Abstract
Familial clustering of malignancies provides a unique opportunity to identify molecular causes of cancer. Polycythemia vera (PV) is a myeloproliferative disorder due to an unknown somatic stem cell defect that leads to clonal myeloid hyperproliferation. We studied 6 families with PV. The familial predisposition to PV appears to follow an autosomal dominant inheritance pattern with incomplete penetrance. All examined females informative for a transcriptional clonality assay had clonal hematopoiesis. We excluded linkage between PV and a number of previously proposed candidate disease loci (c-mpl, EPOR, 20q, 13q, 5q, 9p). Therefore, mutations at these loci are unlikely primary causes of familial PV. The finding of erythropoietin-independent erythroid progenitors in healthy family members indicated the presence of the PV stem cell clone in their hematopoiesis. This finding, together with clonal hematopoiesis in the affected individuals, supports the hypothesis of multiple genetic defects involved in the early pathogenesis of PV. (Blood. 2003;102:3793-3796)
Introduction
Sporadic myeloproliferative disorders (MPDs) are due to an acquired mutation of a single hematopoietic cell resulting in clonal circulating myeloid progeny.1 Accumulation of erythrocytes is a hallmark of polycythemia vera (PV), while the accumulation of platelets, neutrophils, basophils, and eosinophils is variable. The molecular lesion responsible for PV is unknown. The erythroid progenitors in PV form erythropoietin-independent erythroid colonies (EECs) in clonogenic cultures in the absence of exogenous erythropoietin (Epo).2 This unique feature permits the distinction of PV from other primary polycythemias with or without a family history.3,4 The EEC assay also identifies an early PV stage that lacks the full PV phenotype and allows differentiation of PV in those patients initially presenting with thrombocytosis from those individuals having essential thrombocythemia.5 Familial clustering of polycythemia is seen in congenital polycythemic states including primary familial and congenital polycythemia, Chuvash polycythemia, high oxygen-affinity globin mutants, and biphospho-glycerate mutase deficiency.6 However, few case reports of familial occurrence of PV have been reported.7-10
Study design
All affected family members had classical diagnosis of PV based on the PV Study Group criteria.11 All studies were performed under approved institutional review board protocols (Baylor College of Medicine and University of Alabama at Birmingham), and all subjects included in this study provided written consent to perform DNA and cell culture studies on their blood samples.
Results and discussion
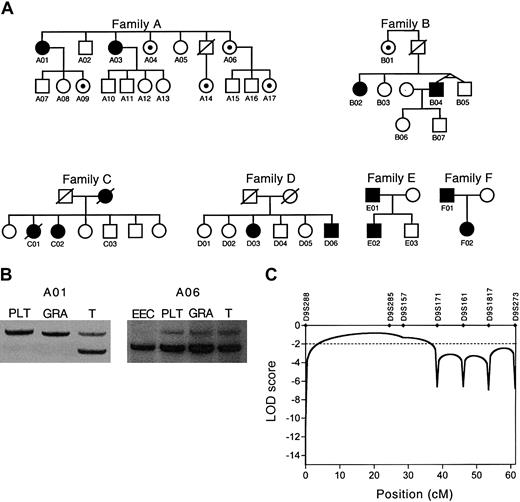
We studied 6 white families of heterogeneous ethnic background, each with multiple members with PV (Figure 1A). We detected EECs in peripheral blood cultures in all the affected family members in all families. The clinical findings are summarized in Table 1; these results of clinical findings represent all of the available patients and these were not preselected. In addition to the affected members with the full PV phenotype, we tested family members without PV for the presence of EECs in their peripheral blood. We identified subjects in families A and B who had no clinical signs or symptoms of PV but EECs were present in their peripheral blood (subjects A04, A06, A09, A14, A17, B01). Clonality was demonstrated in the informative females with full PV phenotype (Table 1; Figure 1B). The members with partial phenotype (EECs only) were polyclonal, indicating the contribution of normal stem cells to productive hematopoiesis, and therefore, lack of clinical symptoms of PV. To examine whether the presence of EECs is due to a somatic mutation or an inherited mutant gene, we isolated the Epo-independent erythroid cell population and analyzed its clonality (this sample consisted of approximately 500-1000 blast-forming units-erythroid [BFUEs] harvested from 10 methylcellulose plates followed by magnetic-activated cell sorting for glycophorin A. Each BFUE consists of approximately 800-2000 proerythroblasts). We detected clonal EECs in one informative female who had otherwise polyclonal hematopoiesis; please note that the T cells' X-chromosome allelic usage appears skewed; however, the observed skewing is within the range that we reported in normal hematopoietic progeny (Figure 1B).16
Analyses of 6 families with PV. (A) Pedigrees of 6 families with polycythemia vera (PV). Filled symbols indicate affected individuals. In families A and B, black dots indicate hematologically normal family members with Epo-independent erythroid cells detectable in their peripheral blood cultures. (B) Clonality analysis of individuals A01 and A06 using the IDS gene exonic polymorphism. RNA isolated from T lymphocytes (T), granulocytes (GRA), platelets (PLT), and Epo-independent erythroid cells (EEC) were used for the analysis. The presence of only one expressed allele of the IDS gene in platelets, granulocytes, and Epo-independent erythroid cells is consistent with clonal origin of cells. (C) Exclusion of linkage between chromosome 9p microsatellite markers and PV. LOD scores less than -2.0 satisfy the criteria of exclusion of linkage. The dotted line depicts the customary evidence for genetic exclusion of linkage. cM indicates centimorgan.
Analyses of 6 families with PV. (A) Pedigrees of 6 families with polycythemia vera (PV). Filled symbols indicate affected individuals. In families A and B, black dots indicate hematologically normal family members with Epo-independent erythroid cells detectable in their peripheral blood cultures. (B) Clonality analysis of individuals A01 and A06 using the IDS gene exonic polymorphism. RNA isolated from T lymphocytes (T), granulocytes (GRA), platelets (PLT), and Epo-independent erythroid cells (EEC) were used for the analysis. The presence of only one expressed allele of the IDS gene in platelets, granulocytes, and Epo-independent erythroid cells is consistent with clonal origin of cells. (C) Exclusion of linkage between chromosome 9p microsatellite markers and PV. LOD scores less than -2.0 satisfy the criteria of exclusion of linkage. The dotted line depicts the customary evidence for genetic exclusion of linkage. cM indicates centimorgan.
Summary of clinical findings
Subject . | Diagnosis . | Sex . | Age at diagnosis, y . | Hct . | Hgb, g/L . | PLT, K/μL . | Splenomegaly . | Karyotype . | Clonality assay . | EEC . |
|---|---|---|---|---|---|---|---|---|---|---|
| A01 | PV | F | 58 | .55 | 180 | 443 | + | 9pLOH | CL | Present |
| A03 | PV | F | 62 | .64 | 197 | 190 | + | Normal | CL | Present |
| A04 | Normal | F | — | .422 | 147 | 310 | − | ND | ND | Present |
| A06 | Normal | F | — | .467 | 162 | 204 | − | ND | PO, EEC-CL | Present |
| B02 | PV | F | 31 | .457 | 127 | 438 | − | 9pLOH | CL | Present |
| B04 | PV | M | 33 | .484 | 162 | 284 | − | Normal | ND | Present |
| C01 | PV | F | 80 | .542 | 177 | 710 | ND | ND | ND | ND |
| C02 | PV | F | 72 | .409 | 136 | 2430 | − | ND | CL | Present |
| C03 | PV | F | 66 | .359 | 120 | 1995 | + | Normal | ND | Present |
| D03 | PV | F | 29 | .46 | 162 | 1190 | − | ND | ND | ND |
| D06 | PV | M | 48 | .55 | 195 | 910 | − | Normal | ND | ND |
| E01 | PV | M | 25 | .573 | 197 | 729 | + | Normal | ND | Present |
| E02 | PV | M | 5 | .61 | 196 | 614 | + | ND | ND | Present |
| F01 | PV | M | 52 | .566 | 182 | 65 | − | ND | ND | Present |
| F02 | PV | F | 30 | .50 | 164 | 168 | − | Normal | CL | Present |
Subject . | Diagnosis . | Sex . | Age at diagnosis, y . | Hct . | Hgb, g/L . | PLT, K/μL . | Splenomegaly . | Karyotype . | Clonality assay . | EEC . |
|---|---|---|---|---|---|---|---|---|---|---|
| A01 | PV | F | 58 | .55 | 180 | 443 | + | 9pLOH | CL | Present |
| A03 | PV | F | 62 | .64 | 197 | 190 | + | Normal | CL | Present |
| A04 | Normal | F | — | .422 | 147 | 310 | − | ND | ND | Present |
| A06 | Normal | F | — | .467 | 162 | 204 | − | ND | PO, EEC-CL | Present |
| B02 | PV | F | 31 | .457 | 127 | 438 | − | 9pLOH | CL | Present |
| B04 | PV | M | 33 | .484 | 162 | 284 | − | Normal | ND | Present |
| C01 | PV | F | 80 | .542 | 177 | 710 | ND | ND | ND | ND |
| C02 | PV | F | 72 | .409 | 136 | 2430 | − | ND | CL | Present |
| C03 | PV | F | 66 | .359 | 120 | 1995 | + | Normal | ND | Present |
| D03 | PV | F | 29 | .46 | 162 | 1190 | − | ND | ND | ND |
| D06 | PV | M | 48 | .55 | 195 | 910 | − | Normal | ND | ND |
| E01 | PV | M | 25 | .573 | 197 | 729 | + | Normal | ND | Present |
| E02 | PV | M | 5 | .61 | 196 | 614 | + | ND | ND | Present |
| F01 | PV | M | 52 | .566 | 182 | 65 | − | ND | ND | Present |
| F02 | PV | F | 30 | .50 | 164 | 168 | − | Normal | CL | Present |
Hct indicates hematocrit; Hgb, hemoglobin; CL, clonal; —, not present; ND, not determined; PO, polyclonal; and EEC, Epo-independent colonies.
The inheritance pattern of familial PV is compatible with an autosomal dominant trait with decreased penetrance. If the finding of EECs is considered as an early sign of the PV phenotype, the penetrance increases. Of the 6 families, 4 (A-D) were used for linkage analysis since these consisted of at least 2 affected siblings. It is possible that the clustering of PV in families E and F could be by chance only and that we could have had a selection bias to recruit these families since families of unusual polycythemic disorders have been referred to us for more than a decade. For these pedigrees, we calculated the power to detect linkage using the SIMLINK software (http://www.sph.umich.edu/group/statgen/software),17 which predicted a maximum logarithm of odds ratio (LOD) score of 4.4 for the given pedigree structures. The simulated LOD score further increased to 5.2 when the subjects positive for EECs were considered as affected (data not shown). Using these families, we could examine a number of candidate loci that were previously proposed to play a role in MPD, or PV in particular. We analyzed the linkage between the PV phenotype and the commonly deleted regions on chromosomes 20q, 13q, and 5q that were found as genetic aberrations in PV.18,19 In addition, the thrombopoietin receptor (c-mpl) and Epo receptor (EPOR) genes have been proposed in the pathogenesis of myeloproliferative disorders including PV.20,21 Recently, we reported the presence of LOH involving chromosome 9p as the most common clonal defect in sporadic PV.12 We detected LOH on chromosome 9p in subjects A01 and B02 (Table 1). We performed linkage analysis using microsatellite markers mapping to these loci. Only the affected subjects with the full PV phenotype were considered “affected” in the linkage analysis. Since LOD scores less than -2.0 are considered exclusion of linkage, we could exclude linkage between the PV phenotype and all the tested loci. LOD scores for the EPOR and c-mpl genes were -3.16 and -2.24, respectively. The commonly deleted regions found in sporadic MPD were also convincingly excluded with LOD scores of -4.40 for 20q, -4.79 for 13q, and -6.27 for 5q. We also fully excluded the chromosome 9p region of LOH (Figure 1C). These results suggest that the frequently observed somatic mutations in MPD involving the chromosomal regions on 20q, 13q, 5q, and 9p are secondary genetic changes and do not target the primary PV locus.
The clinical analysis of the affected family members confirmed that they are phenotypically identical to sporadic PV. Familial occurrence of PV provides a unique insight into the stages of PV since we could identify affected members in an early stage of the disease using the EEC assay. This is not possible in sporadic PV since individuals are identified only when symptomatic. Thrombocythemia was shown to be the first abnormality seen in some PV subjects,5 and interestingly in families C and D, thrombocythemia occurred prior to elevation of hematocrit. In all the affected informative females, we observed clonal circulating myeloid cells as seen in sporadic PV; in some, clonality could not be determined because (1) patients were genotyped for exonic polymorphisms of active X-chromosome genes used for the clonality assays and were not informative, (2) there was failure to get informed consent for this follow-up study, or (3) patients were no longer available or willing to participate in our study.4 Clonal hematopoiesis is a marker of fully developed PV. In families A and B, we observed hematologically normal subjects with EECs present in their peripheral blood. In one of these cases we proved the clonal origin of the Epo-independent cells, but the rest of the myeloid cells were polyclonal. Thus, it is possible that in presymptomatic PV within polyclonal hematopoiesis the PV stem cell clone may be present but its contribution to blood production is limited. At this stage, the progeny of the PV clone can be detectable by the EEC assay. In the symptomatic stage, the PV clone loses regulation and expands, and clonal hematopoiesis appears. It remains to be established if in the individuals with PV who present with thrombocytosis prior to elevation of hematocrit, thrombocytosis precedes (such as seen in subject C02 and in one female with sporadic PV we studied previously) or follows establishment of clonal circulating hematopoietic progeny.
The finding of clonal hematopoiesis suggests that acquired mutations are part of the disease etiology of familial PV. The presence of both inherited and acquired mutations in familial PV allows several interpretations of the disease etiology. As seen in other familial predispositions to cancer (such as retinoblastoma), a mutant nonfunctional copy of the gene is inherited in the families followed by an acquired mutation of the remaining wild-type allele. The disease initiates after both alleles of the gene are mutated (mutation of one allele being inherited and the mutation of the second allele being acquired). Thus, in families with PV predisposition, PV phenotype will be expected to develop at an earlier age than that seen in sporadic PV. This has indeed been the case in families B, E, and F. It is possible that the stem cell clone established in this initial stage undergoes further mutagenesis resulting in acceleration of clonal expansion. An alternative interpretation assumes mutations in 2 or more genes, mutation of one gene being inherited and mutation(s) of other gene(s) being acquired. However, if more than one gene can contribute to the development of a final PV phenotype this will make PV a genetically heterogeneous disorder and some of the positional cloning data interpretation would not be valid. Loss of gene function is a necessary component of the first model, whereas in the second model, gain of function mutations may also be present together with loss of function mutations. In both models, the acquired mutations are responsible for the presence of clonal hematopoiesis. The presence of incomplete penetrance, observed in the families, is compatible with both models.
The chromosomal localization of the “primary PV mutation” remains unknown. The PV phenotype did not show linkage to any of the loci implicated in PV to date. Families with multiple members with PV should prove fundamental in identification of the PV predisposition gene as they offer the possibility for genome-wide linkage analysis and positional cloning.
Prepublished online as Blood First Edition Paper, June 26, 2003; DOI 10.1182/blood-2003-03-0885.
Supported by the Myeloproliferative Disease Foundation (MPD Foundation).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Hanneke Kluin-Nelemans and Dr Rangaswamy Govindarajan for providing blood samples and Valerie Irvin and Yongli Guan for their technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal