to circumvent the undesired induction of allo-HLA-specific T cells may be the transfer of mHag-specific T-cell receptors (TCRs) into immune-competent cells from the donor. If these mHag-specific TCRs can be isolated from T cells exhibiting an immune response in an HLA-identical setting, this will limit the risk of transfer of TCRs that have not only mHag specificity but also affinity for other peptide-HLA-A2 complexes from HLA-A2-positive patients. Previously, the feasibility to redirect the specificity of recipient T lymphocytes by transfer of an antigen-specific TCR has been demonstrated.22-25 Introduction of an HLA class I-restricted TCR into CD8+ peripheral T cells resulted in antigen-specific cytolytic activity and cytokine secretion by these T cells. TCR transfer studies have demonstrated that the avidity and fine specificity of the TCR is maintained upon transfer.26,27 In addition, in a mouse model Kessels et al showed that these TCR-modified T cells were able to eradicate tumor cells in vivo.28
To be able to transfer HA-2-specific TCRs into T cells and to refine our understanding of the interaction between the TCR and its antigenic peptide-HLA complex, we characterized different HA-2-specific TCR complexes derived from HA-2-specific T-cell clones present in the peripheral blood of a chronic myeloid leukemia (CML) patient with an ongoing GVL response after HLA-identical allo-SCT who was subsequently treated with DLI. The mHag HA-2-specific T cells were isolated by cell sorting using HA-2/HLA-A2 tetrameric complexes and clonally expanded. In this study we characterized the TCR α and β repertoire of many individual HA-2-specific T-cell clones by reverse transcription-polymerase chain reaction (RT-PCR) and sequence analysis and revealed whether restricted TCR usage was employed in the anti-HA-2 response. Although the individual HA-2-specific T-cell clones expressed TCR α chains composed of different V segments and N regions that vary in length and amino acid composition, all TCR complexes of the HA-2-specific T-cell clones expressed a common J segment, Jα42. Transfer of various HA-2-specific TCR complexes into HLA-A2-positive HA-2-negative peripheral blood-derived T cells resulted in redirected cytolytic activity against target cells expressing the HA-2 antigen and not against target cells negative for the HA-2 mHag. The TCR-transferred T cells were as efficient in inhibiting the mHag HA-2-expressing CML progenitor cells as were the original HA-2-specific T-cell clones. Transfer of the various HA-2 TCRs into HLA-A2-negative peripheral blood-derived T cells resulted also in cytolytic activity against HLA-A2-positive CML cells expressing the HA-2 mHag. Importantly, no anti-HLA-A2 alloreactivity was observed, since HLA-A2-positive target cells negative for HA-2 expression, including Epstein-Barr virus (EBV)-transformed B cells, CML cells, and nonhematopoietic cells, were not lysed by the HA-2 TCR-modified T cells. These results show the feasibility of the approach to redirect the antigen specificity of peripheral T cells toward antileukemic reactivity, without the induction of allo-HLA reactivity.
Patients, materials, and methods
Isolation and expansion of HA-2-specific T-cell clones
This study was approved by the LUMC institutional review board. Informed consent was provided according to the Declaration of Helsinki. After informed consent, peripheral blood mononuclear cells (PBMCs) were isolated from a patient entering a complete remission after treatment with DLI for relapsed CML after allo-SCT. In this patient, emergence of HA-2-specific T cells has been shown to coincide with the clinical antileukemic immune response.9 The HA-2/HLA-A2 tetramer-positive T cells were directly clonally isolated from the PBMCs collected at 5 weeks after DLI. PBMCs were labeled with HA-2/HLA-A2 tetramers for 2 hours at 4°C in RPMI without phenol supplemented with 2% fetal bovine serum (FBS), washed 3 times, sorted at 4°C, and plated single cell per well using the fluorescence-activated cell sorter (FACS) Vantage (Becton-Dickinson [BD], San Jose, CA). Tetramers were constructed as described29 with minor modifications and consisted of the HA-2 peptide bound to HLA-A2 molecules. Single HA-2/HLA-A2 tetramer-positive T cells were cultured in Iscove modified Dulbecco medium (IMDM) plus 10% human serum and were nonspecifically stimulated every 2 weeks with feeder cell mixture containing irradiated allogeneic PBMCs (30 Gy), irradiated EBV-transformed B cells (EBV-LCLs; 50 Gy), 800 ng/mL phytohemagglutinin (PHA; Murex Diagnostics, Dartford, United Kingdom), and 100 IU/mL interleukin 2 (IL-2; Chiron, Amsterdam, The Netherlands).
Analysis of TCRVα and TCRVβ transcripts and generation of retroviral vectors and virus supernatant
Total RNA from T-cell clones was extracted using Trizol (Gibco, Carlsbad, CA). The mRNA was transcribed into single-strand cDNA in 25 μL reaction mixture by reverse transcriptase using oligo dT as a primer (Pharmacia, Uppsala, Sweden). TCRVα and Vβ gene usage was determined by RT-PCR and sequencing. A panel of 36 TCRAV- and 30 TCRBV-specific upstream primers in combination with one downstream TCRAC or TCRBC primer, respectively, were used for PCR amplification. TCRAV-AC and TCRBV-BC PCR products were purified with the Qiaquick PCR purification kit (Qiagen, Valencia, CA) and sequenced using the dye terminator cycle sequencing kit (ABI-PRISM; PerkinElmer, Foster City, CA), according to the manufacturer's instructions, to obtain a complete identification of the TCRAV and TCRBV genes. The complete TCR α and β chains of 4 different HA-2-specific T-cell clones, HA2.5 (group A), HA2.6 (group H), HA2.19 (group D), and HA2.20 (group C) were then amplified using specific primers and cloned into the retroviral vectors. The Moloney murine leukemia virus based retroviral vector LZRS and packaging cells ϕ-NX-A were used.30 Two bicistronic retroviral vectors per T-cell clone were constructed in which the multiple cloning site is linked to the downstream internal ribosome entry sequence (IRES) and the marker gene enhanced green fluorescent protein (eGFP)31 or truncated form of the nerve growth factor receptor (ΔNGF-R).32 The TCR α chains were cloned into the retroviral vectors in combination with eGFP and the TCR β chains were cloned in combination with the ΔNGF-R. Retroviral vectors encoding eGFP or ΔNGF-R alone were used as control vectors in the experiments. The constructs were transfected into ϕNX-A cells using calcium phosphate (Life Technologies, Gaithersburg, MD), and 2 days later 2 μg/mL puromycin (Clontech Laboratories, Palo Alto, CA) was added. Ten to 14 days after transfection 6 × 106 cells were plated per 10-cm Petri dish in 10 mL IMDM supplemented with 10% FBS without puromycin. The next day the medium was refreshed and the following day retroviral supernatant was harvested, centrifuged, and frozen in aliquots at -70°C.
Retroviral transduction of peripheral blood-derived T lymphocytes and selection of transduced T cells
Unselected TCRαβ-positive T cells derived from peripheral blood were stimulated with 800 ng/mL PHA and 100 IU/mL IL-2 at a concentration of 0.5 × 106 cells/mL. After 2 days of culture the T cells were transduced with retroviral supernatant using recombinant human fibronectin fragments CH-296.31,33 Briefly, 5 × 106 T cells were cultured on CH-296-coated Petri dishes together with 1 mL of thawed retroviral supernatant for 6 hours or overnight at 37°C, washed, and transferred to 24-well culture plates. The transduction efficiency, measured by the expression of the markers eGFP and ΔNGF-R, was analyzed by flow cytometry. ΔNGF-R expression was detected using the monoclonal antibody (mAb) 20.4 (antihuman NGF-R) obtained from American Type Culture Collection (ATCC, Rockville, MD). In addition, flow cytometric analyses were performed using mAbs specific for CD4 and CD8 (BD) in combination with HA-2/HLA-A2 tetramers. HA-2 TCR-transduced T cells were FACS sorted on bases of marker gene expression, cultured at 1 or 25 cells/well in IMDM supplemented with 10% human serum, and nonspecifically stimulated every 2 weeks with feeder cell mixture.
Cytotoxicity assay
Target cells were harvested and labeled with 50 μCi (1.85 MBq) Na251CrO4 for 60 minutes at 37°C, washed, and added to various numbers of effector T cells in 96-well U-bottomed microtiter plates. After 4 hours or 18 hours of incubation of target and effector cells, 25 μL of supernatant was harvested and measured in a luminescence counter (Topcount-NXT; Packard, Meriden, CT). The mean percentage of specific lysis of triplicate wells was calculated as follows: specific lysis = [(experimental release - spontaneous release)/(maximal release - spontaneous release)] × 100.
Cytokine production assay
T cells were cultured at 1 × 104 cells/well with 1 × 104 cells/well of irradiated EBV-LCLs in 96-well U-bottomed microtiter plates in a volume of 200 μL of IMDM supplemented with 10% FBS and 10 IU/mL IL-2. After 24 hours of culture 100 μL of supernatant was harvested and frozen at -20°C. The supernatants harvested after 24 hours were used to determine the cytokine production of the stimulated T cell using the BD human Th1/Th2 cytokine CBA Kit for detection of interferon γ (IFN-γ), tumor necrosis factor α (TNF-α), IL-10, IL-5, and IL-4 (BD), according to the manufacturer's procedure.
Liquid hematopoietic progenitor cell inhibition assay (PIA)
The PIA was performed as previously described.34 T cells were cultured at 1 × 104 cells/well with 1 × 104 cells/well of cell suspensions containing malignant hematopoietic progenitor cells (HPCs) in 96-well U-bottomed microtiter plates in a volume of 200 μL of IMDM supplemented with 10% FBS and multiple hematopoietic growth factors. T cells were irradiated with 15 Gy prior to use preventing their proliferation in the PIA. HPCs collected from different CML patients were used as target cells. After 5 days of culture, the wells were pulsed with 1 μCi (0.037 MBq) of 3H-thymidine for 18 hours. Cells were harvested and the incorporated 3H-thymidine was determined by luminescence counter.
Results
Functional analysis of the HA-2-specific T cells isolated from an antileukemic response
T cells specific for the mHag HA-2 were clonally isolated from the peripheral blood at the start of the antileukemic response by cell sorting using HA-2/HLA-A2 tetrameric complexes and expanded nonspecifically. A total of 53 T-cell clones were expanded to sufficient numbers to determine the functional activity. Of these T-cell clones 52 T cell clones were brightly staining with the HA-2/HLA-A2 tetramer (Table 1) and all T-cell clones were CD8+ (data not shown). Cytolytic activity of these HA-2-specific T-cell clones was measured using the EBV-LCLs derived from the HA-2-positive patient and HA-2-negative donor. Fifty-one T-cell clones exerted cytolytic activity against the patient EBV-LCL and not against the donor EBV-LCL (Table 1). All HA-2-specific cytolytic T-cell clones produced high amounts of IFN-γ after mHag-specific stimulation with patient EBV-LCLs (> 803 pg/mL) and 30 HA-2-specific T-cell clones produced TNF-α ranging from 10 to 86 pg/mL. Furthermore, part of the HA-2-specific T-cell clones produced IL-4, IL-5, and/or IL-10 after HA-2-specific stimulation. One T-cell clone (HA2.30), although clearly positive for HA-2/HLA-A2 tetramer staining, did not produce a significant amount of cytokines after specific stimulation and did not exert specific cytolytic activity.
The functional activity and tetramer staining of HA-2/HLA-A2 tetramer-positive T-cell clones isolated from an antileukemic response
. | . | Lytic activity, 4 h, % lysis . | . | Cytokine production, pg/mL . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HA-2 clone no. . | HLA-A2/HA-2 MFI . | EBV donor . | EBV patient . | IFN-γ . | TNF-α . | IL-10 . | IL-5 . | IL-4 . | |||||
| HA-2.01 | 206 | 0 | 69 | >5000 | 6 | 585 | 93 | 5 | |||||
| HA-2.02 | 735 | 4 | 64 | >5000 | 0 | 22 | 38 | 5 | |||||
| HA-2.03 | 553 | 6 | 61 | 3619 | 10 | 11 | 653 | 7 | |||||
| HA-2.04 | 306 | 0 | 72 | 3944 | 6 | 11 | 104 | 12 | |||||
| HA-2.05 | 466 | 0 | 79 | >5000 | 10 | 98 | 268 | 34 | |||||
| HA-2.06 | 630 | 0 | 53 | 1375 | 6 | 8 | 248 | 6 | |||||
| HA-2.07 | 520 | 0 | 60 | 4100 | 4 | 205 | 42 | 0 | |||||
| HA-2.08 | 256 | 2 | 64 | >5000 | 25 | 20 | 2433 | 72 | |||||
| HA-2.09 | 638 | 4 | 56 | 4709 | 8 | 9 | 638 | 9 | |||||
| HA-2.10 | 545 | 0 | 66 | 3914 | 86 | 13 | 19 | 5 | |||||
| HA-2.11 | 514 | 4 | 61 | 3319 | 3 | 6 | 53 | 4 | |||||
| HA-2.12 | 601 | 4 | 67 | 1900 | 0 | 6 | 13 | 4 | |||||
| HA-2.13 | 640 | 9 | 64 | 1477 | 10 | 5 | 287 | 39 | |||||
| HA-2.15 | 666 | 0 | 61 | 1476 | 9 | 16 | 138 | 7 | |||||
| HA-2.17 | 208 | 4 | 76 | 3412 | 21 | 62 | 2151 | 78 | |||||
| HA-2.19 | 689 | 0 | 71 | >5000 | 15 | 6 | 99 | 44 | |||||
| HA-2.20 | 468 | 2 | 61 | 3293 | 11 | 13 | 1414 | 13 | |||||
| HA-2.21 | 496 | 1 | 59 | 2211 | 6 | 4 | 65 | 2 | |||||
| HA-2.22 | 533 | 0 | 69 | 803 | 7 | 0 | 131 | 3 | |||||
| HA-2.24 | 777 | 0 | 63 | 1188 | 4 | 2 | 13 | 4 | |||||
| HA-2.25 | 327 | 1 | 71 | 3319 | 30 | 197 | 702 | 31 | |||||
| HA-2.27 | 456 | 3 | 70 | 2211 | 25 | 3 | 3761 | 269 | |||||
| HA-2.28 | 646 | 0 | 71 | 2683 | 15 | 1 | 80 | 5 | |||||
| HA-2.30 | 671 | 0 | 14 | 207 | 0 | 2 | 3 | 2 | |||||
| HA-2.31 | 631 | 2 | 56 | 3372 | 81 | 30 | 1262 | 93 | |||||
| HA-2.32 | 362 | 0 | 54 | 3139 | 12 | 15 | 575 | 42 | |||||
| HA-2.33 | 468 | 0 | 48 | 1250 | 13 | 4 | 15 | 4 | |||||
| HA-2.34 | 341 | 7 | 61 | 4818 | 17 | 16 | 85 | 9 | |||||
| HA-2.35 | 24 | 1 | 5 | 263 | 5 | 14 | 5 | 4 | |||||
| HA-2.36 | 555 | 3 | 61 | 2184 | 13 | 0 | 20 | 5 | |||||
| HA-2.37 | 696 | 0 | 75 | 1399 | 18 | 0 | 97 | 30 | |||||
| HA-2.38 | 510 | 0 | 81 | 3372 | 15 | 0 | 206 | 7 | |||||
| HA-2.41 | 228 | 5 | 83 | 2248 | 10 | 17 | 203 | 11 | |||||
| HA-2.43 | 590 | 0 | 69 | 4532 | 7 | 28 | 83 | 4 | |||||
| HA-2.46 | 894 | 0 | 78 | 2968 | 12 | 5 | 2 | 6 | |||||
| HA-2.47 | 450 | 0 | 77 | 1982 | 20 | 102 | 139 | 5 | |||||
| HA-2.48 | 488 | 0 | 73 | 4463 | 5 | 6 | 3 | 4 | |||||
| HA-2.52 | 551 | 1 | 68 | >5000 | 33 | 16 | 167 | 33 | |||||
| HA-2.53 | 580 | 0 | 68 | >5000 | 81 | 11 | 10 | 4 | |||||
| HA-2.54 | 568 | 0 | 67 | 4709 | 49 | 6 | 3 | 0 | |||||
| HA-2.55 | 769 | 5 | 67 | >5000 | 28 | 15 | 22 | 9 | |||||
| HA-2.58 | 364 | 0 | 55 | 1211 | 7 | 7 | 68 | 6 | |||||
| HA-2.59 | 427 | 0 | 60 | 890 | 23 | 10 | 783 | 67 | |||||
| HA-2.60 | 380 | 0 | 74 | 4745 | 4 | 3 | 17 | 7 | |||||
| HA-2.61 | 300 | 0 | 73 | 2324 | 32 | 25 | 63 | 13 | |||||
| HA-2.62 | 564 | 0 | 85 | 3648 | 42 | 12 | 9 | 14 | |||||
| HA-2.65 | 379 | 0 | 60 | 2608 | 3 | 16 | 865 | 62 | |||||
| HA-2.67 | 252 | 5 | 60 | >5000 | 9 | 2 | 16 | 5 | |||||
| HA-2.68 | 562 | 0 | 57 | 2462 | 13 | 8 | 226 | 49 | |||||
| HA-2.72 | 508 | 0 | 69 | 2129 | 3 | 3 | 1234 | 3 | |||||
| HA-2.73 | 394 | 0 | 70 | 3535 | 6 | 5 | 50 | 9 | |||||
| HA-2.74 | 615 | 0 | 75 | 2661 | 4 | 5 | 123 | 2 | |||||
| HA-2.75 | 420 | 3 | 81 | 2344 | 15 | 6 | 72 | 6 | |||||
. | . | Lytic activity, 4 h, % lysis . | . | Cytokine production, pg/mL . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HA-2 clone no. . | HLA-A2/HA-2 MFI . | EBV donor . | EBV patient . | IFN-γ . | TNF-α . | IL-10 . | IL-5 . | IL-4 . | |||||
| HA-2.01 | 206 | 0 | 69 | >5000 | 6 | 585 | 93 | 5 | |||||
| HA-2.02 | 735 | 4 | 64 | >5000 | 0 | 22 | 38 | 5 | |||||
| HA-2.03 | 553 | 6 | 61 | 3619 | 10 | 11 | 653 | 7 | |||||
| HA-2.04 | 306 | 0 | 72 | 3944 | 6 | 11 | 104 | 12 | |||||
| HA-2.05 | 466 | 0 | 79 | >5000 | 10 | 98 | 268 | 34 | |||||
| HA-2.06 | 630 | 0 | 53 | 1375 | 6 | 8 | 248 | 6 | |||||
| HA-2.07 | 520 | 0 | 60 | 4100 | 4 | 205 | 42 | 0 | |||||
| HA-2.08 | 256 | 2 | 64 | >5000 | 25 | 20 | 2433 | 72 | |||||
| HA-2.09 | 638 | 4 | 56 | 4709 | 8 | 9 | 638 | 9 | |||||
| HA-2.10 | 545 | 0 | 66 | 3914 | 86 | 13 | 19 | 5 | |||||
| HA-2.11 | 514 | 4 | 61 | 3319 | 3 | 6 | 53 | 4 | |||||
| HA-2.12 | 601 | 4 | 67 | 1900 | 0 | 6 | 13 | 4 | |||||
| HA-2.13 | 640 | 9 | 64 | 1477 | 10 | 5 | 287 | 39 | |||||
| HA-2.15 | 666 | 0 | 61 | 1476 | 9 | 16 | 138 | 7 | |||||
| HA-2.17 | 208 | 4 | 76 | 3412 | 21 | 62 | 2151 | 78 | |||||
| HA-2.19 | 689 | 0 | 71 | >5000 | 15 | 6 | 99 | 44 | |||||
| HA-2.20 | 468 | 2 | 61 | 3293 | 11 | 13 | 1414 | 13 | |||||
| HA-2.21 | 496 | 1 | 59 | 2211 | 6 | 4 | 65 | 2 | |||||
| HA-2.22 | 533 | 0 | 69 | 803 | 7 | 0 | 131 | 3 | |||||
| HA-2.24 | 777 | 0 | 63 | 1188 | 4 | 2 | 13 | 4 | |||||
| HA-2.25 | 327 | 1 | 71 | 3319 | 30 | 197 | 702 | 31 | |||||
| HA-2.27 | 456 | 3 | 70 | 2211 | 25 | 3 | 3761 | 269 | |||||
| HA-2.28 | 646 | 0 | 71 | 2683 | 15 | 1 | 80 | 5 | |||||
| HA-2.30 | 671 | 0 | 14 | 207 | 0 | 2 | 3 | 2 | |||||
| HA-2.31 | 631 | 2 | 56 | 3372 | 81 | 30 | 1262 | 93 | |||||
| HA-2.32 | 362 | 0 | 54 | 3139 | 12 | 15 | 575 | 42 | |||||
| HA-2.33 | 468 | 0 | 48 | 1250 | 13 | 4 | 15 | 4 | |||||
| HA-2.34 | 341 | 7 | 61 | 4818 | 17 | 16 | 85 | 9 | |||||
| HA-2.35 | 24 | 1 | 5 | 263 | 5 | 14 | 5 | 4 | |||||
| HA-2.36 | 555 | 3 | 61 | 2184 | 13 | 0 | 20 | 5 | |||||
| HA-2.37 | 696 | 0 | 75 | 1399 | 18 | 0 | 97 | 30 | |||||
| HA-2.38 | 510 | 0 | 81 | 3372 | 15 | 0 | 206 | 7 | |||||
| HA-2.41 | 228 | 5 | 83 | 2248 | 10 | 17 | 203 | 11 | |||||
| HA-2.43 | 590 | 0 | 69 | 4532 | 7 | 28 | 83 | 4 | |||||
| HA-2.46 | 894 | 0 | 78 | 2968 | 12 | 5 | 2 | 6 | |||||
| HA-2.47 | 450 | 0 | 77 | 1982 | 20 | 102 | 139 | 5 | |||||
| HA-2.48 | 488 | 0 | 73 | 4463 | 5 | 6 | 3 | 4 | |||||
| HA-2.52 | 551 | 1 | 68 | >5000 | 33 | 16 | 167 | 33 | |||||
| HA-2.53 | 580 | 0 | 68 | >5000 | 81 | 11 | 10 | 4 | |||||
| HA-2.54 | 568 | 0 | 67 | 4709 | 49 | 6 | 3 | 0 | |||||
| HA-2.55 | 769 | 5 | 67 | >5000 | 28 | 15 | 22 | 9 | |||||
| HA-2.58 | 364 | 0 | 55 | 1211 | 7 | 7 | 68 | 6 | |||||
| HA-2.59 | 427 | 0 | 60 | 890 | 23 | 10 | 783 | 67 | |||||
| HA-2.60 | 380 | 0 | 74 | 4745 | 4 | 3 | 17 | 7 | |||||
| HA-2.61 | 300 | 0 | 73 | 2324 | 32 | 25 | 63 | 13 | |||||
| HA-2.62 | 564 | 0 | 85 | 3648 | 42 | 12 | 9 | 14 | |||||
| HA-2.65 | 379 | 0 | 60 | 2608 | 3 | 16 | 865 | 62 | |||||
| HA-2.67 | 252 | 5 | 60 | >5000 | 9 | 2 | 16 | 5 | |||||
| HA-2.68 | 562 | 0 | 57 | 2462 | 13 | 8 | 226 | 49 | |||||
| HA-2.72 | 508 | 0 | 69 | 2129 | 3 | 3 | 1234 | 3 | |||||
| HA-2.73 | 394 | 0 | 70 | 3535 | 6 | 5 | 50 | 9 | |||||
| HA-2.74 | 615 | 0 | 75 | 2661 | 4 | 5 | 123 | 2 | |||||
| HA-2.75 | 420 | 3 | 81 | 2344 | 15 | 6 | 72 | 6 | |||||
The HA-2/HLA-A2 tetramer staining is shown as MFI, the cytokine production upon specific stimulation with HLA-A2-positive HA-2-positive stimulator cells (EBV-patient) is shown. The background cytokine production by the different HA-2-specific T-cell clones upon stimulation with HLA-A2-positive HA-2-negative stimulator cells (EBV-donor) is not shown. The background production of IFN-γ < 200 pg/mL, TNF-α < 5 pg/mL, IL-10 < 20 pg/mL, IL-5 < 100 pg/mL, IL-4 < 20 pg/mL.
Analysis of the TCRAV and BV repertoire of the HA-2-specific T-cell clones
By RT-PCR and sequence analysis the TCR usage of 26 HA-2-specific T-cell clones was determined. We revealed that the T-cell clones could be divided in 12 different groups, each group expressing a particular TCR α and TCR β chain (Table 2). The TCR α usage of the different HA-2-specific T-cell clone groups was diverse. Seven different TCRAV chains were detected. Based on the National Center for Biotechnology Information (NCBI) genebank, the following TCRAV chains were detected: AV26S2, AV21S1, AV35S1, AV20S1, AV5S1, AV25S1, and ADV38S2. Two groups of HA-2-specific T-cell clones, group E and group I, expressed 2 TCRAV chains. The TCR β chain of the different T-cell clone groups was restricted by 3 different TCRBV chains: TCR BV7S8, BV18S1, and BV10S3. One T-cell clone, HA2.7, present in group I expressed 2 TCRBV chains, expressing TCRBV10S3 as well as TCRBV7S8. Since the TCRBV7S8 present in HA2.7 is almost identical with the TCRBV7S8 chains of several other HA-2-specific T-cell clones, except for the CDR3 region, we speculated that this TCRBV chain was exerting the HA-2 specificity. Although there was restricted TCRBV usage in the HA-2-specific T-cell clones, the CDR3β regions composed of the N-D-N region and Jβ region, were very diverse (Table 3). The N-D-N region was variable in amino acid composition and in size (4 to 16 amino acids long). Interestingly, the CDR3 regions of the various TCR α chains all contained a common J region, the Jα42 segment (Table 4). The N regions present in the TCR α chains of the 12 different T-cell groups were variable in size and amino acid composition, although the N-region variation in the individual AV families was restricted, as is shown in Table 5 for all the T-cell clones expressing AV21S1.
The TCRαβ usage of HA-2-specific T-cell clones
. | . | . | TCRAV . | . | . | . | . | . | . | TCR BV . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group . | n . | Clone no. . | AV5S1 AV15S1† . | AV20S1 AV30S1† . | AV21S1 AV23S1† . | AV25S1 AV32S1† . | AV26S2 AV4S1† . | AV35S1 AV25S1† . | ADV38S1 AV14S1† . | BV7S8 BV6S2† . | BV10S3 BV12S1† . | BV18S1 BV18S1† . | ||||||||
| A | 6 | 5, 8, 28, 43, 60, 73 | +* | — | — | — | — | — | — | — | — | + | ||||||||
| B | 4 | 1, 4, 17, 59 | — | — | — | — | +* | — | — | + | — | — | ||||||||
| C | 3 | 20, 21, 32 | — | — | +* | — | — | — | — | + | — | — | ||||||||
| D | 3 | 19, 30, 53 | — | +* | — | — | — | — | — | — | — | + | ||||||||
| E | 2 | 2, 10 | — | — | +* | — | — | — | + | + | — | — | ||||||||
| F | 2 | 13, 52 | — | — | +* | — | — | — | — | + | — | — | ||||||||
| G | 1 | 3 | — | — | +* | — | — | — | — | + | — | — | ||||||||
| H | 1 | 6 | — | — | +* | — | — | — | — | — | — | + | ||||||||
| I | 1 | 7 | — | — | — | + | — | +* | — | + | + | — | ||||||||
| J | 1 | 31 | — | +* | — | — | — | — | — | — | — | + | ||||||||
| K | 1 | 65 | — | — | +* | — | — | — | — | + | — | — | ||||||||
| L | 1 | 58 | — | — | +* | — | — | — | — | + | — | — | ||||||||
. | . | . | TCRAV . | . | . | . | . | . | . | TCR BV . | . | . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group . | n . | Clone no. . | AV5S1 AV15S1† . | AV20S1 AV30S1† . | AV21S1 AV23S1† . | AV25S1 AV32S1† . | AV26S2 AV4S1† . | AV35S1 AV25S1† . | ADV38S1 AV14S1† . | BV7S8 BV6S2† . | BV10S3 BV12S1† . | BV18S1 BV18S1† . | ||||||||
| A | 6 | 5, 8, 28, 43, 60, 73 | +* | — | — | — | — | — | — | — | — | + | ||||||||
| B | 4 | 1, 4, 17, 59 | — | — | — | — | +* | — | — | + | — | — | ||||||||
| C | 3 | 20, 21, 32 | — | — | +* | — | — | — | — | + | — | — | ||||||||
| D | 3 | 19, 30, 53 | — | +* | — | — | — | — | — | — | — | + | ||||||||
| E | 2 | 2, 10 | — | — | +* | — | — | — | + | + | — | — | ||||||||
| F | 2 | 13, 52 | — | — | +* | — | — | — | — | + | — | — | ||||||||
| G | 1 | 3 | — | — | +* | — | — | — | — | + | — | — | ||||||||
| H | 1 | 6 | — | — | +* | — | — | — | — | — | — | + | ||||||||
| I | 1 | 7 | — | — | — | + | — | +* | — | + | + | — | ||||||||
| J | 1 | 31 | — | +* | — | — | — | — | — | — | — | + | ||||||||
| K | 1 | 65 | — | — | +* | — | — | — | — | + | — | — | ||||||||
| L | 1 | 58 | — | — | +* | — | — | — | — | + | — | — | ||||||||
Nucleotide and amino acid sequence of the TCR β chain of HA-2-specific T-cell clones
Clone group . | V beta . | N-D-N region . | J beta . | C beta . |
|---|---|---|---|---|
| A | BV18S1 | 2S1 | 2 | |
| TGT GCC AGC | TCC CGG ACT AGC GGG GCT AAC | AAT GAG CAG TTC TTC GGG CCA GGG ACA CGG CTC ACC GTG CTA | GAG GAC | |
| C A S | S R T S G A N | N E Q F F G P G T R L T V L | E D | |
| B | BV7S8 | 2S7 | 2 | |
| TGT GCC AGC | AGC GCA GTA GCG GGG ACC | TCC TAC GAG CAG TAC TTC GGG CCG GGC ACC AGG CTC ACG GTC ACA | GAG GAC | |
| C A S | S A V A G T | S Y E Q Y F G P G T R L T V T | E D | |
| C | BV7S8 | 1S4 | 1 | |
| TGT GCC AGC | AGC TTA GGC CTT TGG GGG AC GAT AAT | GAA AAA CTG TTT TTT GGC AGT GGA ACC CAG CTC TCT GTC TTG | GAG GAC | |
| C A S | S L G L W G T D N | E K L F F G S G T Q L S V L | E D | |
| D | BV18S1 | 2S2 | 2 | |
| TGT GCC AGC | TCC CCG CCC AGC GGG GGT TCG | ACC GGG GAG CTG TTT TTT GGA GAA GGC TCT AGG CTG ACC GTA CTG | GAG GAC | |
| C A S | S P P S G G S | T G E L F F G E G S R L T V L | E D | |
| E | BV7S8 | 2S2 | 2 | |
| TGT GCC AGC | AGA AAA GCG GGA CAG GAA AAC | ACC GGG GAG CTG TTT TTT GGA GAA GGC TCT AGG CTG ACC GTA CTG | GAG GAC | |
| C A S | R K A G Q E N | T G E L F F G E G S R L T V L | E D | |
| F | BV7S8 | 2S3 | 2 | |
| TGT GCC AGC | TTA TTT CCC GGT CAG GAC AGG GGA CAC GGG ACT AGC GGC AGC ACA GAT | ACG CAG TAT TTT GGC CCA GGC ACC CGG CTG ACA GTG CTC | GAG GAC | |
| C A S | L F P G Q D R G H G T S G S T D | T Q Y F G P G T R L T V L | E D | |
| G | BV7S8 | 2S1 | 2 | |
| TGT GCC AGC | AGC TTA GTA CCG | AAT GAG CAG TTC TTC GGG CCA GGG ACA CGG CTC ACC GTG CTA | GAG GAC | |
| C A S | S L V P | N E Q F F G P G T R L T V L | E D | |
| H | BV18S1 | 2S1 | 2 | |
| TGT GCC AGC | TCA CCA CTG GGC TCC TAC | AAT GAG CAG TTC TTC GGG CCA GGG ACA CGG CTC ACC GTG CTA | GAG GAC | |
| C A S | S P L G S Y | N E Q F F G P G T R L T V L | E D | |
| I | BV7S8 | 2S3 | 2 | |
| TGT GCC AGC | AGC CCC CCT AGC GGG AGG TAT | ACG CAG TAT TTT GGC CCA GGC ACC CGG CTG ACA GTG CTC | GAG GAC | |
| C A S | S P P S G R Y | T Q Y F G P G T R L T V L | E D | |
| BV10S3 | 1S1 | 1 | ||
| TGT GCC ATC | AGT GAG TCG CGA ACA GGG ATC GCT | GAA GCT TTC TTT GGA CAA GGC ACC AGA CTC ACA GTT GTA | GAG GAC | |
| C A I | S E S R T G I A | E A F F G Q G T R L T V V | E D | |
| J | BV18S1 | 1S6 | 1 | |
| TGT GCC AGC | TCA CCG ACA GGG GGG TCG CGA | TCA CCC CTC CAC TTT GGG AAC GGG ACC AGG CTC ACT GTG ACA | GAG GAC | |
| C A S | S P T G G S R | S P L H F G N G T R L T V T | E D | |
| K | BV7S8 | 2S1 | 2 | |
| TGT GCC AGC | AGC TGG CCG GAC ACC TAC | AAT GAG CAG TTC TTC GGG CCA GGG ACA CGG CTC ACC GTG CTA | GAG GAC | |
| C A S | S W P D T Y | N E Q F F G P G T R L T V L | E D | |
| L | BV7S8 | 2S5 | 2 | |
| TGT GCC AGC | AGC TTT CAC AGC | CAA GAG ACC CAG TAC TTC GGG CCA GGC ACG CGG CTC CTG GTG CTC | GAG GAC | |
| C A S | S F H S | Q E T Q Y F G P G T R L L V L | E D | |
| 5H13 | BV18S1 | 2S7 | 2 | |
| TGT GCC AGC | TCA CCT CCT GGG TAC | TCC TAC GAG CAG TAC TTC GGG CCG GGC ACC AGG CTC ACG GTC ACA | GAG GAC | |
| C A S | S P P G Y | S Y E Q Y F G P G T R L T V T | E D |
Clone group . | V beta . | N-D-N region . | J beta . | C beta . |
|---|---|---|---|---|
| A | BV18S1 | 2S1 | 2 | |
| TGT GCC AGC | TCC CGG ACT AGC GGG GCT AAC | AAT GAG CAG TTC TTC GGG CCA GGG ACA CGG CTC ACC GTG CTA | GAG GAC | |
| C A S | S R T S G A N | N E Q F F G P G T R L T V L | E D | |
| B | BV7S8 | 2S7 | 2 | |
| TGT GCC AGC | AGC GCA GTA GCG GGG ACC | TCC TAC GAG CAG TAC TTC GGG CCG GGC ACC AGG CTC ACG GTC ACA | GAG GAC | |
| C A S | S A V A G T | S Y E Q Y F G P G T R L T V T | E D | |
| C | BV7S8 | 1S4 | 1 | |
| TGT GCC AGC | AGC TTA GGC CTT TGG GGG AC GAT AAT | GAA AAA CTG TTT TTT GGC AGT GGA ACC CAG CTC TCT GTC TTG | GAG GAC | |
| C A S | S L G L W G T D N | E K L F F G S G T Q L S V L | E D | |
| D | BV18S1 | 2S2 | 2 | |
| TGT GCC AGC | TCC CCG CCC AGC GGG GGT TCG | ACC GGG GAG CTG TTT TTT GGA GAA GGC TCT AGG CTG ACC GTA CTG | GAG GAC | |
| C A S | S P P S G G S | T G E L F F G E G S R L T V L | E D | |
| E | BV7S8 | 2S2 | 2 | |
| TGT GCC AGC | AGA AAA GCG GGA CAG GAA AAC | ACC GGG GAG CTG TTT TTT GGA GAA GGC TCT AGG CTG ACC GTA CTG | GAG GAC | |
| C A S | R K A G Q E N | T G E L F F G E G S R L T V L | E D | |
| F | BV7S8 | 2S3 | 2 | |
| TGT GCC AGC | TTA TTT CCC GGT CAG GAC AGG GGA CAC GGG ACT AGC GGC AGC ACA GAT | ACG CAG TAT TTT GGC CCA GGC ACC CGG CTG ACA GTG CTC | GAG GAC | |
| C A S | L F P G Q D R G H G T S G S T D | T Q Y F G P G T R L T V L | E D | |
| G | BV7S8 | 2S1 | 2 | |
| TGT GCC AGC | AGC TTA GTA CCG | AAT GAG CAG TTC TTC GGG CCA GGG ACA CGG CTC ACC GTG CTA | GAG GAC | |
| C A S | S L V P | N E Q F F G P G T R L T V L | E D | |
| H | BV18S1 | 2S1 | 2 | |
| TGT GCC AGC | TCA CCA CTG GGC TCC TAC | AAT GAG CAG TTC TTC GGG CCA GGG ACA CGG CTC ACC GTG CTA | GAG GAC | |
| C A S | S P L G S Y | N E Q F F G P G T R L T V L | E D | |
| I | BV7S8 | 2S3 | 2 | |
| TGT GCC AGC | AGC CCC CCT AGC GGG AGG TAT | ACG CAG TAT TTT GGC CCA GGC ACC CGG CTG ACA GTG CTC | GAG GAC | |
| C A S | S P P S G R Y | T Q Y F G P G T R L T V L | E D | |
| BV10S3 | 1S1 | 1 | ||
| TGT GCC ATC | AGT GAG TCG CGA ACA GGG ATC GCT | GAA GCT TTC TTT GGA CAA GGC ACC AGA CTC ACA GTT GTA | GAG GAC | |
| C A I | S E S R T G I A | E A F F G Q G T R L T V V | E D | |
| J | BV18S1 | 1S6 | 1 | |
| TGT GCC AGC | TCA CCG ACA GGG GGG TCG CGA | TCA CCC CTC CAC TTT GGG AAC GGG ACC AGG CTC ACT GTG ACA | GAG GAC | |
| C A S | S P T G G S R | S P L H F G N G T R L T V T | E D | |
| K | BV7S8 | 2S1 | 2 | |
| TGT GCC AGC | AGC TGG CCG GAC ACC TAC | AAT GAG CAG TTC TTC GGG CCA GGG ACA CGG CTC ACC GTG CTA | GAG GAC | |
| C A S | S W P D T Y | N E Q F F G P G T R L T V L | E D | |
| L | BV7S8 | 2S5 | 2 | |
| TGT GCC AGC | AGC TTT CAC AGC | CAA GAG ACC CAG TAC TTC GGG CCA GGC ACG CGG CTC CTG GTG CTC | GAG GAC | |
| C A S | S F H S | Q E T Q Y F G P G T R L L V L | E D | |
| 5H13 | BV18S1 | 2S7 | 2 | |
| TGT GCC AGC | TCA CCT CCT GGG TAC | TCC TAC GAG CAG TAC TTC GGG CCG GGC ACC AGG CTC ACG GTC ACA | GAG GAC | |
| C A S | S P P G Y | S Y E Q Y F G P G T R L T V T | E D |
Only the last 3 residues of the V region are shown, followed by the N-D-N region, the J region, and the first 2 residues of the C region. T-cell clone 5H13, also specific for the mHag HA-2, was derived from an unrelated donor.
Nucleotide and amino acid sequence of the TCR α chain of HA-2-specific T-cell clones
Clone group . | v alpha . | N region . | J alpha . | C alpha . |
|---|---|---|---|---|
| A | AV5S1 | 42 | ||
| TGT GCA GAG | ACT GCG GGT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A E | T A G | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| B | AV26S1 | 42 | ||
| ACT CGC ATC | CTG AGA CCT GGG ATG AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| Y C I | L R P G M N | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| C | AV21S1 | 42 | ||
| TGT GCT GTG | AGG CCG GGG AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R P G N | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| D | AV20S1 | 42 | ||
| TGT GCT GTG | CAG GCA GGC | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | Q A G | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| E | ADV38S2 | 30 | ||
| CTA GTG CTT | ATA GGG GAC | AGA GAT GAC AAG ATC ATC TTT GGA AAA GGG ACA CGA CTT CAT ATT CTC CCC | AAT ATC | |
| L V L | I G D | R D D K I I F G K G T R L H I L P | N I | |
| AV21S1 | 42 | |||
| TGT GCT GTG | AGG CCT AAG AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R P K N | Y G G S Q G N L T F G K G T K L S V K P | N I | |
| F | AV21S1 | 42 | ||
| TGT GCT GTG | AGG TCA GGG | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R S G | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| G | AV21S1 | 42 | ||
| TGT GCT GTG | AGG CCA GGA AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R P G N | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| H | AV21S1 | 42 | ||
| TGT GCT GTG | AGA TAT AAG AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R Y K N | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| I | AV35S1 | 42 | ||
| TTC TGT GCT | GGG CCC ATG CCG ACT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| F C A | G P M P T | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| AV25S1 | 34 | |||
| CTG TGC AGG | ATG AGC TAT GGC | ACC GAC AAG CTC ATC TTT GGG ACT GGG ACC AGA TTA CAA GTC TTT | AAT ATC | |
| N L L | L C R G | T D K L I F G T G T R L Q V F | N I | |
| J | AV20S1 | 42 | ||
| TGT GCT GTG | CAG GCC GGT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | Q A G | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| K | AV21S1 | 42 | ||
| TGT GCT GTG | AGG CCG ATG AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R P M N | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| L | AV21S1 | 42 | ||
| TGT GCT GTG | AGG CGG GGC | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R R G | Y G G S Q G N L I F G R G T K L S V K P | N I | |
| 5H13 | AV35S1 | 42 | ||
| TTC TGT AGG | GGC CCC CCT CGT AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| F C A | G P P R N | Y G G S Q G N L I F G K G T K L S V K P | N I |
Clone group . | v alpha . | N region . | J alpha . | C alpha . |
|---|---|---|---|---|
| A | AV5S1 | 42 | ||
| TGT GCA GAG | ACT GCG GGT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A E | T A G | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| B | AV26S1 | 42 | ||
| ACT CGC ATC | CTG AGA CCT GGG ATG AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| Y C I | L R P G M N | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| C | AV21S1 | 42 | ||
| TGT GCT GTG | AGG CCG GGG AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R P G N | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| D | AV20S1 | 42 | ||
| TGT GCT GTG | CAG GCA GGC | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | Q A G | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| E | ADV38S2 | 30 | ||
| CTA GTG CTT | ATA GGG GAC | AGA GAT GAC AAG ATC ATC TTT GGA AAA GGG ACA CGA CTT CAT ATT CTC CCC | AAT ATC | |
| L V L | I G D | R D D K I I F G K G T R L H I L P | N I | |
| AV21S1 | 42 | |||
| TGT GCT GTG | AGG CCT AAG AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R P K N | Y G G S Q G N L T F G K G T K L S V K P | N I | |
| F | AV21S1 | 42 | ||
| TGT GCT GTG | AGG TCA GGG | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R S G | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| G | AV21S1 | 42 | ||
| TGT GCT GTG | AGG CCA GGA AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R P G N | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| H | AV21S1 | 42 | ||
| TGT GCT GTG | AGA TAT AAG AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R Y K N | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| I | AV35S1 | 42 | ||
| TTC TGT GCT | GGG CCC ATG CCG ACT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| F C A | G P M P T | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| AV25S1 | 34 | |||
| CTG TGC AGG | ATG AGC TAT GGC | ACC GAC AAG CTC ATC TTT GGG ACT GGG ACC AGA TTA CAA GTC TTT | AAT ATC | |
| N L L | L C R G | T D K L I F G T G T R L Q V F | N I | |
| J | AV20S1 | 42 | ||
| TGT GCT GTG | CAG GCC GGT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | Q A G | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| K | AV21S1 | 42 | ||
| TGT GCT GTG | AGG CCG ATG AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R P M N | Y G G S Q G N L I F G K G T K L S V K P | N I | |
| L | AV21S1 | 42 | ||
| TGT GCT GTG | AGG CGG GGC | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| C A V | R R G | Y G G S Q G N L I F G R G T K L S V K P | N I | |
| 5H13 | AV35S1 | 42 | ||
| TTC TGT AGG | GGC CCC CCT CGT AAT | TAT GGA GGA AGC CAA GGA AAT CTC CAT CTT TGG AAA AGG CAC TAA ACT CTC TGT TAA ACC | AAT ATC | |
| F C A | G P P R N | Y G G S Q G N L I F G K G T K L S V K P | N I |
Only the last 3 residues of the V region are shown followed by the N region, the J region, and the first two residues of the C region. T-cell clone 5H13, also specific for the mHag HA-2, was derived from an unrelated donor.
Amino acid sequence of the AV21S1 expressing HA-2 clones
Clone group . | V alpha . | N region . | J region . |
|---|---|---|---|
| C | AV21S1 | J42 | |
| C A V | R P G N | Y G G | |
| E | AV21S1 | J42 | |
| C A V | R P K N | Y G G | |
| F | AV21S1 | J42 | |
| C A V | R S G | Y G G | |
| G | AV21S1 | J42 | |
| C A V | R P G N | Y G G | |
| H | AV21S1 | J42 | |
| C A V | R Y K N | Y G G | |
| K | AV21S1 | J42 | |
| C A V | R P M N | Y G G | |
| L | AV21S1 | J42 | |
| C A V | R R G | Y G G |
Clone group . | V alpha . | N region . | J region . |
|---|---|---|---|
| C | AV21S1 | J42 | |
| C A V | R P G N | Y G G | |
| E | AV21S1 | J42 | |
| C A V | R P K N | Y G G | |
| F | AV21S1 | J42 | |
| C A V | R S G | Y G G | |
| G | AV21S1 | J42 | |
| C A V | R P G N | Y G G | |
| H | AV21S1 | J42 | |
| C A V | R Y K N | Y G G | |
| K | AV21S1 | J42 | |
| C A V | R P M N | Y G G | |
| L | AV21S1 | J42 | |
| C A V | R R G | Y G G |
Only the last 3 residues of the V region are shown, followed by the N region and the first 3 residues of the J region.
To demonstrate that the Jα42 region present in all HA-2-specific TCR complexes was selected on the basis of specific recognition of the HA-2 antigenic peptide in the context of HLA-A2, the TCR α and β usage of 2 HA-1-specific T-cell clones derived from the same CML patient was determined. The 2 HA-1-specific T-cell clones expressed either AV13S1 or AV25S1 in combination with BV7S9 and importantly expressed Jα28 or Jα9, respectively. Furthermore, the TCR usage of other HA-2-specific T cells derived from an unrelated individual was determined. The TCR α and β usage of the HA-2-specific T-cell clone 5H13 derived from a patient during GVHD35 demonstrated to be TCR AV35S1 and BV18S1 (Tables 3-4). Interestingly, also in this HA-2-specific T-cell clone the Jα42 segment was present in the TCR α chain. The TCR α sequence was almost identical to the TCR α sequence of T-cell clone HA2.7 representing group I. Both T-cell clones used the AV35S1 and Jα42 region; only the N region, although similar in length, differed in amino acid composition. The TCR β sequence of clone 5H13 contained the BV18S1 segment present in 33% of the TCR complexes of the HA-2-specific T cells. These results indicated that the TCR usage of the HA-2-specific T-cell clones is diverse as well as restricted.
Redirected mHag-specific reactivity of peripheral blood-derived T cells
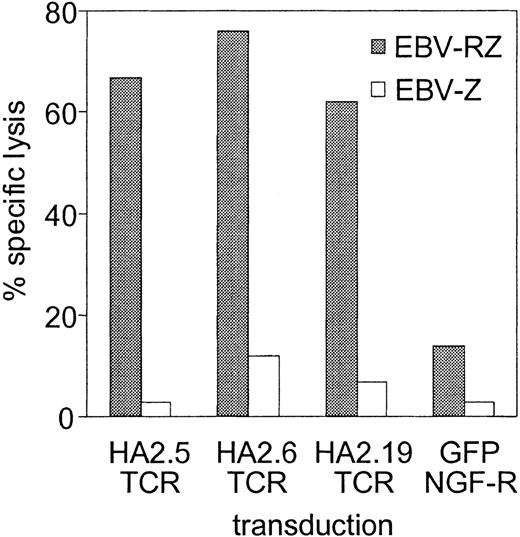
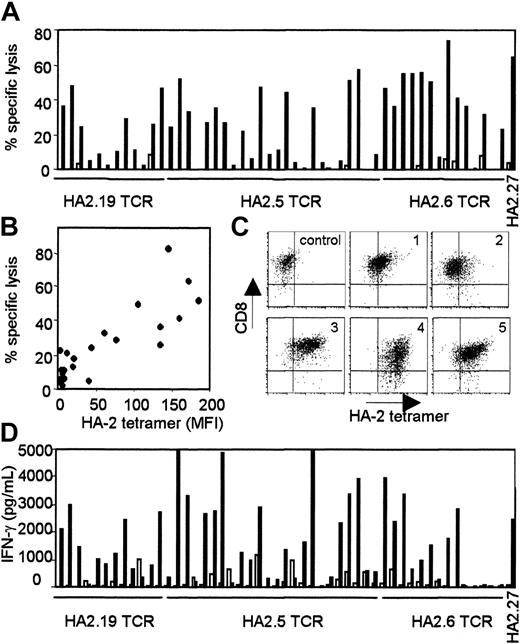
The TCRs of several HA-2-specific T-cell clones belonging to groups A, C, D, and H were cloned into pLZRS retroviral vectors. PBMCs from an HLA-A2-positive HA-2-negative individual were transduced with the different TCRs (transduction efficiencies varied between 25%-45%), sorted on the basis of marker gene expression and expanded as bulk populations or as T-cell clones. Figure 1 demonstrated the functional activity of the TCR-transduced bulk populations against 2 HLA-identical EBV-LCLs with disparity in HA-2 expression. All HA-2 TCR-transduced bulk populations exerted high cytolytic activity against HA-2-expressing EBV-LCLs and not against the HA-2-negative EBV-LCLs. The control-transduced bulk population was not reactive against both target cell populations. From the HA-2 TCR transductions 50 CD8+ T-cell clones were expanding, 24 clones were transduced with the HA2.5-TCR (group A),14 with the HA2.6-TCR (group H) and 12 with the HA2.19-TCR (group D). Figure 2A showed that 50% of HA2.5 and HA2.19 TCR-transduced T-cell clones lysed the HA-2-expressing target cells efficiently (more than 20% specific lysis), and 78% of the HA2.6 TCR-transduced T-cell clones lysed the HA-2-expressing EBV-LCL efficiently. Most HA-2 TCR-transduced cytolytic T-cell clones were positive for HA-2/HLA-A2 tetramer staining (Figure 2B), and staining was homogeneous on the individual T-cell clones (Figure 2C). A clear correlation between tetramer staining and cytolytic activity of the HA-2 TCR-transduced T-cell clones was observed. Although the cytolytic activity of one of the original HA-2-specific T-cell clones, HA2.27, was comparable with part of the TCR-transduced T-cell clones, the tetramer staining of HA2.27 was markedly higher (mean fluorescence intensity [MFI] of 474). Furthermore, approximately 70% of the HA-2 TCR-transferred T-cell clones produced IFN-γ (Figure 2D), and 25% to 30% of the T-cell clones produced TNF-α after mHag-specific stimulation (data not shown). Most of the HA-2 TCR-transduced T-cell clones that exerted cytolytic activity and stained with HA-2/HLA-A2 tetramers, also produced IFN-γ upon mHag stimulation.
HA-2-specific cytotoxic response of the various HA-2 TCR-modified T-cell lines. HA-2 TCR-modified T cells were tested in a cytotoxicity assay against HLA-A2-positive HA-2-positive target cells (EBV-RZ; ▦) and HLA-A2-positive HA-2-negative target cells (EBV-Z; □) at an effector-target (E/T) ratio of 10:1. As negative control, T cells transduced with retroviral vectors encoding for marker genes only were used. The cytotoxicity assay was performed for 4 hours.
HA-2-specific cytotoxic response of the various HA-2 TCR-modified T-cell lines. HA-2 TCR-modified T cells were tested in a cytotoxicity assay against HLA-A2-positive HA-2-positive target cells (EBV-RZ; ▦) and HLA-A2-positive HA-2-negative target cells (EBV-Z; □) at an effector-target (E/T) ratio of 10:1. As negative control, T cells transduced with retroviral vectors encoding for marker genes only were used. The cytotoxicity assay was performed for 4 hours.
Functional analyses of the various HA-2 TCR-modified T-cell clones. (A) HA-2 TCR-modified T-cell clones were tested in a cytotoxicity assay against HLA-A2-positive HA-2-positive target cells (EBV-RZ; ▪) and HLA-A2-positive HA-2-negative target cells (EBV-Z; □) at an E/T ratio of 10:1. The cytotoxicity assay was performed for 4 hours. (B) HA-2/HLA-A2 tetramer staining of the HA-2 TCR-modified T-cell clones, shown as MFI, correlated with the lytic activity against HLA-A2-positive HA-2-positive target cells (EBV-RZ). (C) HA-2/HLA-A2 tetramer staining and CD8 expression of 5 representative HA-2 TCR-transferred T-cell clones1-5 and one GFP/NGFR transduced T-cell clone is shown. (D) IFN-γ production of the HA-2 TCR-modified T-cell clones upon stimulation with HLA-A2-positive HA-2-positive stimulator cells (EBV-RZ; ▪) and HLA-A2-positive HA-2-negative stimulator cells (EBV-Z; □) was measured after 24 hours of stimulation. The original HA-2-specific T-cell clone HA2.27 was used as positive control in these experiments. The HA-2/HLA-A2 tetramer staining of HA2.27 was also determined (MFI = 474).
Functional analyses of the various HA-2 TCR-modified T-cell clones. (A) HA-2 TCR-modified T-cell clones were tested in a cytotoxicity assay against HLA-A2-positive HA-2-positive target cells (EBV-RZ; ▪) and HLA-A2-positive HA-2-negative target cells (EBV-Z; □) at an E/T ratio of 10:1. The cytotoxicity assay was performed for 4 hours. (B) HA-2/HLA-A2 tetramer staining of the HA-2 TCR-modified T-cell clones, shown as MFI, correlated with the lytic activity against HLA-A2-positive HA-2-positive target cells (EBV-RZ). (C) HA-2/HLA-A2 tetramer staining and CD8 expression of 5 representative HA-2 TCR-transferred T-cell clones1-5 and one GFP/NGFR transduced T-cell clone is shown. (D) IFN-γ production of the HA-2 TCR-modified T-cell clones upon stimulation with HLA-A2-positive HA-2-positive stimulator cells (EBV-RZ; ▪) and HLA-A2-positive HA-2-negative stimulator cells (EBV-Z; □) was measured after 24 hours of stimulation. The original HA-2-specific T-cell clone HA2.27 was used as positive control in these experiments. The HA-2/HLA-A2 tetramer staining of HA2.27 was also determined (MFI = 474).
HA-2-specific recognition by chimeric HA-2-TCR complexes
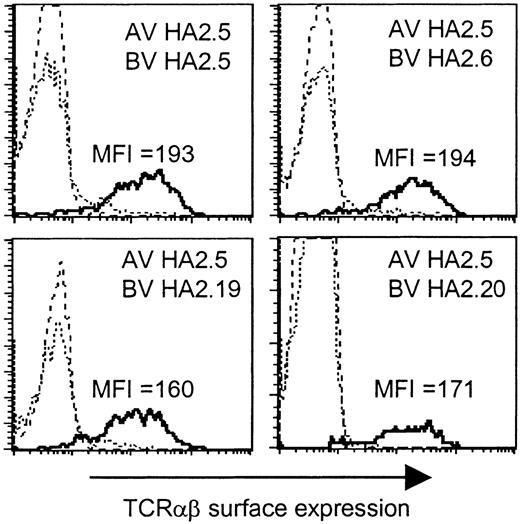
Based on the presence of the Jα42 gene segment in all TCR α chains, we analyzed whether this region would determine the HA-2-specific recognition. To determine whether all TCR α and β chains were able to pair with each other and therefore would be able to form stable TCRαβ complexes at the cell surface, we generated a Jurkat T-cell clone, designated clone 76, which is deficient for both TCR α and β chains, and transduced clone 76 with the different chimeric TCR combinations consisting of the TCR α chain derived from one HA-2-specific T-cell clone and the TCR β chain from another HA-2-specific T-cell clone. In Figure 3 the TCRαβ surface expression is shown of clone 76 transduced with the TCR α chain of clone HA2.5 in combination with the TCR β chain of clone HA2.5, HA2.6, HA2.19, or HA2.20, as representative examples of all transferred TCR combinations. All chimeric TCRs were able to form stable cell surface TCRαβ complexes. No difference in the MFI of TCRαβ expression was observed between clone 76 transduced with the original TCR combinations (MFI 169-199) and the chimeric TCR combinations (MFI 160-225). These results demonstrated that all chimeric TCRs composed of the different TCR α and β chains were able to pair with each other as efficiently as the original TCR combinations.
TCRαβ surface expression of the TCRα/β-deficient Jurkat clone 76 transduced with chimeric TCRs. Jurkat clone 76 was transduced with the TCR α chain of clone HA2.5 in combination with the TCR β chain of clone HA2.5, HA2.6, HA2.19, or HA2.20. The different subpopulations (dashed line indicates double marker gene negative; stippled line, single marker gene positive; or solid line, double marker gene positive) were gated and the TCRαβ expression was shown as histogram plots.
TCRαβ surface expression of the TCRα/β-deficient Jurkat clone 76 transduced with chimeric TCRs. Jurkat clone 76 was transduced with the TCR α chain of clone HA2.5 in combination with the TCR β chain of clone HA2.5, HA2.6, HA2.19, or HA2.20. The different subpopulations (dashed line indicates double marker gene negative; stippled line, single marker gene positive; or solid line, double marker gene positive) were gated and the TCRαβ expression was shown as histogram plots.
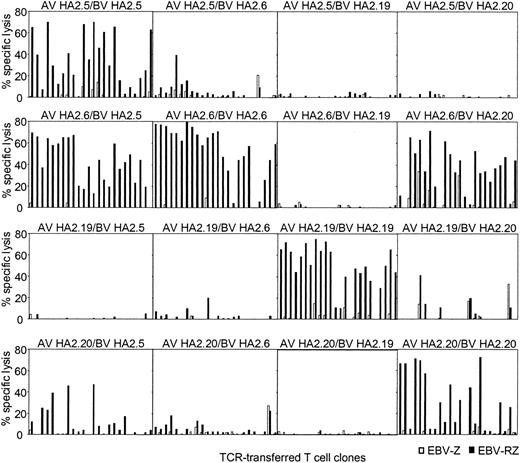
Peripheral T cells were transduced with all possible chimeric TCR combinations and the functionality of the transferred chimeric TCR was compared with the original TCR combinations. From all these HA-2 TCR transductions 24 different T-cell clones were generated that were selected on the basis of marker gene expression, expanded, and tested for HA-2-specific cytolytic activity (Figure 4). From the original HA-2-TCR combinations 50% to 87% of the T-cell clones exerted more than 20% specific cytolytic activity against HA-2-expressing target cells. The TCR-transduced T-cell clones from 2 chimeric TCR combinations were functionally as active as the original TCR combinations, since 75% to 87% of the T-cell clones were able to specifically lyse HA-2-expressing target cells. These 2 chimeric TCR combinations were composed of the TCRAV21S1 from HA2.6 (group H) in combination with the TCRBV18S1 from HA2.5 (group A) or TCRBV7S8 from HA2.20 (group C). The chimeric TCR combination of AV21S1 from HA2.20 and BV18S1 from HA2.5 also lead to a functional HA-2-specific TCR complex, although less efficient, since 5 of 24 T-cell clones were able to exert cytolytic activity against the HA-2-expressing target cells. In all other chimeric TCR combinations no or only a few clones were able to exert HA-2-specific recognition. These experiments using the chimeric TCR combinations demonstrate that the HA-2 specificity is not only determined by the Jα42 region but in addition by the N region of the TCR α chain and the CDR3 region of the TCR β chain, since differences in the N region of the TCR α chain or the N-D-N region of the TCR β chain were enough to abrogate the HA-2 specificity. The AV21S1 of group C (HA2.20) and H (HA2.6), for example, are almost identical; only 2 amino acids were different in the N region, however, pairing of the AV21S1 of group H (HA2.6) with the BV7S8 of group C (HA2.20) resulted in a HA-2-specific TCR complex, whereas the TCR complex composed of the AV21S1 of group C (HA2.20) and the BV18S1 of group H (HA2.6) was not HA-2 specific. In addition, the BV18S1 of group A (HA2.5) and group H (HA2.6) have the same V, J, and C usage but have a different N-D-N region. Pairing of the AV21S1 of group H (HA2.6) with the BV18S1 of group A (HA2.5) resulted in a HA-2-specific TCR complex; however, the TCR complex composed of the AV5S1 of group A (HA2.5) and the BV18S1 of group H (HA2.6) exerted low HA-2 specificity.
HA-2-specific cytolytic reactivity of chimeric TCRs. T-cell clones transduced with chimeric TCR complexes, consisting of the TCR α chain derived from one HA-2-specific T-cell clone and the TCR β chain from another HA-2-specific T-cell clone, were tested in a cytotoxicity assay against HLA-A2-positive HA-2-positive target cells (EBV-RZ; ▪) and HLA-A2-positive HA-2-negative target cells (EBV-Z; □) at an E/T ratio of 10:1. For every chimeric TCR combination, 24 TCR-transferred T-cell clones were tested. The composition of the chimeric TCR combinations are indicated above each panel. The cytotoxicity assay was performed for 4 hours.
HA-2-specific cytolytic reactivity of chimeric TCRs. T-cell clones transduced with chimeric TCR complexes, consisting of the TCR α chain derived from one HA-2-specific T-cell clone and the TCR β chain from another HA-2-specific T-cell clone, were tested in a cytotoxicity assay against HLA-A2-positive HA-2-positive target cells (EBV-RZ; ▪) and HLA-A2-positive HA-2-negative target cells (EBV-Z; □) at an E/T ratio of 10:1. For every chimeric TCR combination, 24 TCR-transferred T-cell clones were tested. The composition of the chimeric TCR combinations are indicated above each panel. The cytotoxicity assay was performed for 4 hours.
Antileukemic reactivity of HA-2 TCR-modified T cells
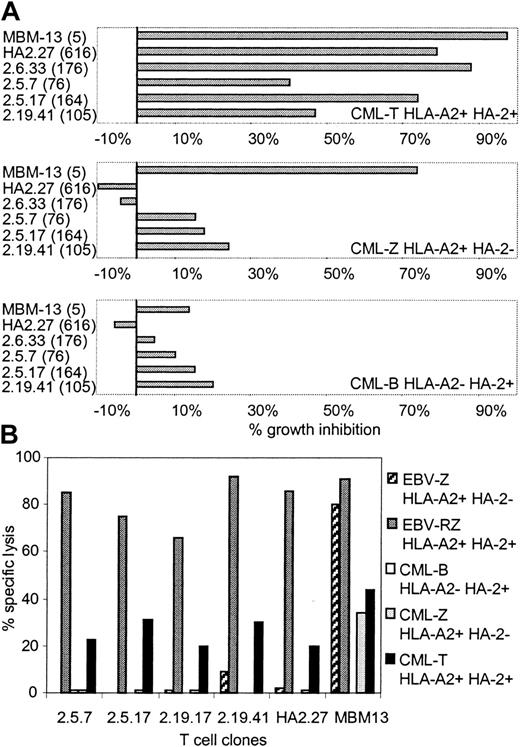
To investigate whether these HA-2 TCR-transduced T-cell clones generated from PBMCs of HLA-A2-positive HA-2-negative individuals exerted cytolytic activity against CML cells, several HA-2 TCR-modified T-cell clones were tested in the liquid hematopoietic PIA and in cytotoxicity assays. Figure 5A illustrates the specific growth inhibition of HLA-A2-positive CML progenitor cells expressing the HA-2 antigen by the HA-2 TCR-transferred T-cell clones. The inhibitory capacity of the HA-2 TCR-modified T-cell clones correlated with the level of cell surface HA-2-TCR complexes measured by HA-2/HLA-A2 tetramer staining. HA-2 TCR-modified T cells with MFI of HA-2/HLA-A2 tetramer staining above 160 were as efficient as the original HA-2-specific T-cell clone HA-2.27. HA-2 TCR-modified T cells with intermediate HA-2 TCR expression (MFI < 160) exerted intermediate growth inhibition of the HA-2-expressing CML cells. The TCR-modified T cells did not specifically inhibit the growth of HLA-A2-negative or HLA-A2-positive HA-2-negative CML progenitor cells. These results were confirmed by the cytotoxicity assays, demonstrating that the HA-2 TCR-modified T cells exerted specific cytolytic activity against the HLA-A2-positive CML cells expressing the HA-2 antigen (Figure 5B).
Antileukemic reactivity of HA-2 TCR-modified T-cell clones. HA-2 TCR-modified T-cell clones were tested in a (A) liquid hematopoietic progenitor cell inhibition assay (PIA) and in a (B) cytotoxicity assay against HLA-A2-positive HA-2-positive CML cells (CML-T), HLA-A2-positive HA-2-negative CML cells (CML-Z), and HLA-A2-negative HA-2-positive CML cells (CML-B). As positive controls, the original HA-2-specific T-cell clone HA2.27 and the allo-HLA-A2-specific T-cell clone MBM13 were used. Between parentheses the MFI of HA-2/HLA-A2 tetramer staining is indicated. In the cytotoxicity assay HLA-A2-positive HA-2-positive EBV-LCLs (EBV-RZ) and HLA-A2-positive HA-2-negative EBV-LCLs (EBV-Z) were included. The effector T cells were tested in the PIA at an E/T ratio of 1:1 and in the cytotoxicity assay at an E/T ratio of 7:1. The cytotoxicity assay was performed for 18 hours.
Antileukemic reactivity of HA-2 TCR-modified T-cell clones. HA-2 TCR-modified T-cell clones were tested in a (A) liquid hematopoietic progenitor cell inhibition assay (PIA) and in a (B) cytotoxicity assay against HLA-A2-positive HA-2-positive CML cells (CML-T), HLA-A2-positive HA-2-negative CML cells (CML-Z), and HLA-A2-negative HA-2-positive CML cells (CML-B). As positive controls, the original HA-2-specific T-cell clone HA2.27 and the allo-HLA-A2-specific T-cell clone MBM13 were used. Between parentheses the MFI of HA-2/HLA-A2 tetramer staining is indicated. In the cytotoxicity assay HLA-A2-positive HA-2-positive EBV-LCLs (EBV-RZ) and HLA-A2-positive HA-2-negative EBV-LCLs (EBV-Z) were included. The effector T cells were tested in the PIA at an E/T ratio of 1:1 and in the cytotoxicity assay at an E/T ratio of 7:1. The cytotoxicity assay was performed for 18 hours.
To verify whether TCR-redirected T cells with mHag HA-2 specificity but without HLA-A2 alloreactivity could be generated from HLA-A2-negative individuals, PBMCs from an HLA-A2-negative donor were transduced with the different HA-2-TCR complexes, sorted on bases of HA-2/HLA-A2 tetramer staining, and the sorted cell lines were tested against different target cells. As demonstrated in Figure 6, transfer of the different HA-2 TCRs into HLA-A2-negative PBMCs resulted in cytolytic activity against HA-2 peptide-loaded HLA-A2-positive EBV-LCLs as well as HLA-A2-positive EBV-LCLs endogenously expressing HA-2. In addition, HLA-A2-positive CML cells expressing HA-2 were efficiently lysed by the HA-2 TCR-modified T cells. Importantly, no anti-HLA-A2 alloreactivity was observed, since HLA-A2-positive target cells negative for HA-2, including EBV-LCLs, CML, cells and the HFF fibroblast cell line, were not lysed by the HA-2 TCR-modified T cells, whereas the allo-HLA-A2-specific T-cell clone MBM13 lysed these HLA-A2-expressing target cells efficiently. These results illustrate that redirection of antileukemic reactivity by transfer of mHag-specific TCRs into peripheral T cells without the occurrence of allo-HLA reactivity is feasible.
HA-2-specific cytolytic activity of HA-2 TCR-modified T cells without HLA-A2 alloreactivity. PBMCs transduced with the different HA-2-TCR complexes were tested in a 51Cr-release assay against HLA-A2-positive HA-2-positive EBV-LCLs (EBV-RZ), HLA-A2-positive HA-2-negative EBV-LCLs (EBV-Z), EBV-Z loaded with the HA-2 peptide (EBV-Z + HA-2pep), HLA-A2-positive HA-2-positive CML cells (CML-T), HLA-A2-positive HA-2-negative CML cells (CML-Z), HLA-A2-negative HA-2-positive CML cells (CML-B), and HLA-A2-positive HFF fibroblast cells. As positive controls the original HA-2-specific T-cell clone HA2.27 and the allo-HLA-A2-specific T-cell clone MBM13 were used. The effector T cells were tested at an E/T ratio of 5:1 (HA-2.5 TCR-modified T cells, GFP/NGF-R-transduced T cells, HA-2.27 and MBM13) or 2:1 (HA-2.6 TCR-modified T cells) or 15:1 (HA-2.19 TCR-modified T cells). The cytotoxicity assay was performed for 18 hours.
HA-2-specific cytolytic activity of HA-2 TCR-modified T cells without HLA-A2 alloreactivity. PBMCs transduced with the different HA-2-TCR complexes were tested in a 51Cr-release assay against HLA-A2-positive HA-2-positive EBV-LCLs (EBV-RZ), HLA-A2-positive HA-2-negative EBV-LCLs (EBV-Z), EBV-Z loaded with the HA-2 peptide (EBV-Z + HA-2pep), HLA-A2-positive HA-2-positive CML cells (CML-T), HLA-A2-positive HA-2-negative CML cells (CML-Z), HLA-A2-negative HA-2-positive CML cells (CML-B), and HLA-A2-positive HFF fibroblast cells. As positive controls the original HA-2-specific T-cell clone HA2.27 and the allo-HLA-A2-specific T-cell clone MBM13 were used. The effector T cells were tested at an E/T ratio of 5:1 (HA-2.5 TCR-modified T cells, GFP/NGF-R-transduced T cells, HA-2.27 and MBM13) or 2:1 (HA-2.6 TCR-modified T cells) or 15:1 (HA-2.19 TCR-modified T cells). The cytotoxicity assay was performed for 18 hours.
Discussion
In this study, we show that HA-2 TCR-transduced T cells exert HA-2-specific HLA-A2-restricted cytolytic activity and specifically inhibit the proliferation of HA-2-positive leukemic cells as efficient as the original HA-2-specific T-cell clones. This illustrates that HA-2 TCR transfer can redirect T cells to antileukemic reactivity. The antileukemic reactivity could be observed in HA-2 TCR-modified T cells derived from HLA-A2-positive as well as HLA-A2-negative individuals, without any signs of anti-HLA-A2 alloreactivity. The HA-2 TCR-modified T cells exerted HA-2-specific cytotoxicity, could be visualized using HA-2/HLA-A2 tetrameric complexes, and produced IFN-γ and TNF-α upon mHag-specific stimulation.
The HA-2 TCRs were derived from HA-2-specific T-cell clones that were isolated from a CML patient with an ongoing antileukemic immune response. Twelve different groups of HA-2-specific T-cell clones could be identified in this individual, having differential usage of TCR α and β chains. The BV usage of the HA-2-specific T-cell clones was restricted to 2 different V gene segments, BV7S8 and BV18S1; however, the N-D-N regions were diverse as well as the J regions. Although the AV usage of the HA-2-specific T-cell clones was diverse, the J gene segment usage was similar for all the HA-2-specific TCR complexes. Since there are 60 different Jα gene segments, it is striking that all HA-2-specific T-cell clones expressed the same Jα42 gene segment. Based on the absence of the Jα42 gene segment in mHag HA-1-specific T-cell clones selected from the same individual and the presence of the Jα42 gene segment in the TCR complex of an HA-2-specific T-cell clone derived from an unrelated CML patient, our results imply that the Jα42 gene segment is important for HA-2-specific peptide recognition. Based on the restricted amino acid length and composition of the CDR3α region of the T-cell clones expressing the AV21S1, we speculate that this region also influences the HA-2-specific peptide recognition (Table 5). We propose that especially the C-terminal amino acids of the Jα42 segment in combination with the N region will be responsible for peptide-specific recognition, since the C-terminal part of the J sequence is located at the interface of the TCR major histocompatibility complex (MHC)/peptide ligand interaction.36 The fact that the highest heterogeneity in amino acid composition is located at the C-terminal part of the J segments is in agreement with this assumption. In addition, based on the HA-2-specific recognition of part of the chimeric TCR complexes consisting of the TCR α chain derived from one HA-2-specific T-cell clone and the TCR β chain from another HA-2-specific T-cell clone, we demonstrated that the TCR β chain also contributed to the HA-2-specific recognition.
The relative dependency on TCR α chain in antigen recognition has been described in several systems. HIV glycoprotein (gp) 160-specific RT-1 TCR α or β single transgenic mice demonstrated that a single TCR α chain in combination with various TCR β chains can generate functional antigen-specific T cells, indicating a predominant role of the TCR α chain in this peptide specificity.37 Thus far no data are available that demonstrate that different T-cell clones reactive against a single antigenic peptide all display a common Jα or Jβ region. Only the mHag HA-1-specific T-cell clones have been reported to contain interindividual conservation of the TCR β chain V region BV7S938,39 (nomenclature according to Arden et al40 is BV6S4). Also the HA-1-specific T-cell clones presented in this study expressed TCR β chains containing the BV7S9.
Refining our understanding of interactions between the TCR and its antigenic peptide will enable us to engineer antigen-specific and high-affinity TCRs for future gene transfer purposes. Thus far it was still unclear whether the TCR repertoire usage of antigen-specific T cells against single antigenic epitope was diverse or limited and whether there are common rules for specific antigen recognition. The first and probably the main reason for this uncertainty is the difficulty to isolate and expand antigen-specific T cells. With the recent developed tetramer technology in combination with single-cell sorting it is currently possible to isolate antigen-specific T cells directly from peripheral blood or tumor infiltrating sites.41 With the use of tetramer-positive single-cell sorting in combination with a short nonspecific expansion, it is now possible to visualize an antigen-specific TCR repertoire, which is likely to represent the TCR repertoire of the in vivo antigen-specific immune response. Second, most human repertoire studies thus far reported primarily analyzed the TCRBV chain repertoire. Third, since approximately 30% of human T cells express 2 TCR α chains,42 sequencing of only one of the TCR α chains without functional analysis can lead to loss of correlation between antigen specificity and expression of a certain TCR α region.
The HA-2-TCR complexes were derived from HA-2-specific T-cell clones generated following HLA-identical allo-SCT, implying that the T cells were reactive against the allogeneic mHag HA-2 expressed by the patient in the context of self-HLA-A2. Transfer of these TCR complexes into primary T cells from HLA-A2-negative individuals, and subsequently adoptive transfer into patients positive for HLA-A2 and the mHag HA-2, may result in a new therapeutic strategy. HA-2 TCR-transferred T cells from HLA-A2-negative donors will be able to recognize the hematopoietic cells of the patient, including the malignant cells, leading to the eradication of the hematologic malignancy. In contrast, donor-derived hematopoietic cells will not be attacked, since these cells are HLA-A2 negative, and nonhematopoietic cells from the patient will not be attacked, since these cells are HA-2 negative. Following allo-SCT of an HLA-A2-positive patient with a hematopoietic T-cell-depleted graft from an HLA-A2-negative donor transfer of HA-2 TCR into donor-derived T cells that are tolerant to patient tissues will enable us to generate mHag HA-2-specific T cells that recognize the hematopoietic tissues from the HLA-A2/HA-2-positive patient, leading to eradication of the hematologic tumor, without the recognition of HLA-A2 molecules presenting other HLA-A2 binding peptides, thereby minimizing GVHD.
In conclusion, in this study we demonstrated that transfer of HA-2-specific TCR genes into peripheral CD8+ T cells of HLA-A2-positive HA-2-negative individuals resulted in antileukemic reactivity. This indicates that HA-2 TCR transfer may be used as an alternative strategy to generate HA-2-specific T cells to treat hematologic malignancies of HLA-A2-positive HA-2-expressing patients. Based on the antileukemic reactivity of the HA-2 TCR-transferred T cells, without the occurrence of alloreactivity against other peptides presented in HLA-A2 molecules and the fact that a high percentage of all individuals express the HA-2 mHag, HA-2 TCR transfer can also be an attractive strategy of performing immunotherapy of relapsed hematologic malignancies after allogeneic-SCT in HLA-A2-positive patients that receive stem cells from HLA-A2-negative donors.
Prepublished online as Blood First Edition Paper, July 17, 2003; DOI 10.1182/blood-2003-05-1524.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Reinier van der Linden, Maarten van de Keur, Marian van de Meent, and Linda Cox for expert technical assistance.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal