Abstract
Posttransplantation lymphoproliferative disorders (PTLDs) represent a serious complication of solid organ transplantation. This study assessed the molecular histogenesis of 52 B-cell monoclonal PTLDs, including 12 polymorphic PTLDs (P-PTLDs), 36 diffuse large B-cell lymphomas (DLBCLs), and 4 Burkitt/Burkitt-like lymphomas (BL/BLLs). Somatic hypermutation (SHM) of immunoglobulin variable (IgV) genes documented that most monoclonal B-cell PTLDs (75% P-PTLDs, 91.3% DLBCLs, 100% BL/BLLs) derive from germinal center (GC)-experienced B cells. B-cell lymphoma 6 (BCL6) mutations occurred in 25% P-PTLDs, 60.6% DLBCLs, and 75.0% BL/BLLs. A first histogenetic category of PTLDs (31.2% DLBCLs) express the BCL6+/multiple myeloma oncogene-1 protein (MUM1-/+)/CD138- profile and mimic B cells experiencing the GC reaction, as also suggested by ongoing SHM in a fraction of these cases. A second subset of PTLDs (66.7% P-PTLDs and 31.2% DLBCLs) display the BCL6-/MUM1+/CD138- phenotype and mimic B cells that have concluded the GC reaction. A third histogenetic category of PTLDs (25.0% P-PTLDs and 31.2% DLBCLs) shows the BCL6-/MUM1+/CD138+ profile, consistent with preterminally differentiated post-GC B cells. Crippling mutations of IgV heavy chain (IgVH) and/or IgV light chain (IgVL) genes, leading to sterile rearrangements and normally preventing cell survival, occur in 4 DLBCLs and 1 BL/BLL that may have been rescued from apoptosis through expression of Epstein-Barr virus (EBV)-encoded latent membrane protein 1 (LMP1). Overall, the histogenetic diversity of monoclonal B-cell PTLDs may help define biologically homogeneous categories of the disease. (Blood. 2003;102: 3775-3785)
Introduction
Posttransplantation lymphoproliferative disorders (PTLDs) represent a major complication of solid organ transplantation and are related to the chronic administration of iatrogenic immunosuppression.1-4 Most PTLDs are of B-cell origin, frequently arise in extranodal sites, and display a marked clinical aggressiveness.1-4 Despite these common features, PTLDs are histologically and molecularly heterogeneous and may arise at different times after transplantation.1-10 Early onset PTLDs are mainly regarded as Epstein-Barr virus (EBV)-driven lymphoproliferations that are frequently, although not always, polyclonal or oligoclonal, whereas most late onset PTLDs are true monoclonal lymphoid malignancies that are not necessarily associated with EBV infection.1-10 The predominant pathologic categories of PTLDs include plasmacytic hyperplasia, polymorphic PTLDs (P-PTLDs), and monomorphic B-cell lymphoma, comprising diffuse large B-cell lymphoma (DLBCL) and Burkitt/Burkitt-like lymphoma (BL/BLL).3,4 Correlative studies of the morphologic and molecular features of PTLDs have contributed to the recognition of specific disease categories and have provided prognostic indicators for these disorders.5,6,11,12
Molecular histogenetic studies have contributed significantly to the understanding of the heterogeneity of lymphoid malignancies in both immunocompetent and immunocompromised hosts. During the last few years, the understanding of B-cell lymphoma histogenesis has received impulse by the growing number of histogenetic markers allowing the distinction of mature B cells into different compartments, namely virgin B cells, germinal center (GC) B cells, and post-GC B cells.13-15 Genotypic markers of B-cell histogenesis are mainly represented by somatic hypermutation (SHM) of immunoglobulin variable (IgV) genes, which takes place in the course of T-cell-dependent immune reactions in the GC microenvironment.13-15 Positivity for IgV mutations indicates that a given lymphoma derives from GC or post-GC B cells. In particular, the presence of ongoing IgV mutations, leading to intraclonal heterogeneity, suggests that the lymphoma clone reflects centroblasts experiencing the GC reaction, whereas the absence of intraclonal heterogeneity is consistent with derivation from late centrocytes or post-GC B cells that have terminated the GC reaction.13-15 Mutations of the BCL6 proto-oncogene, which are physiologically acquired by B cells at the time of GC transit, are also regarded as a complementary marker of histogenesis.16,17
Phenotypic markers of B-cell lymphoma histogenesis are exemplified by the B-cell lymphoma 6 (BCL6), multiple myeloma oncogene-1 protein (MUM1), and CD138 proteins and help refine the distinction between GC and post-GC B cells. In fact, expression of BCL6 clusters with the GC stage of differentiation, MUM1 positivity clusters with B cells exiting the GC and with post-GC B cells, and CD138 is a marker of preterminal B-cell differentiation.18-20 On these bases, B-cell lymphomas arising in other immunodeficiency contexts have been schematically distinguished into lymphomas devoid of IgV SHM and related to pre-GC B cells; lymphomas associated with IgV SHM and BCL6 expression, which closely reflect GC B cells; and lymphomas associated with IgV SHM and MUM1 and/or CD138 positivity, representing lymphomas of post-GC B cells.20-23
Although some types of EBV-positive PTLDs have been recently shown to associate with IgV SHM,24 a systematic investigation of molecular and phenotypic markers of histogenesis in both EBV-positive and EBV-negative PTLDs is currently lacking. The aim of this study was a comprehensive analysis of the molecular histogenesis of the pathologic spectrum of monoclonal B-cell PTLDs. By applying a wide panel of B-cell lymphoma histogenetic markers, both genetic and phenotypic, we report that virtually all monoclonal B-cell PTLDs originate from B cells that have experienced the GC reaction and reflect different stages of mature B-cell differentiation.
Patients, materials, and methods
Patients and pathologic specimens
The basis of this study was formed by 52 specimens of monoclonal B-cell PTLDs, collected from 51 solid organ transplant recipients. Paraffin-embedded biopsy specimens were available from 32 cases and frozen specimens were available from 20 cases. There were 26 cases that had been referred to the Divisions of Hematology and Pathology, Niguarda Hospital, Milan, Italy; 16 cases had been referred to the Division of Hematology, Ospedali Riuniti, Bergamo, Italy; and 10 cases had been referred to the Division of Pathology, Policlinico S. Matteo, Pavia, Italy. Samples of monoclonal B-cell PTLDs were derived from involved organs and were obtained in the course of routine diagnostic procedures before specific therapy. In 5 cases, PTLD was diagnosed at autopsy. Cases had been selected for being of B-cell origin and of proven monoclonality by immunohistochemical and/or molecular studies. The fraction of malignant cells in the pathologic specimen was 60% or more, as determined by morphologic and immunophenotypic studies. Based on the World Health Organization classification of hematopoietic tumors,4 PTLDs were classified into polymorphic PTLDs (P-PTLD; n = 12), and monomorphic lymphoma, namely diffuse large B-cell lymphoma (DLBCL; n = 36) and Burkitt/Burkitt-like lymphoma (BL/BLL; n = 4). Cases of plasmacytic hyperplasia were not included in this study. Clinical features have been previously reported for a fraction of cases.8,10 Genomic DNA from PTLD samples was isolated using a commercial kit (QIAamp DNA mini Kit; QIAGEN, Milano, Italy) according to the manufacturer's instructions. Approval was obtained from the Amedeo Avogadro University of Eastern Piedmont institutional review board for these studies. Informed consent was provided according to the Declaration of Helsinki.
Molecular analysis of IgVH genes
IgV heavy chain (IgVH) gene rearrangements were amplified with a set of 6 VH gene family-specific primers that hybridize to sequences in the VH leader region in conjunction with a JH degenerated primer in separate reactions for each VH primer.25,26 Samples for which no clonal IgVH rearrangement was obtained with leader primers were amplified with a set of 6 VH gene family-specific primers that hybridize to sequences in framework region (FR) 1 or FR2 in conjunction with a JH degenerated primer.27 The sequences of leader and FR1 VH primers have been reported previously.25-27 The sequences of FR2 VH primers are as follows: 5′-GGA CAA RGG CTT GAG TGG AT-3′ (VH1.1), 5′-GGA MAA SGS CTT GAG TGG AT -3′ (VH1.2), 5′-GGG AAR GGV CTG GAG TGG AT-3′ (VH4-5), 5′-GDT CCG CCA GGC TCC AG-3′ (VH3.11), 5′-GGT CCG SCA AGC TCC AG -3′ (VH3.12), 5′-GAT CCG TCA GCC CCC AG -3′ (VH2), 5′-GGA AAA GGT CTG GAG TGG GT -3′ (VH3.21), 5′-GGG AAG GGT CTG GAG TGG GT -3′ (VH3.22), 5′-GGG AAA GGG CTG GAG TGG GT -3′ (VH3.22a), and 5′-TCG AGA GGC CTT GAG TGG -3′ (VH6). Sequence of the degenerated JH primer is as follows: 5′-CTY ACC TGA RGA GAC RGT GAC C-3′. Polymerase chain reaction (PCR) was performed for 35 cycles (40 for paraffin-embedded biopsy specimens) with an annealing temperature of 60°C.
PCR products were separated by agarose gel electrophoresis, purified using the Perfectprep Gel Cleanup kit (Eppendorf, Hamburg, Germany), and directly sequenced using a commercially available kit (ThermoSequenase; Amersham Life Sciences, Amersham, United Kingdom) as reported.28 Sequences obtained were analyzed and initially aligned to the database of sequences obtained in our laboratory, to exclude possibility of contamination. Subsequently, sequences were aligned to the V-BASE sequence directory (MRC Centre for Protein Engineering, Cambridge, United Kingdom) using MacVector 6.0.1 software (Oxford Molecular Group, Oxford, United Kingdom) and DNA-Plot software (http://www.dnaplot.de/input/human_v.html; Science Company Accelerys, San Diego, CA). Sequences were aligned to the closest germ-line IgVH genes and the number of somatic mutations was determined. Mutations occurring at the last nucleotide position of the IgVH fragment were excluded from the mutational analysis because they might result from nucleotide deletion at the joining sites. The IgVH gene sequences were considered mutated if deviation from the corresponding germ-line gene was more than 2%. Criteria for the identification of D elements in the complementarity determining region 3 (CDR3) were either (1) 100% homology with a D element over a stretch of at least 7 base pair (bp), or (2) a single base-pair difference within a stretch of at least 8 bp.29
Analysis of intraclonal heterogeneity of IgVH genes
To evaluate the presence of ongoing SHM of IgVH genes, clonal IgVH rearrangements were amplified with appropriate primers using the high-fidelity PfuTurbo DNA polymerase (Stratagene, La Jolla, CA) and cloned into the pCR4-TOPO plasmid vector (Invitrogen, Paisley, United Kingdom). At least 20 randomly picked bacterial clones were sequenced on an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA) using ABI Prism Big Dye Terminator Kit version 2.0 (Applied Biosystems). Sequences were analyzed using MacVector 6.0.1 software (H.-H. Althaus and W. Muller, University of Cologne, Germany) and Multiple Sequence Alignment Software (F. Corpet, Centre Inra de Toulouse, France).32 For evaluation of ongoing SHM of IgVH genes, only clones with identical or near identical CDR3 were considered. The following definitions were used:33 unconfirmed mutation—a substitution mutation observed in only one clone; confirmed mutation—a mutation observed in more than one clone. Only confirmed mutations were considered as an evidence of ongoing SHM, whereas unconfirmed mutations were disregarded. To determine the PfuTurbo DNA polymerase error rate of our experimental strategy, 20 clones of c-MYC exon 2 (nucleotides +4486 to +5068) were amplified from normal fibroblasts and sequenced.33 These clones were generated according to the same PCR and cloning procedures used for IgVH genes. The error rate in our laboratory was 0.01%, which amounts to about 0.04 mutations per IgVH clone.
Molecular analysis of IgVL genes
In selected cases, rearrangements of IgV light chain (IgVL) genes were amplified with a set of IgVκ and IgVλ gene family-specific primers that hybridize to sequences in the Vκ and Vλ leader regions in conjunction with the appropriate Jκ or Jλ degenerated primers.25,26 Samples for which no clonal IgVL gene rearrangement was obtained with leader primers were amplified with a set of Vκ and Vλ gene family-specific primers hybridizing to sequences in FR1 in conjunction with the appropriate Jκ or Jλ degenerated primers.25-27,34 PCRs were performed for 35 cycles (40 for paraffin-embedded biopsy specimens) with an annealing temperature of 60°C. DNA sequencing of PCR amplimers and analysis of the obtained sequences were performed as described in “Molecular analysis of IgVH genes.”
Analysis of BCL6 gene
Mutations of BCL6 5′ noncoding regions (GenBank accession number AY189709) were assessed by PCR DNA direct sequencing of a 739-bp region located downstream of the first BCL6 noncoding exon and harboring more than 95% of BCL6 mutations in B-cell lymphoma. The sequence of oligonucleotides used as primers, as well as PCR conditions, have been reported in detail previously.28 PCR products were purified and sequenced on both strands from independent PCR reactions.
Analysis of viral infection
Infection by EBV was investigated by EBV-encoded RNA (EBER) in situ hybridization (ISH).20 For EBER-positive cases, immunostaining for latent membrane protein 1 (LMP1) and EBV nuclear antigen 2 (EBNA2) was performed with specific antibodies directed against LMP1 and EBNA2 (CS1-4 and PE2; Dakopatts A/S, Glostrup, Denmark).20 Human herpesvirus 8 (HHV-8) infection was assessed by PCR analysis as previously reported.20
Immunohistochemical studies of BCL6, MUM1, and CD138
Immunohistochemistry was performed by the avidin-biotin-peroxidase complex (ABC-px) or alkaline phosphatase antialkaline phosphatase (APAAP) methods.35,36 The BCL6 protein was detected by the PG-B6 monoclonal antibody (MoAb; Dakopatts A/S).37 Expression of MUM1 was investigated with the ICSAT/M-17 polyclonal goat antibody20 (Santa Cruz Biotechnology, Santa Cruz, CA). CD138 expression was assessed using the B-B4 MoAb (Serotec, Oxford, England).38 All antigens were tested on paraffin-embedded tissue sections. For MUM1 and BCL6 assessment, paraffin-embedded sections were treated in a microwave oven at 250 W for 30 minutes in EGTA (ethylene glycol tetraacetic acid) solution (1 mM, pH 8). Immunostaining for MUM1 and BCL6 was performed on an automated immunostainer (Nexes; Ventana Medical Systems, Tucson, AZ) according to a modified version of the company protocols. Immunostaining for CD138 was performed using the APAAP method.36 Only definite and unambiguous staining on unequivocal malignant cells was accepted as positive. In the case of P-PTLDs, staining was evaluated on atypical immunoblasts, medium sized lymphoid cells, and on cells with irregular nuclei resembling centrocytes.
Results
Characteristics of the patient panel
The clinical characteristics of monoclonal B-cell PTLD patients included in the study are reported in Table 1. Of the patients, 41 were male and 10 were women. The median age was 45.5 years (range, 1-67 years). PTLD patients had been subjected to transplantation of heart (n = 31), kidney (n = 11), liver (n = 7), and lung (n = 2). Most patients received cyclosporine A and azathioprine as immunosuppressive regimen (Table 1). Median time from transplantation to PTLD was 72.0 months (range, 2-158 months). According to the definition of Armitage et al,39 8 PTLDs were classified as early onset (≤ 12 months from transplantation) and 44 as late onset (≥ 12 months). Of the cases, 14 were diagnosed as stage I disease, 14 as stage II, 8 as stage III, and 15 as stage IV. All PTLDs included were of B-cell origin and comprised 12 P-PTLDs, 36 DLBCLs, and 4 BL/BLLs. DLBCL cases displayed features of centroblastic lymphoma (n = 16), immunoblastic lymphoma (n = 17), and CD30+ anaplastic lymphoma (n = 3). One patient (case 2913) developed 2 subsequent PTLDs, including a P-PTLD (identified as 2913A) and a DLBCL (identified as 2913B), both diagnosed as stage I. Data on treatment modalities and outcome, when available, are detailed in Table 1.
Clinical and virologic features of monoclonal B-cell PTLDs
Case . | Histology . | Sex . | TX . | Age at TX, y . | Immune suppression . | Interval from TX, mo . | Tumor site . | Stage . | EBV . | Treatment modalities* . | Response . | Outcome, mo from PTLD . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2614 | P-PTLD | M | Lung | 46 | C + A | 5 | Lung | IIE | + | CT | SD | Death at 1 mo |
| 2618 | P-PTLD | M | Heart | 67 | C + A | 6 | Lung | IIE | + | No treatment | CR | Alive at 76 mo |
| 2913A | P-PTLD | M | Heart | 38 | C + A | 152 | Tonsil | IE | - | CT + RT | CR | Death at 16 mo |
| 2916 | P-PTLD | M | Heart | 54 | C + A | 6 | Lung | IE | + | S | CR | Alive at 54 mo |
| 3462 | P-PTLD | M | Liver | 61 | C + A | 7 | Liver | IE | + | NA | NA | Autoptic diagnosis |
| 3464 | P-PTLD | M | Liver | 59 | C + A | 5 | Liver | IE | + | S + anti-CD20 + HDIg | CR | Alive at 27 mo |
| 3468 | P-PTLD | M | Heart | 53 | C + A | 2 | Tongue | IE | + | No treatment | CR | Alive at 23 mo |
| 3471 | P-PTLD | M | Liver | 33 | C + A | 7 | Liver | IV | + | CT + HDIg + RF | CR | Alive at 31 mo |
| 3515 | P-PTLD | M | Heart | 13 | C + A | 79 | Lymph node | II | - | CT | PD | Death at 18 mo |
| 3516 | P-PTLD | M | Heart | 18 | C + A | 22 | Lymph node | II | + | CT | NE | Death at 20 mo |
| 3519 | P-PTLD | M | Heart | 59 | C + A | 84 | Lymph node | III | - | CT | NE | Death at 6 mo |
| 3521 | P-PTLD | F | Heart | 14 | C + A | 51 | Lymph node | I | + | S | CR | Alive at 41 mo |
| 2798 | DLBCL, CB | F | Kidney | 58 | C + A | 103 | Lymph node | II | - | CT + HDIg | CR | Alive at 45 mo |
| 2898 | DLBCL, CB | M | Heart | 34 | C + A | 81 | Lymph node | III | - | CT + S + HDIg | CR | Alive at 52 mo |
| 2909 | DLBCL, CB | M | Heart | 59 | C + A | 44 | Spleen | III | - | No treatment | PD | Death at 5 mo |
| 2912 | DLBCL, CB | M | Heart | 43 | C + A | 113 | Mesenteric mass | II | - | CT | PR | Death at 15 mo |
| 2914 | DLBCL, CB | M | Heart | 48 | C + A | 110 | Stomach | IV | - | CT | CR | Alive at 70 mo |
| 3459 | DLBCL, CB | M | Lung | 27 | C + A | 128 | Lymph node | IV | + | CT + anti-CD20 + HDIg | CR | Alive at 25 mo |
| 3465 | DLBCL, CB | F | Liver | 55 | C + A | 95 | Lymph node | III | - | CT | CR | Alive at 21 mo |
| 3466 | DLBCL, CB | M | Liver | 45 | C + A | 104 | Skin | IE | - | CT + S | CR | Alive at 22 mo |
| 3467 | DLBCL, CB | M | Heart | 52 | C + A | 128 | Lymph node | IV | + | P | NA | Death at 1 d |
| 3518 | DLBCL, CB | M | Heart | 55 | C + A | 60 | Lymph node | II | + | CT + RT | PD | Death at 8 mo |
| 3522 | DLBCL, CB | M | Liver | 1 | FK506 | 29 | Jejunum | IIE | - | CT + S + anti-CD20 | CR | Alive at 41 mo |
| 3523 | DLBCL, CB | F | Heart | 12 | C + A | 102 | Lymph node | I | - | RT + anti-CD20 | CR | Alive at 29 mo |
| 3524 | DLBCL, CB | M | Heart | 21 | C + A | 78 | Skin | IV | - | CT | CR | Death at 12 mo |
| 3527 | DLBCL, CB | M | Kidney | 28 | C + A | 51 | Lymph node | III | - | CT | NE | Death at 3 mo |
| 3528 | DLBCL, CB | M | Heart | 16 | C + A | 95 | Lymph node | II | - | CT + anti-CD20 | PR | Death at 4 mo |
| 3530 | DLBCL, CB | M | Heart | 58 | C + A | 144 | Lymph node | II | - | RT + anti-CD20 | CR | Alive at 15 mo |
| 2616 | DLBCL, IB | F | Liver | 39 | C + A | 63 | Peritoneum | IV | + | CT + HDIg | CR | Alive at 60 mo |
| 2620 | DLBCL, IB | M | Kidney | 64 | C + A | 36 | Liver | IV | + | NA | NA | Autoptic diagnosis |
| 2621 | DLBCL, IB | M | Heart | 56 | C + A | 5 | Lung | IV | + | NA | NA | Autoptic diagnosis |
| 2799 | DLBCL, IB | M | Heart | 56 | C + A | 43 | Skin | IV | + | CT + HDIg | SD | Death at 4 months |
| 2802 | DLBCL, IB | M | Heart | 45 | C + A | 28 | Mediastinum | IV | + | CT + HDIg | PR | Death at 3 mo |
| 2803 | DLBCL, IB | F | Kidney | 49 | C + A | 64 | Stomach | IIE | - | CT + S | CR | Death at 16 mo |
| 2892 | DLBCL, IB | M | Heart | 44 | FK506 + A | 42 | Retroperitoneum | I | + | IFN + HDIg | CR | Alive at 137 mo |
| 2895 | DLBCL, IB | M | Kidney | 46 | C + A | 72 | Lymph node | III | + | NA | NA | Autoptic diagnosis |
| 2911 | DLBCL, IB | M | Heart | 42 | C + A | 90 | Heart | IE | + | CT | PR | Death at 3 mo |
| 2915 | DLBCL, IB | M | Heart | 56 | C + A | 56 | Lymph node | II | + | CT | PR | Death at 1 mo |
| 3461 | DLBCL, IB | M | Kidney | 30 | na | 72 | Tonsil | IIE | + | NK | NK | NK |
| 3469 | DLBCL, IB | F | Kidney | 26 | na | 90 | Jejunum | IIE | - | NK | NK | NK |
| CT + S + RT + HDIg + | ||||||||||||
| 3476 | DLBCL, IB | M | Heart | 53 | C + A | 108 | Skin | IE | + | CTL | CR | Death at 38 mo |
| 3517 | DLBCL, IB | M | Heart | 49 | C + A | 84 | Lymph node | IV | - | NA | NA | Autoptic diagnosis |
| 3520 | DLBCL, IB | M | Kidney | 15 | C + P | 156 | Kidney | IE | + | CT + S + RT | PD | Death at 4 mo |
| 3525 | DLBCL, IB | F | Kidney | 21 | C + A | 108 | Spleen | IV | - | CT | NE | Death at 1 mo |
| 3529 | DLBCL, IB | F | Heart | 6 | C + A | 72 | Lymph node | III | + | CT + anti-CD20 | CR | Alive at 21 mo |
| 2861 | DLBCL, AP | M | Kidney | 47 | C + A | 77 | Lymph node | III | - | CT | PD | Death at 6 mo |
| 2913B | DLBCL, AP | M | Heart | 38 | C + A | 158 | Tonsil | IE | + | CT + RT | PD | Death at 10 mo |
| 3458 | DLBCL, AP | F | Kidney | 60 | C + A | 14 | Skin | IV | + | CT + anti-CD20 + HDIg | PR | Death at 6 mo |
| 2617 | BL/BLL | M | Heart | 51 | C + A | 38 | Lung | IV | + | NK | PD | Death at 10 d |
| 2890 | BL/BLL | M | Heart | 27 | C + A | 106 | Tonsil | IE | - | CT + RT | PD | Death at 22 mo |
| 3463 | BL/BLL | M | Heart | 50 | C + A | 92 | Skin | IV | + | CT + HDIg | CR | Alive at 57 mo |
| 3526 | BL/BLL | M | Heart | 5 | C + A | 94 | Jaw | IE | + | CT + RT | PR | Death at 18 mo |
Case . | Histology . | Sex . | TX . | Age at TX, y . | Immune suppression . | Interval from TX, mo . | Tumor site . | Stage . | EBV . | Treatment modalities* . | Response . | Outcome, mo from PTLD . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2614 | P-PTLD | M | Lung | 46 | C + A | 5 | Lung | IIE | + | CT | SD | Death at 1 mo |
| 2618 | P-PTLD | M | Heart | 67 | C + A | 6 | Lung | IIE | + | No treatment | CR | Alive at 76 mo |
| 2913A | P-PTLD | M | Heart | 38 | C + A | 152 | Tonsil | IE | - | CT + RT | CR | Death at 16 mo |
| 2916 | P-PTLD | M | Heart | 54 | C + A | 6 | Lung | IE | + | S | CR | Alive at 54 mo |
| 3462 | P-PTLD | M | Liver | 61 | C + A | 7 | Liver | IE | + | NA | NA | Autoptic diagnosis |
| 3464 | P-PTLD | M | Liver | 59 | C + A | 5 | Liver | IE | + | S + anti-CD20 + HDIg | CR | Alive at 27 mo |
| 3468 | P-PTLD | M | Heart | 53 | C + A | 2 | Tongue | IE | + | No treatment | CR | Alive at 23 mo |
| 3471 | P-PTLD | M | Liver | 33 | C + A | 7 | Liver | IV | + | CT + HDIg + RF | CR | Alive at 31 mo |
| 3515 | P-PTLD | M | Heart | 13 | C + A | 79 | Lymph node | II | - | CT | PD | Death at 18 mo |
| 3516 | P-PTLD | M | Heart | 18 | C + A | 22 | Lymph node | II | + | CT | NE | Death at 20 mo |
| 3519 | P-PTLD | M | Heart | 59 | C + A | 84 | Lymph node | III | - | CT | NE | Death at 6 mo |
| 3521 | P-PTLD | F | Heart | 14 | C + A | 51 | Lymph node | I | + | S | CR | Alive at 41 mo |
| 2798 | DLBCL, CB | F | Kidney | 58 | C + A | 103 | Lymph node | II | - | CT + HDIg | CR | Alive at 45 mo |
| 2898 | DLBCL, CB | M | Heart | 34 | C + A | 81 | Lymph node | III | - | CT + S + HDIg | CR | Alive at 52 mo |
| 2909 | DLBCL, CB | M | Heart | 59 | C + A | 44 | Spleen | III | - | No treatment | PD | Death at 5 mo |
| 2912 | DLBCL, CB | M | Heart | 43 | C + A | 113 | Mesenteric mass | II | - | CT | PR | Death at 15 mo |
| 2914 | DLBCL, CB | M | Heart | 48 | C + A | 110 | Stomach | IV | - | CT | CR | Alive at 70 mo |
| 3459 | DLBCL, CB | M | Lung | 27 | C + A | 128 | Lymph node | IV | + | CT + anti-CD20 + HDIg | CR | Alive at 25 mo |
| 3465 | DLBCL, CB | F | Liver | 55 | C + A | 95 | Lymph node | III | - | CT | CR | Alive at 21 mo |
| 3466 | DLBCL, CB | M | Liver | 45 | C + A | 104 | Skin | IE | - | CT + S | CR | Alive at 22 mo |
| 3467 | DLBCL, CB | M | Heart | 52 | C + A | 128 | Lymph node | IV | + | P | NA | Death at 1 d |
| 3518 | DLBCL, CB | M | Heart | 55 | C + A | 60 | Lymph node | II | + | CT + RT | PD | Death at 8 mo |
| 3522 | DLBCL, CB | M | Liver | 1 | FK506 | 29 | Jejunum | IIE | - | CT + S + anti-CD20 | CR | Alive at 41 mo |
| 3523 | DLBCL, CB | F | Heart | 12 | C + A | 102 | Lymph node | I | - | RT + anti-CD20 | CR | Alive at 29 mo |
| 3524 | DLBCL, CB | M | Heart | 21 | C + A | 78 | Skin | IV | - | CT | CR | Death at 12 mo |
| 3527 | DLBCL, CB | M | Kidney | 28 | C + A | 51 | Lymph node | III | - | CT | NE | Death at 3 mo |
| 3528 | DLBCL, CB | M | Heart | 16 | C + A | 95 | Lymph node | II | - | CT + anti-CD20 | PR | Death at 4 mo |
| 3530 | DLBCL, CB | M | Heart | 58 | C + A | 144 | Lymph node | II | - | RT + anti-CD20 | CR | Alive at 15 mo |
| 2616 | DLBCL, IB | F | Liver | 39 | C + A | 63 | Peritoneum | IV | + | CT + HDIg | CR | Alive at 60 mo |
| 2620 | DLBCL, IB | M | Kidney | 64 | C + A | 36 | Liver | IV | + | NA | NA | Autoptic diagnosis |
| 2621 | DLBCL, IB | M | Heart | 56 | C + A | 5 | Lung | IV | + | NA | NA | Autoptic diagnosis |
| 2799 | DLBCL, IB | M | Heart | 56 | C + A | 43 | Skin | IV | + | CT + HDIg | SD | Death at 4 months |
| 2802 | DLBCL, IB | M | Heart | 45 | C + A | 28 | Mediastinum | IV | + | CT + HDIg | PR | Death at 3 mo |
| 2803 | DLBCL, IB | F | Kidney | 49 | C + A | 64 | Stomach | IIE | - | CT + S | CR | Death at 16 mo |
| 2892 | DLBCL, IB | M | Heart | 44 | FK506 + A | 42 | Retroperitoneum | I | + | IFN + HDIg | CR | Alive at 137 mo |
| 2895 | DLBCL, IB | M | Kidney | 46 | C + A | 72 | Lymph node | III | + | NA | NA | Autoptic diagnosis |
| 2911 | DLBCL, IB | M | Heart | 42 | C + A | 90 | Heart | IE | + | CT | PR | Death at 3 mo |
| 2915 | DLBCL, IB | M | Heart | 56 | C + A | 56 | Lymph node | II | + | CT | PR | Death at 1 mo |
| 3461 | DLBCL, IB | M | Kidney | 30 | na | 72 | Tonsil | IIE | + | NK | NK | NK |
| 3469 | DLBCL, IB | F | Kidney | 26 | na | 90 | Jejunum | IIE | - | NK | NK | NK |
| CT + S + RT + HDIg + | ||||||||||||
| 3476 | DLBCL, IB | M | Heart | 53 | C + A | 108 | Skin | IE | + | CTL | CR | Death at 38 mo |
| 3517 | DLBCL, IB | M | Heart | 49 | C + A | 84 | Lymph node | IV | - | NA | NA | Autoptic diagnosis |
| 3520 | DLBCL, IB | M | Kidney | 15 | C + P | 156 | Kidney | IE | + | CT + S + RT | PD | Death at 4 mo |
| 3525 | DLBCL, IB | F | Kidney | 21 | C + A | 108 | Spleen | IV | - | CT | NE | Death at 1 mo |
| 3529 | DLBCL, IB | F | Heart | 6 | C + A | 72 | Lymph node | III | + | CT + anti-CD20 | CR | Alive at 21 mo |
| 2861 | DLBCL, AP | M | Kidney | 47 | C + A | 77 | Lymph node | III | - | CT | PD | Death at 6 mo |
| 2913B | DLBCL, AP | M | Heart | 38 | C + A | 158 | Tonsil | IE | + | CT + RT | PD | Death at 10 mo |
| 3458 | DLBCL, AP | F | Kidney | 60 | C + A | 14 | Skin | IV | + | CT + anti-CD20 + HDIg | PR | Death at 6 mo |
| 2617 | BL/BLL | M | Heart | 51 | C + A | 38 | Lung | IV | + | NK | PD | Death at 10 d |
| 2890 | BL/BLL | M | Heart | 27 | C + A | 106 | Tonsil | IE | - | CT + RT | PD | Death at 22 mo |
| 3463 | BL/BLL | M | Heart | 50 | C + A | 92 | Skin | IV | + | CT + HDIg | CR | Alive at 57 mo |
| 3526 | BL/BLL | M | Heart | 5 | C + A | 94 | Jaw | IE | + | CT + RT | PR | Death at 18 mo |
TX indicates transplantation; P-PTLD, polymorphic PTLD; M, male; C, cyclosporine A; A, azathioprine; CT, chemotherapy (the regimen varied and was tailored to the patient's age, stage, and performance status); SD, stable disease; CR, complete remission; RT, radiotherapy; S, surgery; NA, not applicable; anti-CD20, rituximab; HDIg, high-dose immunoglobulin; RF, radiofrequency; PD, progressive disease; P, prednisone; NE, not evaluable; F, female; DLBCL, diffuse large B-cell lymphoma; CB, centroblastic; PR, partial remission; P, prednisone; FK506, tacrolimus; IB, immunoblastic; IFN, interferon; na, not available; NK, not known; CTL, cytotoxic T lymphocytes; AP, anaplastic; and BL/BLL, Burkitt lymphoma/Burkitt-like lymphoma.
All patients underwent reduction of immunosuppression, with the exception of cases 3528 and 3529 and cases in whom the diagnosis of PTLD was autoptic.
Sequence analysis of IgVH gene rearrangements in monoclonal B-cell PTLDs
By combining the results of 3 strategies used for IgVH analysis, a clonal IgVH rearrangement could be identified in 41 (78.8%) of 52 PTLD samples. Failure to detect clonal IgVH rearrangements in 11 cases may result from somatic mutations in the region annealing to PCR primers, thus leading to false-negative results. Alternatively, absent detection of IgVH rearrangements may be due to aberrant rearrangements or to loss of IgVH sequences.40
Among PTLDs yielding IgVH amplimers, a functional rearrangement was obtained from 33 (80.5%) of 41 cases (Table 2). In 8 (19.5%) of 41 PTLDs, the only rearrangement obtained was nonfunctional because of an out-of-frame IgVH rearrangement (5 cases) or because of crippling mutations leading to the introduction of stop codons within the originally productive rearrangement (3 cases) (Table 3).
Analysis of functional IgVH gene rearrangements in monoclonal B-cell PTLDs
Case . | Histology . | IgVH family . | Closest IgVH germline gene . | % of mutation . | D . | JH . |
|---|---|---|---|---|---|---|
| 2614 | P-PTLD | VH3 | 3-74 | GL | NA | JH4b/JH5b |
| 2618 | P-PTLD | VH3 | 3-30.3 | 5.64 | D4-23 | JH6b |
| 2916 | P-PTLD | VH5 | 5-51 | 2.77 | NA | JH3b |
| 3464 | P-PTLD | VH3 | 3-15 | GL | NA | JH3b |
| 3468 | P-PTLD | VH3 | 3-07 | 12.3 | D5-18/D5-5 | JH6a |
| 3471 | P-PTLD | VH3 | 3-30 | 12.1 | D3-3 | JH6b |
| 3515 | P-PTLD | VH3 | 3-15 | 10.3 | NA | JH4b |
| 3519 | P-PTLD | VH3 | 3-30 | 10.3 | D3-22 | JH4b |
| 2798 | DLBCL, centroblastic | VH4 | 4-34 | 23.2 | NA | JH3a/JH6 |
| 2898 | DLBCL, centroblastic | VH4 | 4-34 | 11.8 | NA | JH4b |
| 2912 | DLBCL, centroblastic | VH5 | 5-51 | 4.06 | D6-19 | JH4b |
| 2914 | DLBCL, centroblastic | VH3 | 3-74 | 8.64 | D1-7 | JH4b |
| 3459 | DLBCL, centroblastic | VH1 | 1-69 | 17.6 | D3-10 | JH4d |
| 3466 | DLBCL, centroblastic | VH3 | 1-f | 3.73 | D6-19 | JH4b |
| 3467 | DLBCL, centroblastic | VH2 | S12-7 | GL | D6-25 | JH3b |
| 3518 | DLBCL, centroblastic | VH3 | 3-33 | 4.41 | D5-5/D5-18 | JH4/JH5 |
| 3522 | DLBCL, centroblastic | VH4 | 4-34 | 6.87 | D3-3 | JH3b |
| 3523 | DLBCL, centroblastic | VH1 | 1-46 | 2.71 | D4-23 | JH4b |
| 3524 | DLBCL, centroblastic | VH3 | 3-15 | 14.9 | NA | JH2 |
| 3528 | DLBCL, centroblastic | VH1 | 1-18 | 8.11 | D3-16 | JH4b |
| 3530 | DLBCL, centroblastic | VH3 | 3-07 | 24.1 | NA | JH4b |
| 2616 | DLBCL, immunoblastic | VH3 | 3-30 | 9.68 | NA | JH4b |
| 2621 | DLBCL, immunoblastic | VH3 | 3-30 | GL | D3-10 | JH6b |
| 2799 | DLBCL, immunoblastic | VH4 | 4-34 | 11.6 | D6-13 | JH4b |
| 2802 | DLBCL, immunoblastic | VH3 | LSG6.1 | 5.28 | D4-17 | JH6b |
| 2803 | DLBCL, immunoblastic | VH3 | 3-11 | 2.80 | D3-3 | JH6c |
| 2911 | DLBCL, immunoblastic | VH3 | 3-15 | 2.25 | D2-21 | JH3b |
| 3469 | DLBCL, immunoblastic | VH3 | 3-15 | 17.2 | NA | JH4b |
| 3476 | DLBCL, immunoblastic | VH4 | 4-04 | 8.62 | D4-23/D1 | JH6b |
| 3517 | DLBCL, immunoblastic | VH3 | 3-23 | 7.80 | D3-9 | JH1/JH4b |
| 3525 | DLBCL, immunoblastic | VH4 | 4-34 | 2.12 | D3-10 | JH6b |
| 2617 | BL/BLL | VH4 | 4-59 | 5.45 | D3-3 | JH6b |
| 2890 | BL/BLL | VH4 | 4-61 | 12.0 | NA | JH4b |
Case . | Histology . | IgVH family . | Closest IgVH germline gene . | % of mutation . | D . | JH . |
|---|---|---|---|---|---|---|
| 2614 | P-PTLD | VH3 | 3-74 | GL | NA | JH4b/JH5b |
| 2618 | P-PTLD | VH3 | 3-30.3 | 5.64 | D4-23 | JH6b |
| 2916 | P-PTLD | VH5 | 5-51 | 2.77 | NA | JH3b |
| 3464 | P-PTLD | VH3 | 3-15 | GL | NA | JH3b |
| 3468 | P-PTLD | VH3 | 3-07 | 12.3 | D5-18/D5-5 | JH6a |
| 3471 | P-PTLD | VH3 | 3-30 | 12.1 | D3-3 | JH6b |
| 3515 | P-PTLD | VH3 | 3-15 | 10.3 | NA | JH4b |
| 3519 | P-PTLD | VH3 | 3-30 | 10.3 | D3-22 | JH4b |
| 2798 | DLBCL, centroblastic | VH4 | 4-34 | 23.2 | NA | JH3a/JH6 |
| 2898 | DLBCL, centroblastic | VH4 | 4-34 | 11.8 | NA | JH4b |
| 2912 | DLBCL, centroblastic | VH5 | 5-51 | 4.06 | D6-19 | JH4b |
| 2914 | DLBCL, centroblastic | VH3 | 3-74 | 8.64 | D1-7 | JH4b |
| 3459 | DLBCL, centroblastic | VH1 | 1-69 | 17.6 | D3-10 | JH4d |
| 3466 | DLBCL, centroblastic | VH3 | 1-f | 3.73 | D6-19 | JH4b |
| 3467 | DLBCL, centroblastic | VH2 | S12-7 | GL | D6-25 | JH3b |
| 3518 | DLBCL, centroblastic | VH3 | 3-33 | 4.41 | D5-5/D5-18 | JH4/JH5 |
| 3522 | DLBCL, centroblastic | VH4 | 4-34 | 6.87 | D3-3 | JH3b |
| 3523 | DLBCL, centroblastic | VH1 | 1-46 | 2.71 | D4-23 | JH4b |
| 3524 | DLBCL, centroblastic | VH3 | 3-15 | 14.9 | NA | JH2 |
| 3528 | DLBCL, centroblastic | VH1 | 1-18 | 8.11 | D3-16 | JH4b |
| 3530 | DLBCL, centroblastic | VH3 | 3-07 | 24.1 | NA | JH4b |
| 2616 | DLBCL, immunoblastic | VH3 | 3-30 | 9.68 | NA | JH4b |
| 2621 | DLBCL, immunoblastic | VH3 | 3-30 | GL | D3-10 | JH6b |
| 2799 | DLBCL, immunoblastic | VH4 | 4-34 | 11.6 | D6-13 | JH4b |
| 2802 | DLBCL, immunoblastic | VH3 | LSG6.1 | 5.28 | D4-17 | JH6b |
| 2803 | DLBCL, immunoblastic | VH3 | 3-11 | 2.80 | D3-3 | JH6c |
| 2911 | DLBCL, immunoblastic | VH3 | 3-15 | 2.25 | D2-21 | JH3b |
| 3469 | DLBCL, immunoblastic | VH3 | 3-15 | 17.2 | NA | JH4b |
| 3476 | DLBCL, immunoblastic | VH4 | 4-04 | 8.62 | D4-23/D1 | JH6b |
| 3517 | DLBCL, immunoblastic | VH3 | 3-23 | 7.80 | D3-9 | JH1/JH4b |
| 3525 | DLBCL, immunoblastic | VH4 | 4-34 | 2.12 | D3-10 | JH6b |
| 2617 | BL/BLL | VH4 | 4-59 | 5.45 | D3-3 | JH6b |
| 2890 | BL/BLL | VH4 | 4-61 | 12.0 | NA | JH4b |
P-PTLD indicates polymorphic PTLD; GL, germ line; NA, not able to be aligned (the sequence between the VH and the JH gene segment could not be aligned to any known D gene); DLBCL; diffuse large B-cell lymphoma; and BL/BLL, Burkitt lymphoma/Burkitt-like lymphoma.
Molecular characteristics of monoclonal B cell PTLD carrying nonfunctional IgVH gene rearrangements
. | . | IgVH . | . | . | . | . | . | IgVL . | . | . | . | . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case . | Histology . | IgVH family . | Closest IgVH germ-line gene . | % of mutations . | D . | JH . | Functional status . | IgVL family . | Closest IgVL germline gene . | % of mutation . | JL . | Functional status . | |||||||||
| 3521 | P-PTLD | VH3 | 3-30 | GL | D3-9 | JH4b | Out of frame | Vk2 | A18 | GL | NA | Pseudogene | |||||||||
| 3527 | DLBCL, centroblastic | VH4 | 4-30.1 | 7.12 | NA | JH3b | Out of frame | — | — | — | — | — | |||||||||
| 2892 | DLBCL, immunoblastic | VH3 | 3-66 | 8.59 | NA | JH6b | Out of frame | Vk3 | L2 | 5.42 | Jk1 | Crippled | |||||||||
| 2895 | DLBCL, immunoblastic | VH3 | 3-07 | 7.20 | NA | JH6b | Crippled | Vk1 | L12 | 6.32 | Jk1 | Crippled | |||||||||
| 2915 | DLBCL, immunoblastic | VH3 | 3-30.3 | 11.8 | D1-26 | JH6b | Crippled | Vλ 1 | 1e | 7.67 | Jλ2 | Crippled | |||||||||
| 3461 | DLBCL, immunoblastic | VH4 | 4-34 | 7.42 | D4-17 | JH6b | Crippled | Vk1 | L12 | 6.9 | Jk1 | Crippled | |||||||||
| 3520 | DLBCL, immunoblastic | VH4 | 4-30.2 | 10.7 | D3-10 | JH5b | Out of frame | Vk4 | B3 | GL | Jk3 | Functional | |||||||||
| 3526 | BL/BLL | VH1 | hv1263 | GL | NA | JH6c | Out of frame | Vλ2 | 2c | 4.95 | Jλ2 | Crippled | |||||||||
. | . | IgVH . | . | . | . | . | . | IgVL . | . | . | . | . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case . | Histology . | IgVH family . | Closest IgVH germ-line gene . | % of mutations . | D . | JH . | Functional status . | IgVL family . | Closest IgVL germline gene . | % of mutation . | JL . | Functional status . | |||||||||
| 3521 | P-PTLD | VH3 | 3-30 | GL | D3-9 | JH4b | Out of frame | Vk2 | A18 | GL | NA | Pseudogene | |||||||||
| 3527 | DLBCL, centroblastic | VH4 | 4-30.1 | 7.12 | NA | JH3b | Out of frame | — | — | — | — | — | |||||||||
| 2892 | DLBCL, immunoblastic | VH3 | 3-66 | 8.59 | NA | JH6b | Out of frame | Vk3 | L2 | 5.42 | Jk1 | Crippled | |||||||||
| 2895 | DLBCL, immunoblastic | VH3 | 3-07 | 7.20 | NA | JH6b | Crippled | Vk1 | L12 | 6.32 | Jk1 | Crippled | |||||||||
| 2915 | DLBCL, immunoblastic | VH3 | 3-30.3 | 11.8 | D1-26 | JH6b | Crippled | Vλ 1 | 1e | 7.67 | Jλ2 | Crippled | |||||||||
| 3461 | DLBCL, immunoblastic | VH4 | 4-34 | 7.42 | D4-17 | JH6b | Crippled | Vk1 | L12 | 6.9 | Jk1 | Crippled | |||||||||
| 3520 | DLBCL, immunoblastic | VH4 | 4-30.2 | 10.7 | D3-10 | JH5b | Out of frame | Vk4 | B3 | GL | Jk3 | Functional | |||||||||
| 3526 | BL/BLL | VH1 | hv1263 | GL | NA | JH6c | Out of frame | Vλ2 | 2c | 4.95 | Jλ2 | Crippled | |||||||||
All PTLDs included in this table failed to express Ig molecules.
P-PTLD indicates polymorphic PTLD; GL, germ line; DLBCL, diffuse large B-cell lymphoma; NA, not able to be aligned; —, no product found; and BL/BLL, Burkitt lymphoma/Burkitt-like lymphoma.
In PTLDs yielding only nonfunctional IgVH rearrangements (n = 8) with the leader, FR1 or FR2 primers described previously, additional molecular strategies were also used. In particular, alternative FR1 primers (http://www.mrc-cpe.cam.ac.uk/PRIMERS.php?menu=901) were used and the degenerated JH primer was substituted with JH primers specific for each JH family.27 However, despite this extensive molecular analysis, no productive IgVH rearrangement was detected in these 8 PTLD samples.
The detailed characterization of functional and nonfunctional IgVH rearrangements identified in PTLDs is reported in Table 2 and Table 3, respectively. Overall, the distribution of IgVH families used by PTLDs reflects the complexity of the rearranged IgVH repertoire of mature B cells, since VH3, the largest and most frequently used family, was found most often (23/41; 56.1%), followed by VH4 (11/41; 26.8%) and VH1 (4/41; 9.76%). Overall, the use of IgVH genes by PTLDs apparently did not show any bias toward the preferential use of specific genes; also, use of JH segments appeared to reflect the normal representation of peripheral antigen-experienced B cells (Tables 2, 3).41,42 According to the criteria adopted (“Patients, materials, and methods”), 26 D genes could be assigned (Tables 2, 3).
Analysis of IgVH gene mutations in monoclonal B-cell PTLDs
Results of the presence of IgVH SHM in PTLDs are illustrated in detail in Tables 2 and 3. Among PTLDs showing functional IgVH rearrangements, SHM of IgVH genes was detected in 29 (87.9%) of 33 PTLDs, while 4 cases displayed IgVH genes with less than 2% difference from the most similar germ-line gene. Among mutated cases, the average mutation frequency was 9.25 ± 5.91% (median, 8.62; range, 2.12%-24.1%). With respect to histology, SHM of functionally rearranged IgVH genes occurred in 6 (75.0%) of 8 P-PTLDs, in 12 (92.3%) of 13 DLBCLs centroblastic, in 9 (90.0%) of 10 DLBCLs immunoblastic, and in 2 of 2 BL/BLLs (Table 2). The average mutation frequency did not differ significantly among the various clinico-pathologic categories of PTLDs (Table 2). With respect to PTLDs showing nonfunctional IgVH rearrangements, SHM of IgVH genes occurred in 6 (75.0%) of 8 cases (Table 3).
The distribution of replacement (R) and silent (S) mutations within functional IgVH gene rearrangements has been used to determine whether the tumor cells were selected to conserve FR sequences and maintain antigen binding (indicated by a lower than expected number of R mutations in the FR) and has been taken as an indicator for a possible selection of the tumor antibody molecule for high-affinity binding (indicated by a higher than expected number of R mutations in the CDR).30,31 The distribution of SHM was analyzed by the binomial and the multinomial distribution models on all PTLDs carrying functionally rearranged and somatically mutated IgVH genes (n = 29).30,31 The results of the binomial and the multinomial statistical methods were superimposable in all but 3 cases (Table 4). A lower than expected number of R mutations in the FR, suggesting pressure to preserve antigen binding, was observed in 12 (41.4%) of 29 PTLDs, while a higher than expected number of R mutations in the CDR, suggesting antigen selection, was observed in 9 (31.0%) of 29 cases (Table 4).
Analysis of the mutational pattern of functionally rearranged IgVH genes in monoclonal B-cell PTLDs
. | . | IgVH Germ-line gene . | % of mutation . | . | Observed R/S* . | Expected R/S† . | P‡ . | . | |
|---|---|---|---|---|---|---|---|---|---|
| Case . | Histology . | . | . | FR/CDR . | . | . | PB . | PM . | |
| 2618 | P-PTLD | 3-30.3 | 5.64 | FR | 1.33 | 2.97 | <.001 | .028 | |
| CDR | 2.50 | 3.87 | .109 | .109 | |||||
| 2916 | P-PTLD | 5-51 | 2.77 | FR | 0.95 | 2.96 | .001 | .002 | |
| CDR | 1.43 | 4.92 | .079 | .841 | |||||
| 3468 | P-PTLD | 3-07 | 12.3 | FR | 1.60 | 3.10 | .069 | .089 | |
| CDR | 1.00 | 4.11 | .187 | .685 | |||||
| 3471 | P-PTLD | 3-30 | 12.1 | FR | 0.57 | 3.17 | .001 | <.001 | |
| CDR | 1.00 | 3.44 | .200 | .453 | |||||
| 3515 | P-PTLD | 3-15 | 10.3 | FR | 0.60 | 2.90 | <.001 | <.001 | |
| CDR | 8.00 | 3.45 | .029 | .027 | |||||
| 3519 | P-PTLD | 3-30 | 10.3 | FR | 2.50 | 3.13 | .107 | .123 | |
| CDR | 2.50 | 3.87 | .083 | .113 | |||||
| 2798 | DLBCL, CB | 4-34 | 23.2 | FR | 1.30 | 2.74 | <.001 | <.001 | |
| CDR | 18.0 | 4.39 | .001 | <.001 | |||||
| 2898 | DLBCL, CB | 4-34 | 11.8 | FR | 1.22 | 2.74 | .059 | .087 | |
| CDR | 0.40 | 4.39 | .390 | .97 | |||||
| 2912 | DLBCL, CB | 5-51 | 4.06 | FR | 3.00 | 3.35 | .155 | .211 | |
| CDR | 4.00 | 3.58 | .150 | .170 | |||||
| 2914 | DLBCL, CB | 3-74 | 8.64 | FR | 1.50 | 2.99 | .045 | .048 | |
| CDR | 8.00 | 3.69 | .023 | .022 | |||||
| 3459 | DLBCL, CB | 1-69 | 17.6 | FR | 0.95 | 2.96 | <.001 | <.001 | |
| CDR | 1.80 | 3.55 | .145 | .502 | |||||
| 3466 | DLBCL, CB | 1-f | 3.73 | FR | 4.00 | 3.12 | .122 | .115 | |
| CDR | Infinite | 3.56 | .039 | .048 | |||||
| 3518 | DLBCL, CB | 3-33 | 4.41 | FR | 5.00 | 2.97 | .156 | .191 | |
| CDR | 2.00 | 4.28 | .151 | .176 | |||||
| 3522 | DLBCL, CB | 4-34 | 6.87 | FR | 1.40 | 2.76 | .054 | .517 | |
| CDR | 1.00 | 4.39 | .244 | .060 | |||||
| 3523 | DLBCL, CB | 1-46 | 2.71 | FR | Infinite | 2.98 | .246 | .328 | |
| CDR | 3.00 | 4.40 | .125 | .104 | |||||
| 3524 | DLBCL, CB | 3-15 | 14.9 | FR | 0.94 | 2.90 | <.001 | <.001 | |
| CDR | 1.80 | 3.44 | .143 | .393 | |||||
| 3528 | DLBCL, CB | 1-18 | 8.11 | FR | 1.67 | 2.93 | .046 | .059 | |
| CDR | 7.00 | 4.05 | .073 | .089 | |||||
| 3530 | DLBCL, CB | 3-07 | 24.1 | FR | 2.00 | 3.07 | .077 | .216 | |
| CDR | 6.00 | 4.79 | .033 | .095 | |||||
| 2616 | DLBCL, IB | 3-30 | 9.68 | FR | 1.20 | 3.13 | .002 | .002 | |
| CDR | 6.33 | 3.87 | .013 | .013 | |||||
| 2799 | DLBCL, IB | 4-34 | 11.6 | FR | 1.00 | 2.82 | .062 | .092 | |
| CDR | 3.00 | 4.39 | .077 | .919 | |||||
| 2802 | DLBCL, IB | LSG6.1 | 5.28 | FR | 1.00 | 3.23 | .223 | .208 | |
| CDR | Infinite | 2.95 | .293 | .307 | |||||
| 2803 | DLBCL, IB | 3-11 | 2.80 | FR | Infinite | 2.95 | .118 | .095 | |
| CDR | 6.00 | 4.34 | .036 | .025 | |||||
| 2911 | DLBCL, IB | 3-15 | 2.25 | FR | 0 | 3.18 | .0549 | .027 | |
| CDR | Infinite | 2.95 | .421 | .421 | |||||
| 3469 | DLBCL, IB | 3-15 | 17.2 | FR | 1.22 | 3.23 | .070 | .092 | |
| CDR | 2.00 | 2.95 | .145 | .705 | |||||
| 3476 | DLBCL, IB | 4-04 | 8.62 | FR | 1.60 | 2.70 | .032 | .0543 | |
| CDR | 3.50 | 4.09 | .103 | .119 | |||||
| 3517 | DLBCL, IB | 3-23 | 7.80 | FR | 0.80 | 2.99 | <.001 | <.001 | |
| CDR | 6.00 | 3.55 | <.001 | <.001 | |||||
| 3525 | DLBCL, IB | 4-34 | 2.12 | FR | 4.00 | 2.76 | .296 | .655 | |
| CDR | 0 | 4.40 | .312 | .844 | |||||
| 2617 | BL/BLL | 4-59 | 5.45 | FR | 2.00 | 3.12 | .235 | .333 | |
| CDR | 2.00 | 2.98 | .305 | .504 | |||||
| 2890 | BL/BLL | 4-61 | 12.0 | FR | 2.00 | 2.75 | .053 | .066 | |
| CDR | Infinite | 4.39 | .011 | .011 | |||||
. | . | IgVH Germ-line gene . | % of mutation . | . | Observed R/S* . | Expected R/S† . | P‡ . | . | |
|---|---|---|---|---|---|---|---|---|---|
| Case . | Histology . | . | . | FR/CDR . | . | . | PB . | PM . | |
| 2618 | P-PTLD | 3-30.3 | 5.64 | FR | 1.33 | 2.97 | <.001 | .028 | |
| CDR | 2.50 | 3.87 | .109 | .109 | |||||
| 2916 | P-PTLD | 5-51 | 2.77 | FR | 0.95 | 2.96 | .001 | .002 | |
| CDR | 1.43 | 4.92 | .079 | .841 | |||||
| 3468 | P-PTLD | 3-07 | 12.3 | FR | 1.60 | 3.10 | .069 | .089 | |
| CDR | 1.00 | 4.11 | .187 | .685 | |||||
| 3471 | P-PTLD | 3-30 | 12.1 | FR | 0.57 | 3.17 | .001 | <.001 | |
| CDR | 1.00 | 3.44 | .200 | .453 | |||||
| 3515 | P-PTLD | 3-15 | 10.3 | FR | 0.60 | 2.90 | <.001 | <.001 | |
| CDR | 8.00 | 3.45 | .029 | .027 | |||||
| 3519 | P-PTLD | 3-30 | 10.3 | FR | 2.50 | 3.13 | .107 | .123 | |
| CDR | 2.50 | 3.87 | .083 | .113 | |||||
| 2798 | DLBCL, CB | 4-34 | 23.2 | FR | 1.30 | 2.74 | <.001 | <.001 | |
| CDR | 18.0 | 4.39 | .001 | <.001 | |||||
| 2898 | DLBCL, CB | 4-34 | 11.8 | FR | 1.22 | 2.74 | .059 | .087 | |
| CDR | 0.40 | 4.39 | .390 | .97 | |||||
| 2912 | DLBCL, CB | 5-51 | 4.06 | FR | 3.00 | 3.35 | .155 | .211 | |
| CDR | 4.00 | 3.58 | .150 | .170 | |||||
| 2914 | DLBCL, CB | 3-74 | 8.64 | FR | 1.50 | 2.99 | .045 | .048 | |
| CDR | 8.00 | 3.69 | .023 | .022 | |||||
| 3459 | DLBCL, CB | 1-69 | 17.6 | FR | 0.95 | 2.96 | <.001 | <.001 | |
| CDR | 1.80 | 3.55 | .145 | .502 | |||||
| 3466 | DLBCL, CB | 1-f | 3.73 | FR | 4.00 | 3.12 | .122 | .115 | |
| CDR | Infinite | 3.56 | .039 | .048 | |||||
| 3518 | DLBCL, CB | 3-33 | 4.41 | FR | 5.00 | 2.97 | .156 | .191 | |
| CDR | 2.00 | 4.28 | .151 | .176 | |||||
| 3522 | DLBCL, CB | 4-34 | 6.87 | FR | 1.40 | 2.76 | .054 | .517 | |
| CDR | 1.00 | 4.39 | .244 | .060 | |||||
| 3523 | DLBCL, CB | 1-46 | 2.71 | FR | Infinite | 2.98 | .246 | .328 | |
| CDR | 3.00 | 4.40 | .125 | .104 | |||||
| 3524 | DLBCL, CB | 3-15 | 14.9 | FR | 0.94 | 2.90 | <.001 | <.001 | |
| CDR | 1.80 | 3.44 | .143 | .393 | |||||
| 3528 | DLBCL, CB | 1-18 | 8.11 | FR | 1.67 | 2.93 | .046 | .059 | |
| CDR | 7.00 | 4.05 | .073 | .089 | |||||
| 3530 | DLBCL, CB | 3-07 | 24.1 | FR | 2.00 | 3.07 | .077 | .216 | |
| CDR | 6.00 | 4.79 | .033 | .095 | |||||
| 2616 | DLBCL, IB | 3-30 | 9.68 | FR | 1.20 | 3.13 | .002 | .002 | |
| CDR | 6.33 | 3.87 | .013 | .013 | |||||
| 2799 | DLBCL, IB | 4-34 | 11.6 | FR | 1.00 | 2.82 | .062 | .092 | |
| CDR | 3.00 | 4.39 | .077 | .919 | |||||
| 2802 | DLBCL, IB | LSG6.1 | 5.28 | FR | 1.00 | 3.23 | .223 | .208 | |
| CDR | Infinite | 2.95 | .293 | .307 | |||||
| 2803 | DLBCL, IB | 3-11 | 2.80 | FR | Infinite | 2.95 | .118 | .095 | |
| CDR | 6.00 | 4.34 | .036 | .025 | |||||
| 2911 | DLBCL, IB | 3-15 | 2.25 | FR | 0 | 3.18 | .0549 | .027 | |
| CDR | Infinite | 2.95 | .421 | .421 | |||||
| 3469 | DLBCL, IB | 3-15 | 17.2 | FR | 1.22 | 3.23 | .070 | .092 | |
| CDR | 2.00 | 2.95 | .145 | .705 | |||||
| 3476 | DLBCL, IB | 4-04 | 8.62 | FR | 1.60 | 2.70 | .032 | .0543 | |
| CDR | 3.50 | 4.09 | .103 | .119 | |||||
| 3517 | DLBCL, IB | 3-23 | 7.80 | FR | 0.80 | 2.99 | <.001 | <.001 | |
| CDR | 6.00 | 3.55 | <.001 | <.001 | |||||
| 3525 | DLBCL, IB | 4-34 | 2.12 | FR | 4.00 | 2.76 | .296 | .655 | |
| CDR | 0 | 4.40 | .312 | .844 | |||||
| 2617 | BL/BLL | 4-59 | 5.45 | FR | 2.00 | 3.12 | .235 | .333 | |
| CDR | 2.00 | 2.98 | .305 | .504 | |||||
| 2890 | BL/BLL | 4-61 | 12.0 | FR | 2.00 | 2.75 | .053 | .066 | |
| CDR | Infinite | 4.39 | .011 | .011 | |||||
FR indicates framework region; CDR, complementarity determining region; R, replacement mutations; S, silent mutations; P-PTLD, polymorphic PTLD; DLBCL, diffuse large B-cell lymphoma; CB, centroblastic; IB, immunoblastic; and BL/BLL, Burkitt lymphoma/Burkitt-like lymphoma.
Observed R/S ratio in the FR and CDR.
Expected R/S ratio in the FR and CDR.
P is the probability calculated to evaluate whether the excess or the scarcity of R mutations in CDR and FR, respectively, was due to chance alone; PB, P value calculated according to the binomial distribution model; PM, P value calculated according to the multinomial distribution model. Differences were considered statistically significant when P <.05.
Analysis of intraclonal heterogeneity of SHM of IgVH genes
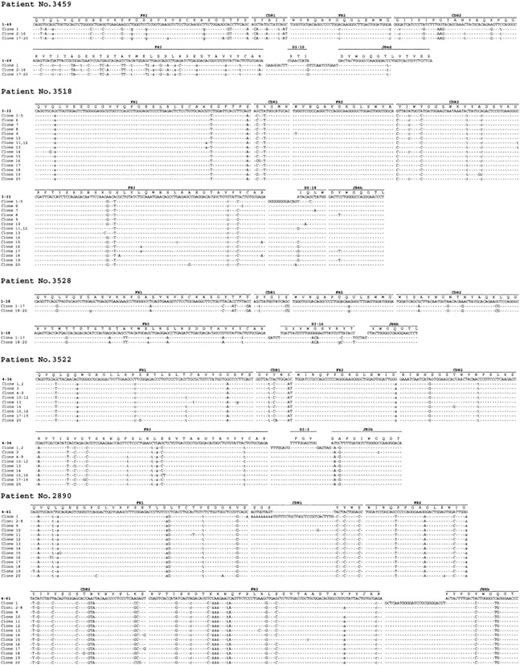
Intraclonal variation of IgVH genes was assessed by extensive molecular cloning in 13 IgVH gene isolates derived from 13 different PTLD specimens (Table 5). In all cases, the clonal IgVH sequence had been previously established by direct DNA sequencing of the PCR product and had been demonstrated to be somatically mutated (mutation range, 2.12%-24.1%). In 8 (61.5%) of 13 PTLDs, the clonal IgVH isolates did not show intraclonal heterogeneity, indicating absence of ongoing IgVH mutations (Table 5). Conversely, 5 of 13 PTLDs showed the presence of more than one mutation, which were not detectable by DNA direct sequencing and that recurred in more than one clone (Table 5; Figure 1). Based on the adopted criteria,33 the presence of such confirmed mutations is consistent with intraclonal heterogeneity of IgVH genes and with ongoing SHM activity of IgVH genes. With respect to histology, ongoing SHM activity of IgVH genes was detected in 4 (57.1%) of 7 DLBCLs centroblastic and in 1 of 1 BL/BLL (Table 5; Figure 1).
Analysis of ongoing SHM of IgVh genes in monoclonal B-cell PTLDs
. | . | Phenotype . | . | . | . | Closest IgVh germ-line gene . | % of mutations . | No. of unconfirmed mutations . | No. of confirmed ongoing mutations . | No. of clones with ongoing mutations . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case . | Histology . | BCL6 . | MUM1 . | CD138 . | EBV . | . | . | . | . | . | ||
| 3515 | P-PTLD | − | − | − | − | 3-15 | 10.3 | 1 | 0 | 0 | ||
| 3519 | P-PTLD | − | + | − | − | 3-30 | 10.3 | 0 | 0 | 0 | ||
| 2898 | DLBCL, centroblastic | + | − | − | − | 4-34 | 11.8 | 0 | 0 | 0 | ||
| 3459 | DLBCL, centroblastic | + | + | − | + | 1-69 | 17.6 | 0 | 4 | 20 | ||
| 3518 | DLBCL, centroblastic | + | − | − | + | 3-33 | 4.41 | 13 | 9 | 20 | ||
| 3522 | DLBCL, centroblastic | + | + | − | − | 4-34 | 6.87 | 6 | 4 | 17 | ||
| 3524 | DLBCL, centroblastic | − | − | − | − | 3-15 | 14.9 | 0 | 0 | 0 | ||
| 3528 | DLBCL, centroblastic | + | − | − | − | 1-18 | 8.11 | 0 | 2 | 3 | ||
| 3530 | DLBCL, centroblastic | − | + | + | − | 3-07 | 24.1 | 0 | 0 | 0 | ||
| 3476 | DLBCL, immunoblastic | − | + | + | + | 4-04 | 8.62 | 0 | 0 | 0 | ||
| 3517 | DLBCL, immunoblastic | ND | ND | ND | − | 3-23 | 7.80 | 0 | 0 | 0 | ||
| 3525 | DLBCL, immunoblastic | − | + | − | − | 4-34 | 2.12 | 0 | 0 | 0 | ||
| 2890 | BL/BLL | ND | ND | ND | − | 4-61 | 12.0 | 12 | 3 | 20 | ||
. | . | Phenotype . | . | . | . | Closest IgVh germ-line gene . | % of mutations . | No. of unconfirmed mutations . | No. of confirmed ongoing mutations . | No. of clones with ongoing mutations . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case . | Histology . | BCL6 . | MUM1 . | CD138 . | EBV . | . | . | . | . | . | ||
| 3515 | P-PTLD | − | − | − | − | 3-15 | 10.3 | 1 | 0 | 0 | ||
| 3519 | P-PTLD | − | + | − | − | 3-30 | 10.3 | 0 | 0 | 0 | ||
| 2898 | DLBCL, centroblastic | + | − | − | − | 4-34 | 11.8 | 0 | 0 | 0 | ||
| 3459 | DLBCL, centroblastic | + | + | − | + | 1-69 | 17.6 | 0 | 4 | 20 | ||
| 3518 | DLBCL, centroblastic | + | − | − | + | 3-33 | 4.41 | 13 | 9 | 20 | ||
| 3522 | DLBCL, centroblastic | + | + | − | − | 4-34 | 6.87 | 6 | 4 | 17 | ||
| 3524 | DLBCL, centroblastic | − | − | − | − | 3-15 | 14.9 | 0 | 0 | 0 | ||
| 3528 | DLBCL, centroblastic | + | − | − | − | 1-18 | 8.11 | 0 | 2 | 3 | ||
| 3530 | DLBCL, centroblastic | − | + | + | − | 3-07 | 24.1 | 0 | 0 | 0 | ||
| 3476 | DLBCL, immunoblastic | − | + | + | + | 4-04 | 8.62 | 0 | 0 | 0 | ||
| 3517 | DLBCL, immunoblastic | ND | ND | ND | − | 3-23 | 7.80 | 0 | 0 | 0 | ||
| 3525 | DLBCL, immunoblastic | − | + | − | − | 4-34 | 2.12 | 0 | 0 | 0 | ||
| 2890 | BL/BLL | ND | ND | ND | − | 4-61 | 12.0 | 12 | 3 | 20 | ||
P-PTLD indicates polymorphic PTLD; DLBCL, diffuse large B-cell lymphoma; ND, not determined; and BL/BLL, Burkitt lymphoma/Burkitt-like lymphoma.
Evidence of intraclonal heterogeneity in IgVH genes of monoclonal B-cell PTLDs. Sequence alignments of the IgVH clones derived from PTLD cases 3459, 3518, 3528, 3522, and 2890. The sequences of clones were aligned and compared with the most homologous germ-line IgVH, D, and JH sequences. Identity with the most homologous germ-line sequence is indicated by dashes. Each mutation is indicated by the appropriate nucleotide: replacements mutations by uppercase letters and silent mutations by lowercase letters; Δ symbol indicates nucleotide deletion. N-additions between IgVH and D, as well as between D and JH segments, are shown for each sequence. CDR indicates complementarity determining region; FR, framework region.
Evidence of intraclonal heterogeneity in IgVH genes of monoclonal B-cell PTLDs. Sequence alignments of the IgVH clones derived from PTLD cases 3459, 3518, 3528, 3522, and 2890. The sequences of clones were aligned and compared with the most homologous germ-line IgVH, D, and JH sequences. Identity with the most homologous germ-line sequence is indicated by dashes. Each mutation is indicated by the appropriate nucleotide: replacements mutations by uppercase letters and silent mutations by lowercase letters; Δ symbol indicates nucleotide deletion. N-additions between IgVH and D, as well as between D and JH segments, are shown for each sequence. CDR indicates complementarity determining region; FR, framework region.
Sequence analysis of IgVL gene rearrangements in monoclonal B-cell PTLDs
Analysis of IgVL genes was performed in PTLDs showing only nonfunctional rearrangements of IgVH genes (n = 8). By combining the results of 3 strategies used for IgVL analysis, a clonal IgVL rearrangement could be identified in 7 (87.5%) of 8 samples (Table 3). Of the cases, 4 used a Vk gene, 2 used a Vλ gene, and 1 used a previously described Vk pseudogene (Table 3).43 Notably, all PTLDs that had been demonstrated to harbor crippled IgVH genes (cases 2895, 2915, and 3461) displayed crippling mutations in their IgVL genes (Table 3). Also, a crippling mutation in the IgVL gene was detected in 2 additional PTLDs carrying an out-of-frame IgVH rearrangement (cases 2892 and 3526; Table 3).
Frequency and molecular features of BCL6 mutations in monoclonal B-cell PTLDs
All 52 samples of PTLDs were subjected to DNA sequence analysis of BCL6 5′ noncoding sequences. Results are summarized in Table 6. Overall, mutations were detected in 26 (50.0%) of 52 PTLDs, including 3 (25.0%) of 12 P-PTLDs, 10 (62.5%) of 16 DLBCLs centroblastic, 10 (58.8%) of 17 DLBCLs immunoblastic, and 3 of 4 BL/BLLs.
Frequency and molecular profile of BCL-6 mutations in monoclonal B-cell PTLDs
Case . | Histology . | Mutation . |
|---|---|---|
| 2916 | P-PTLD | 449T>C, 645G>C, 823T>A, 978G>A |
| 3519 | P-PTLD | 445C>G, 477T>C, 564T>C, 863A>G |
| 3521 | P-PTLD | 443A>T, 506A>G, 668A>G, 802A>G, 803C>G, 837T>G |
| 2909 | DLBCL, centroblastic | 464T>A, 707T>G, 724T>A, 977A>G, 1102T>G |
| 3459 | DLBCL, centroblastic | 855G>A |
| 3466 | DLBCL, centroblastic | 479G>A |
| 3467 | DLBCL, centroblastic | 453A>C, 459G>C, 506A>C, 692G>C |
| 3518 | DLBCL, centroblastic | 523C>T, 1101T>C |
| 3522 | DLBCL, centroblastic | 802A>G, 831G>C |
| 3523 | DLBCL, centroblastic | 441G>A, 826G>A |
| 3524 | DLBCL, centroblastic | 441G>A |
| 3527 | DLBCL, centroblastic | 680C>G, 876C>T |
| 3528 | DLBCL, centroblastic | 445C>G |
| 2616 | DLBCL, immunoblastic | 1031G>C |
| 2802 | DLBCL, immunoblastic | 863A>G, 1060T>G |
| 2892 | DLBCL, immunoblastic | 479G>C |
| 2895 | DLBCL, immunoblastic | 759G>C, 917C>G, 937T>C |
| 2911 | DLBCL, immunoblastic | 469A>T, 669T>A |
| 2915 | DLBCL, immunoblastic | 480C>G, 837T>G, 975G>A, 1007T>C |
| 3469 | DLBCL, immunoblastic | 820G>C, 821G>T |
| 3476 | DLBCL, immunoblastic | 567T>G, 570T>C, 608C>T, 624T>G, 626T>A, 889Δ17 bp |
| 3520 | DLBCL, immunoblastic | 542A>G, 656G>A, 821G>A, 876C>T, 946C>T, 948T>C, 974G>A, 975G>A, 1052A>C, 1065A>G, 1096T>A |
| 3529 | DLBCL, immunoblastic | 1026T>A |
| 2617 | BL/BLL | 734G>A, 747A>C, 748G>A |
| 3463 | BL/BLL | 476C>G, 496T>C, 519G>C, 537T>A, 635T>C, 702C>G, 724T>C |
| 3526 | BL/BLL | 957A>T |
Case . | Histology . | Mutation . |
|---|---|---|
| 2916 | P-PTLD | 449T>C, 645G>C, 823T>A, 978G>A |
| 3519 | P-PTLD | 445C>G, 477T>C, 564T>C, 863A>G |
| 3521 | P-PTLD | 443A>T, 506A>G, 668A>G, 802A>G, 803C>G, 837T>G |
| 2909 | DLBCL, centroblastic | 464T>A, 707T>G, 724T>A, 977A>G, 1102T>G |
| 3459 | DLBCL, centroblastic | 855G>A |
| 3466 | DLBCL, centroblastic | 479G>A |
| 3467 | DLBCL, centroblastic | 453A>C, 459G>C, 506A>C, 692G>C |
| 3518 | DLBCL, centroblastic | 523C>T, 1101T>C |
| 3522 | DLBCL, centroblastic | 802A>G, 831G>C |
| 3523 | DLBCL, centroblastic | 441G>A, 826G>A |
| 3524 | DLBCL, centroblastic | 441G>A |
| 3527 | DLBCL, centroblastic | 680C>G, 876C>T |
| 3528 | DLBCL, centroblastic | 445C>G |
| 2616 | DLBCL, immunoblastic | 1031G>C |
| 2802 | DLBCL, immunoblastic | 863A>G, 1060T>G |
| 2892 | DLBCL, immunoblastic | 479G>C |
| 2895 | DLBCL, immunoblastic | 759G>C, 917C>G, 937T>C |
| 2911 | DLBCL, immunoblastic | 469A>T, 669T>A |
| 2915 | DLBCL, immunoblastic | 480C>G, 837T>G, 975G>A, 1007T>C |
| 3469 | DLBCL, immunoblastic | 820G>C, 821G>T |
| 3476 | DLBCL, immunoblastic | 567T>G, 570T>C, 608C>T, 624T>G, 626T>A, 889Δ17 bp |
| 3520 | DLBCL, immunoblastic | 542A>G, 656G>A, 821G>A, 876C>T, 946C>T, 948T>C, 974G>A, 975G>A, 1052A>C, 1065A>G, 1096T>A |
| 3529 | DLBCL, immunoblastic | 1026T>A |
| 2617 | BL/BLL | 734G>A, 747A>C, 748G>A |
| 3463 | BL/BLL | 476C>G, 496T>C, 519G>C, 537T>A, 635T>C, 702C>G, 724T>C |
| 3526 | BL/BLL | 957A>T |
P-PTLD indicates polymorphic PTLD; DLBCL, diffuse large B-cell lymphoma; and BL/BLL, Burkitt lymphoma/Burkitt-like lymphoma.
The overwhelming majority of mutations included single base-pair substitutions (n = 78), and only one deletion was observed (Table 6). The average frequency of mutation, calculated taking into account only mutated cases, ranged from 0.676 × 10-3 bp to 7.44 × 10-3 bp. The mean frequency of mutation was similar throughout the clinico-pathologic categories of monoclonal B-cell PTLDs. Of the 78 single base-pair substitutions observed, 38 were transitions and 40 were transversions, yielding a transition/transversion ratio of 0.95.
Expression of BCL6, MUM1, and CD138 in monoclonal B-cell PTLDs
There were 44 monoclonal B-cell PTLDs, including 12 P-PTLDs, 14 DLBCLs centroblastic, 15 DLBCLs immunoblastic, and 3 DLBCLs anaplastic were analyzed for the expression pattern of BCL6, MUM1, and CD138. Representative examples are shown in Figure 2. Expression of BCL6 was detected in 10 (22.7%) of 44 PTLDs and clustered with DLBCLs centroblastic (10/14; 71.4%; P < .01), whereas it was consistently absent in P-PTLDs (0/12), DLBCLs immunoblastic (0/15), and DLBCLs anaplastic (0/3). Expression of MUM1 was detected in 34 (75.6%) of 45 PTLDs and preferentially clustered with P-PTLDs (11/12; 91.7%), DLBCLs immunoblastic (15/15; 100%), and DLBCLs anaplastic (3/3; 100%), whereas MUM1 expression was restricted to a fraction of DLBCLs centroblastic (5/14; 35.7%). Expression of CD138 was detected in 13 (29.5%) of 44 PTLDs, including 3 (25.0%) of 12 P-PTLDs, 8 (53.3%) of 15 DLBCLs immunoblastic, 1 (7.14%) of 14 DLBCLs centroblastic, and 1 of 3 DLBCLs anaplastic.
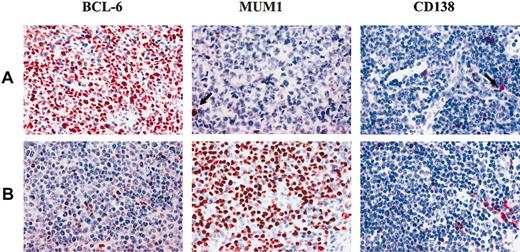
Expression of BCL6, MUM1, and CD138 in monoclonal B-cell PTLDs. (A) Diffuse large B-cell lymphoma with centroblastic morphology displaying the BCL6+/MUM1-/CD138- phenotypic pattern. Medium-sized tumor cells show a nuclear staining pattern with the anti-BCL6 MoAb. Residual plasma cells show nuclear brown staining for anti-MUM1 antibody and cytoplasmic red staining for anti-CD138 MoAb (arrows). (B) Diffuse large B-cell lymphoma with centroblastic morphology displaying the BCL-6-/MUM1+/CD138- phenotypic pattern. Most neoplastic cells show strong nuclear immunoreactivity with the anti-MUM1 antibody. No BCL6 expression is detectable in the neoplastic population; residual plasma cells show cytoplasmic staining for anti-CD138 MoAb. Paraffin-embedded tissue sections, immunoperoxidase method (BCL6, MUM1), APAAP method (CD138), hematoxylin counterstain. Original magnification, × 250.
Expression of BCL6, MUM1, and CD138 in monoclonal B-cell PTLDs. (A) Diffuse large B-cell lymphoma with centroblastic morphology displaying the BCL6+/MUM1-/CD138- phenotypic pattern. Medium-sized tumor cells show a nuclear staining pattern with the anti-BCL6 MoAb. Residual plasma cells show nuclear brown staining for anti-MUM1 antibody and cytoplasmic red staining for anti-CD138 MoAb (arrows). (B) Diffuse large B-cell lymphoma with centroblastic morphology displaying the BCL-6-/MUM1+/CD138- phenotypic pattern. Most neoplastic cells show strong nuclear immunoreactivity with the anti-MUM1 antibody. No BCL6 expression is detectable in the neoplastic population; residual plasma cells show cytoplasmic staining for anti-CD138 MoAb. Paraffin-embedded tissue sections, immunoperoxidase method (BCL6, MUM1), APAAP method (CD138), hematoxylin counterstain. Original magnification, × 250.
Table 7 summarizes the frequency of the phenotypic profiles identified in monoclonal B-cell PTLDs. Overall, the BCL6+/MUM1-/+/CD138- profile selectively associated with DLBCLs (10/32; 31.2%), and in particular with DLBCLs centroblastic (10/14; 71.4%) (Table 7). Conversely, the BCL6-/MUM1+/CD138- profile associated with 8 (66.7%) of 12 P-PTLDs and with 10 (31.2%) of 32 DLBCLs, which included 7 (46.7%) of 15 DLBCLs immunoblastic. Finally, the BCL6-/MUM1+/CD138+ profile was expressed by 3 (25.0%) of 12 P-PTLDs and by 10 (31.2%) of 32 DLBCLs, comprising 8 (53.3%) of 15 DLBCLs immunoblastic (Table 7). There were 3 cases that did not express BCL6, MUM1, or CD138 (Table 7).
Expression of BCL6, MUM1, and CD138 in monoclonal B-cell PTLDs
Histology . | BCL6+/MUM1−/+/CD138− positive/tested (%) . | BCL6−/MUM1+/CD138− positive/tested (%) . | BCL6−/MUM1+/CD138+ positive/tested (%) . | BCL6−/MUM1−/CD138− positive/tested (%) . |
|---|---|---|---|---|
| All PTLDs | 10/44 (22.7) | 18/44 (40.9) | 13/44 (29.5) | 3/44 (6.82) |
| P-PTLD | 0/12 | 8/12 (66.7) | 3/12 (25.0) | 1/12 (8.33) |
| DLBCL (all variants) | 10/32 (31.2) | 10/32 (31.2) | 10/32 (31.2) | 2/32 (6.25) |
| DLBCL, centroblastic | 10/14 (71.4)* | 1/14 (7.14) | 1/14 (7.14) | 2/14 (14.3) |
| DLBCL, immunoblastic | 0/15 | 7/15 (46.7) | 8/15 (53.3) | 0/15 |
| DLBCL, anaplastic | 0/3 | 2/3 (66.7) | 1/3 (33.3) | 0/3 |
Histology . | BCL6+/MUM1−/+/CD138− positive/tested (%) . | BCL6−/MUM1+/CD138− positive/tested (%) . | BCL6−/MUM1+/CD138+ positive/tested (%) . | BCL6−/MUM1−/CD138− positive/tested (%) . |
|---|---|---|---|---|
| All PTLDs | 10/44 (22.7) | 18/44 (40.9) | 13/44 (29.5) | 3/44 (6.82) |
| P-PTLD | 0/12 | 8/12 (66.7) | 3/12 (25.0) | 1/12 (8.33) |
| DLBCL (all variants) | 10/32 (31.2) | 10/32 (31.2) | 10/32 (31.2) | 2/32 (6.25) |
| DLBCL, centroblastic | 10/14 (71.4)* | 1/14 (7.14) | 1/14 (7.14) | 2/14 (14.3) |
| DLBCL, immunoblastic | 0/15 | 7/15 (46.7) | 8/15 (53.3) | 0/15 |
| DLBCL, anaplastic | 0/3 | 2/3 (66.7) | 1/3 (33.3) | 0/3 |
P-PTLD indicates polymorphic PTLD; DLBCL, diffuse large B-cell lymphoma; and BL/BLL, Burkitt lymphoma/Burkitt-like lymphoma.
Of the cases, 8 displayed the BCL6+/MUM1−/CD138− phenotype and 2 displayed the BCL6+/MUM1+/CD138− phenotype.
Viral infection in monoclonal B-cell PTLDs
By EBER ISH, EBV infection was detected in 30 (57.7%) of 52 monoclonal B-cell PTLDs, including 9 (75.0%) of 12 P-PTLDs, 18 (50.0%) of 36 DLBCLs, and 3 (75.0%) of 4 BL/BLLs (Table 1). Among DLBCLs, EBV infection occurred in 3 (18.8%) of 16 DLBCLs centroblastic, 13 (76.5%) of 17 DLBCLs immunoblastic, and 2 (66.7.0%) of 3 DLBCLs anaplastic (Table 1). Comparison of EBV positivity with the presence of antigen stimulation and selection revealed that viral infection occurred both in PTLDs stimulated/selected by antigen (44.4%) and in PTLDs without evidence of antigen stimulation and selection (46.2%) (Tables 1,4).
Among EBER-positive PTLDs, most P-PTLDs (6/8; 75.0%) displayed the LMP1+/EBNA2+ (latency III) phenotype, whereas the remaining 2 P-PTLDs were LMP1+/EBNA2- (latency II). Among EBV-positive DLBCLs, all (n = 3) DLBCLs centroblastic and 4 of 12 DLBCLs immunoblastic displayed the LMP1-/EBNA2- (latency I) phenotype. The remaining cases of DLBCLs immunoblastic and DLBCLs anaplastic associated either with the LMP1+/EBNA2+ phenotype (3/12 DLBCLs immunoblastic and 1/2 DLBCLs anaplastic) or with the LMP1+/EBNA2- pattern (5/12 DLBCLs immunoblastic and 1/2 DLBCLs anaplastic). In all monoclonal B-cell PTLD categories, the expression of LMP1 and/or EBNA2 was mutually exclusive with expression of BCL6 (not shown).
HHV-8 DNA sequences were scored negative in all cases tested (not shown).
Discussion
This study aimed at a comprehensive investigation of the molecular histogenesis of both EBV-positive and EBV-negative monoclonal B-cell PTLDs. By applying a consolidated panel of genetic and phenotypic markers of B-cell lymphoma histogenesis to a series of 52 monoclonal B-cell PTLDs, we show that most of these lymphomas derive from GC-experienced B cells. Despite this common origin, monoclonal B-cell PTLDs reflect heterogeneous stages of B-cell maturation and different degrees of immunologic competence of B cells. These results expand our knowledge of PTLD histogenesis and pathogenesis and may be of relevance for a proper understanding of disease heterogeneity.
In accordance with a recent report on EBV-positive PTLDs,22 our results show that SHM of IgVH genes occurs in approximately 90% monoclonal B-cell PTLDs, indicating that malignant transformation targets GC B cells and their descendants both in EBV-positive and EBV-negative monoclonal B-cell PTLDs. These same cellular subsets also give rise to most other B-cell lymphomas in immunocompetent hosts and in immunodeficiency settings other than posttransplantation, including AIDS and primary immunodeficiencies.13-15,20-22 Among monoclonal B-cell PTLDs, derivation from GC-related B cells occurs independent of type of transplanted organ, interval between transplantation and lymphoma, histology, and site of origin of the lymphoma.
Despite a common derivation from GC-experienced B cells, the precise histogenesis of single cases of monoclonal B-cell PTLDs displays a certain degree of molecular and phenotypic heterogeneity that may be distinguished into 3 main categories. PTLDs belonging to the first histogenetic category conceivably reflect B cells residing within the GC and actively experiencing the GC reaction. These PTLDs associate with ongoing activity of the SHM process and are morphologically classified as DLBCLs centroblastic or as BL/BLLs. Although the GC-derivation of these PTLDs is further reinforced by expression of the BCL6 protein, we cannot formally exclude that BCL6 expression may be secondary to genomic aberrations of the BCL6 gene. A second category of monoclonal B-cell PTLDs reflects the BCL6-/MUM1+/CD138- phenotype and comprises the P-PTLD and DLBCL immunoblastic morphotypes. The BCL6-/MUM1+/CD138- profile suggests that this PTLD subset is related to B cells that have concluded the GC reaction but have not yet undergone terminal differentiation. Remarkably, this histogenetic profile is the most common among both early- and late-onset PTLDs, but is rare among AIDS-related lymphomas, reinforcing the notion that monoclonal B-cell PTLDs may be biologically different from lymphomas of similar histologies arising in HIV-positive hosts.20,22 The third histogenetic category of monoclonal B-cell PTLDs is reminiscent of post-GC and preterminally differentiated B cells that show the BCL6-/MUM1+/CD138+ phenotype and, if EBV-positive, express the LMP1 antigen. These PTLDs are morphologically represented by either P-PTLDs or DLBCLs immunoblastic and mimic a histogenetic profile frequently found in lymphomas arising in the context of AIDS.20,22 The post-GC origin of a significant fraction of PTLDs is also documented by expression of the Src homology 2-containing protein phosphatase 1, which associates with post-GC B cells.44
A sizeable subset of monoclonal B-cell PTLDs arising from GC-related B cells is characterized by detection solely of nonfunctional rearrangements of IgVH genes and/or IgVL genes. Crippling mutations, introducing a stop codon in a previously functional rearrangement, account for the majority of these sterile rearrangements in IgVH and/or IgVL genes of PTLDs and abrogate Ig expression in the lymphoma clone. Because a functional B-cell receptor (BCR) is required for survival of normal B cells during GC transit and may be necessary also for many lymphomas, it is conceivable that PTLD cells are rescued from apoptosis through mechanisms independent of antigen priming.13-15 Such rescue may imply one or more antiapoptotic pathways. First, all PTLDs carrying sterile IgVH and/or IgVL rearrangements express the EBV-encoded LMP1 antigen, which inhibits apoptosis through up-regulation of BCL-2.45 Indeed, BCL-2 was expressed by this PTLD subset (not shown). Apoptotic rescue of EBV-positive PTLDs with crippling IgV mutations might also potentially occur through virus-encoded LMP-2A, which allows normal B-cell developmental checkpoints to be bypassed and is capable of providing B cells with survival signals in the absence of normal BCR signaling.46-48 An additional pathway promoting survival of PTLDs with crippling IgV mutations may involve inactivation of the death-associated protein kinase (DAP-k) gene, which occurs in almost 90% of monoclonal B-cell PTLDs (D.R. et al, unpublished observation, July 2003). DAP-k is a proapoptotic serine-threonine kinase involved in the extrinsic pathway of apoptosis initiated by gamma interferon (INFγ), tumor necrosis factor α (TNFα), and Fas ligand, and its inactivation through promoter hypermethylation prevents apoptosis triggered by death receptors.49,50
A small group of monoclonal B-cell PTLDs lack clues of IgVH SHM. These cases tend to arise early after transplantation, may belong to both P-PTLD and DLBCL morphotypes, consistently carry EBV infection, and mimic a post-GC phenotypic profile. According to conventional models of B-cell lymphoma histogenesis, monoclonal B-cell PTLDs with germ-line IgVH genes would derive from truly pre-GC B cells.13-15 However, the view that all B-cell lymphoproliferations with germ-line IgVH genes derive from pre-GC B cells has been recently challenged by gene profiling analysis of B-cell chronic lymphocytic leukemias devoid of IgVH mutations.51,52 Therefore, similar to a fraction of B-cell chronic lymphocytic leukemias, an alternative histogenetic origin of PTLDs with germ-line IgVH genes may be represented by B cells that have transited through the GC but have been impaired in exerting a full GC reaction, and consequently, in their acquisition of IgVH somatic mutations.51,52 Notably, EBV-positive lymphomas derived from immunologically naive B cells but mimicking a post-GC phenotype are also found in the context of AIDS, whereas they are extremely rare in other settings, suggesting that development of EBV-positive lymphoproliferations displaying an immunologically naive/phenotypically differentiated B-cell profile may be specifically related to the host's immune disfunction.20-23 Their survival during GC transit might be attributed to LMP1, which in fact is expressed by this subset of PTLDs, or to other molecular lesions preventing apoptosis.14
Molecular prognostic markers, including mutations of the BCL6 proto-oncogene, have proved useful in refining the prognostication of PTLDs.11,12 In this respect, knowledge of the molecular histogenesis of PTLDs may potentially further contribute to refine the distinction of PTLDs into more homogeneous categories with prognostic relevance. An appropriate response to this issue will come from studies of prospective series of PTLDs that have been treated homogeneously. Also, because this study focused on monoclonal B-cell PTLDs, future investigations are required to clarify the histogenesis of other PTLD categories, namely polyclonal and oligoclonal PTLDs.
Prepublished online as Blood First Edition Paper, August 7, 2003; DOI 10.1182/blood-2003-05-1683.
Supported by Cofin 2002—Ministero dell'Istruzione, Università e Ricerche (MIUR), Rome, Italy; Ricerca Finalizzata 2002, Ministero della Salute, Rome, Italy; Progetto Strategico Oncologia, Consiglio Nazionale delle Ricerche (CNR)-MIUR; Programma Nazionale di Ricerca sull'AIDS—Progetto Patologia, Clinica e Terapia dell'AIDS, Istituto Superiore di Sanità (ISS), Rome, Italy; and by Fondazione Cassa di Risparmio di Torino (CRT), Torino, Italy. D.C., M.C., E.B., and C.D. have been supported by fellowships from Fondazione “Piera Pietro Giovanni Ferrero,” Alba, Italy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal