Abstract
To define specific pathways important in the multistep transformation process of normal plasma cells (PCs) to monoclonal gammopathy of uncertain significance (MGUS) and multiple myeloma (MM), we have applied microarray analysis to PCs from 5 healthy donors (N), 7 patients with MGUS, and 24 patients with newly diagnosed MM. Unsupervised hierarchical clustering using 125 genes with a large variation across all samples defined 2 groups: N and MGUS/MM. Supervised analysis identified 263 genes differentially expressed between N and MGUS and 380 genes differentially expressed between N and MM, 197 of which were also differentially regulated between N and MGUS. Only 74 genes were differentially expressed between MGUS and MM samples, indicating that the differences between MGUS and MM are smaller than those between N and MM or N and MGUS. Differentially expressed genes included oncogenes/tumor-suppressor genes (LAF4, RB1, and disabled homolog 2), cell-signaling genes (RAS family members, B-cell signaling and NF-κB genes), DNA-binding and transcription-factor genes (XBP1, zinc finger proteins, forkhead box, and ring finger proteins), and developmental genes (WNT and SHH pathways). Understanding the molecular pathogenesis of MM by gene expression profiling has demonstrated sequential genetic changes from N to malignant PCs and highlighted important pathways involved in the transformation of MGUS to MM. (Blood. 2003;102:4504-4511)
Introduction
Long-term follow-up of individuals with monoclonal gammopathy of uncertain significance (MGUS) has shown that approximately 1% of cases progress to symptomatic multiple myeloma (MM) annually.1 In addition to the classical clinical features, it is well recognized that toward the end of MM's natural history a more aggressive phase with peripheral blood involvement develops in a proportion of patients. This disease phase is associated with interleukin-6 (IL-6) independence and alteration in adhesion molecule expression.2 Molecular models attempting to describe the pathogenesis of myeloma combine the clinical data from the different disease phases with data from cytogenetics, immunoglobulin gene mutational analysis, and recurrent chromosomal abnormalities to generate a multistep model.2 Using this approach it is possible to recognize the transition of normal plasma cells through MGUS to myeloma and to define genetic changes associated with each of these steps.2-5
Sequencing of the immunoglobulin variable region has demonstrated that plasma cells (PCs) from individuals with MGUS show intraclonal variation, whereas no such variation is seen in myeloma.3 This finding is consistent with MGUS PCs being continually exposed to somatic hypermutation, whereas in myeloma a single PC clone becomes dominant and expands to replace the marrow. Studies using polymerase chain reaction (PCR) and fluorescent in situ hybridization (FISH) have demonstrated that translocations involving the switch region of chromosome 14 occur with equal frequency in MGUS and myeloma. It has, therefore, been suggested that class switch recombination events occur early in the disease process,6-8 with the most common translocations involving t(4;14), t(11;14), and t(14;16). The timing of interstitial deletions of chromosome 13 is more difficult to define because of the technical difficulties of chromosome analysis. FISH studies suggest that 13q deletions and aneuploidy are present in a proportion of MGUS cases; however, the exact timing of these abnormalities is unknown.7,8
Recent technological advances have enabled researchers to address changes in global gene expression by using microarrays, with the ultimate aim of developing molecular profiles associated with malignancy. The application of this approach to the analysis of the multistep pathogenesis of myeloma will allow us to understand how global gene expression patterns change during these distinct disease stages and may highlight pathways previously not thought to be important in the transformation process. We have applied microarray expression analysis to PCs isolated from healthy controls and malignant PCs isolated from individuals with MGUS and myeloma. We report that most gene expression changes occur during the transformation of normal PCs to malignant MGUS PCs, and we highlight a number of genes important in this progression including members of the Wingless (WNT) and Sonic pathways, which are developmental pathways not previously implicated in myeloma pathogenesis.
Patients, materials, and methods
Samples and plasma cell selection
Bone marrow aspirate samples were obtained after informed consent from healthy donors (N), individuals with MGUS, and patients with newly diagnosed MM. For gene array studies a series of bone marrow aspirates (N = 5, MGUS = 7, MM = 24) was analyzed. A further series (N = 6, MGUS = 6, MM = 7) was used for validation studies by reverse transcription-PCR (RT-PCR). After red blood cell lysis of bone marrow with 0.86% ammonium chloride, PCs were positively selected using CD138 microbeads and magnet-assisted cell sorting (MACS; Miltenyi Biotech, Bergisch Gladbach, Germany), according to the manufacturer's instructions. Purity (more than 95%) was assessed by morphology and flow cytometry (FACSort; BD Biosciences, San Jose, CA). Plasma cells were identified by CD38 and CD45 expression and by forward and side scatter characteristics. Flow cytometric analysis of MGUS cases demonstrated that more than 70% of plasma cells had a malignant phenotype (CD19-CD56+).9
RNA extraction, quantification, and amplification
RNA was extracted using commercially available kits (Qiagen [Valencia, CA] and Stratagene [La Jolla, CA]), according to the manufacturers' instructions. After DNAse treatment, an Abl PCR was performed to confirm the absence of DNA contamination. The amount of RNA isolated was estimated using a real-time quantitative PCR assay with a Pre-Designed Assay Reagent (PDAR; Applied Biosystems, Foster City, CA) and a standard curve. To have adequate material for hybridization, the starting RNA was amplified using a modified switch mechanism at the 5′ end of RNA transcripts (SMART) PCR protocol (BD Biosciences, San Jose, CA), in which a 5′ T7 polymerase promoter site was incorporated to create amplified cDNA compatible with downstream processing for the GeneChip system (Affymetrix, Santa Clara, CA). Briefly 0.05 to 1 μg total RNA was combined with an oligo dT-T7 RT primer and a template switch oligonucleotide before first-strand synthesis (20 nmol dithiothreitol, 10 nmol dNTP with PowerScript reverse transcriptase; BD Biosciences) at 42°C for 1 hour. The resultant first-strand SMART cDNA was then combined in a reaction containing 0.4 mM dNTPs, 1.5 mM MgCl2, 1 × PCR buffer, 0.1 μM T7 PCR primer, and 0.1 μM SMART PCR primer, according to the manufacturer's instructions. After a hot start (TaKaRa LA Taq, 1 minute, 95°C), thermal cycling conditions for the appropriate number of cycles were 95°C for 5 seconds, 65°C for 5 seconds, and 68°C for 6 minutes. Since PCR inevitably favors short sequences over long ones, it was necessary to assess the optimum number of PCR cycles (15, 18, 21, 24, or 27) for each sample so that the reaction could be terminated before overcycling of the shorter sequences. In accordance with the manufacturer's instructions, the optimum cycle number was determined to be one cycle less than when the products became visible on 1.2% agarose EtBr gel. After thermal cycling, the PCR products were cleaned using the QiaQuick PCR Purification Kit (Qiagen), according to the manufacturer's instructions. An additional Abl PCR was performed to reconfirm the absence of contaminating genomic DNA. Control experiments (quantitative RT-PCR and gene expression analysis) using cell lines to assess the reproducibility of the modified SMART method demonstrated linear amplification (Figure 1).
Comparison of unamplified and SMART-amplified expression profiles. The expression levels of 12 600 genes between unamplified and SMART-amplified myeloma cell line H929 demonstrated linear amplification.
Comparison of unamplified and SMART-amplified expression profiles. The expression levels of 12 600 genes between unamplified and SMART-amplified myeloma cell line H929 demonstrated linear amplification.
Gene expression profiling
Human genome U95AV2 GeneChip arrays (Affymetrix, Santa Clara, CA) containing probes for 12 600 expressed sequences were used for mRNA expression profiling. Biotinylated RNA was synthesized using the BioArray RNA transcript labeling kit (Enzo, Farmingdale, NY) with biotinylated ribonucleotides for 5 hours at 37°C. In vitro transcription products were purified using RNeasy columns (Qiagen). Biotinylated RNA was fragmented for 35 minutes at 94°C in 40 mM Tris acetate (pH 8.1), 100 mM potassium acetate, and 30 mM magnesium acetate. Arrays were hybridized with biotinylated in vitro transcription products for 16 hours at 45°C according to the manufacturer's instructions. The Fluidic Station 400 (Affymetrix) was used for washing, and a 3-step staining protocol was used to enhance detection of the hybridized biotinylated RNA—incubation with streptavidin-phycoerythrin conjugate, labeling with an antistreptavidin goat biotinylated antibody (Vector Laboratories, Burlingame, CA), and staining with streptavidin-phycoerythrin conjugate. Arrays were scanned by the Affymetrix fluorescence reader (Hewlett Packard). The excitation source was an argon ion laser, and emission was detected by a photomultiplier tube through a 570-nm long-pass filter. The raw image DAT data files were initially processed using Affymetrix GeneChip software (version 4) to create CEL files, and DNA-Chip Analyzer (dChip)10 was used for data quality checking and high-level analyses.
RT-PCR
SYBR Green real-time PCR (Applied Biosystems) was performed on a further series of selected plasma cell cDNA from 6 N, 6 MGUS, and 7 MM for frizzled related protein (FRZB) and selenoprotein P (SEPP1), genes that were shown to be differentially expressed in the N, MGUS, and MM groups in the microarray experiments. Primer sequences designed for intron/exon boundaries for FRZB and SEPP1, respectively, were F 5'-GGGCTATGAAGATGAGGAACGT, R 5'-ACCGAGTCGATCCTTCCACTT and F 5'-CATATGTAGAAGAAGCCATTAAGATTGC, R 5'-GCCAAAGATACACGTTTACAAAAGTC (Invitrogen, Paisley, United Kingdom). Thermal cycling conditions were 2 minutes at 50°C, 10 minutes at 95°C, and 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. RT-PCR was performed to determine the relative expression of XBP1 and XBP1s on the same patient samples and a series of MM cell lines. Primer sequences were F 5'-CCTTGTAGTTGAGAACCAGG, R 5'-GGGGCTTGGTATATATGTGG, and PCR was performed at an annealing temperature of 60°C for 36 cycles. Products were run on a 3% agarose gel containing ethidium bromide.
Statistical analysis
Array normalization and expression value calculation were performed using dChip,10 which is freely available to academic users at www.dchip.org. Invariant set normalization was used to normalize arrays at the probe cell level to make them comparable, and the model-based method was used for probe-selection and computing expression values.10,11 Model-based expression values with attached standard errors were used to compute group means of N, MGUS, and MM and the 90% lower confidence bound of -fold change between groups.11 Previous publications have highlighted concerns regarding errors in gene expression because of technical variation and batch differences12 ; therefore, initial analysis was based on a group of samples comprising 5 N, 5 MGUS, and 9 MM run in a single batch. Further analysis incorporating an additional 2 MGUS and 15 MM was in keeping with these initial results, though the gene lists were slightly smaller and compatible with a number of genes being related to technical rather than true disease-related differences (data not shown). The genes and gene lists discussed in this paper were identified in both sets of analyses.
Results
Unsupervised hierarchical clustering identifies 2 main groups
Experiments were initially performed in triplicate (n = 5) or duplicate (n = 7). In hierarchical clustering, replicate samples always lay together within the clustering tree (data not shown). This suggests that experiments were reproducible, and subsequently the remaining samples were run as single chip experiments. Within dChip arrays were opened as a single group. The initial data set (5 N, 5 MGUS, and 5 MM) and the expanded data set (5 N, 7 MGUS, and 24 MM) were analyzed in separate dChip sessions. After normalization and model-based expression to handle array outliers, the dChip filter genes function identified 125 genes with large variation across the samples (filtering criteria: [1.10 < SD/mean between < 10.00] P call % in the array used, 20% or greater). Hierarchical clustering using this gene list identified 2 main branches; one contained N samples only, and the other contained MGUS and MM samples. The MGUS/MM branch was further subdivided into 2 groups, one containing mainly MGUS samples, and the other comprising mainly MM samples (Figure 2A). Of the genes differentially expressed between the sample groups, approximately 70% were highly expressed in N, 20% were highly expressed in MGUS, and only 10% were highly expressed in MM.
Gene array analysis demonstrates the differences between normal and malignant PCs. (A) “Filter gene” function within dChip identified 125 genes with a large variation across samples (filtering criteria: [1.10 < SD/mean between < 10.00] P call % in the array used, 20% or greater). Unsupervised clustering using this list separates the samples into 2 trees: normal samples in one tree versus MGUS and MM samples in the other. The second tree further separates into 2 branches, one containing mainly MM samples and the other containing mainly MGUS samples. (B) To identify specific genes differentially expressed between N and MGUS, arrays were grouped according to sample type, and their mean expression levels were compared using the dChip compare samples facility (criteria, lower bound -fold change [E/B > 1.1] or [B/E > 1.3], mean difference [E - B > 50] or [B - E > 150], t test P < .05). Two hundred sixty-three genes were identified, most of which were down-regulated. (C) Using this gene list to hierarchically cluster all the samples defines 2 branches: one contains the N samples, and the other contains MGUS and MM samples.
Gene array analysis demonstrates the differences between normal and malignant PCs. (A) “Filter gene” function within dChip identified 125 genes with a large variation across samples (filtering criteria: [1.10 < SD/mean between < 10.00] P call % in the array used, 20% or greater). Unsupervised clustering using this list separates the samples into 2 trees: normal samples in one tree versus MGUS and MM samples in the other. The second tree further separates into 2 branches, one containing mainly MM samples and the other containing mainly MGUS samples. (B) To identify specific genes differentially expressed between N and MGUS, arrays were grouped according to sample type, and their mean expression levels were compared using the dChip compare samples facility (criteria, lower bound -fold change [E/B > 1.1] or [B/E > 1.3], mean difference [E - B > 50] or [B - E > 150], t test P < .05). Two hundred sixty-three genes were identified, most of which were down-regulated. (C) Using this gene list to hierarchically cluster all the samples defines 2 branches: one contains the N samples, and the other contains MGUS and MM samples.
Supervised analysis demonstrates that most genes differentially expressed between N and MGUS/MM PCs are down-regulated
To identify specific genes differentially expressed between N, MGUS, and MM, arrays were grouped according to sample type, and their mean expression levels were compared using the dChip compare samples function. Unsupervised analysis had shown that most genes were down-regulated in MGUS and MM PCs compared with N PCs, and -fold changes for genes down-regulated were far greater than those for up-regulated genes. To retain the up-regulated genes with smaller yet statistically significant -fold changes that might have biologic significance and at the same time identify the most down-regulated genes, comparison criteria were made more stringent for genes more highly expressed in the baseline group (N) than in either experiment group (MGUS or MM). This resulted in a more balanced list of genes underexpressed and overexpressed in the experiment groups, which proved more reliable for hierarchical clustering. Comparison criteria used were lower bound -fold change [E/B > 1.1] or [B/E > 1.3], mean difference [E - B > 50] or [B - E > 150], t test P < .05, where B is the mean expression of the baseline group (N PCs) and E is the mean expression of the experiment group (MGUS or MM).
Using these criteria 263 genes were differentially expressed between 5 N (B) and 5 MGUS (E) (172 genes down-regulated, 91 genes up-regulated) (Figure 2B; Table 1). Hierarchical clustering of the samples (5 N, 5 MGUS, 9 MM) using this gene list defined 2 branches: one contained all the N samples, and the other contained the MGUS and MM samples (Figure 2C; Table 1). When the 5 N (B) were compared to the 9 MM (E), 380 genes were found to be differentially expressed between the 2 groups (252 genes down-regulated, 128 genes up-regulated), of which 197 genes were also differentially regulated in the N versus MGUS comparison. When the 5 MGUS (B) were compared to the 9 MM (E) samples, only a few genes were identified using the comparison criteria above. Therefore, to identify a larger group of genes, a less stringent and more balanced set of criteria were used (lower bound -fold change [E/B or B/E > 1.1] and mean difference [E-BorB-E > 50] t test P < .05). Seventy-four genes were identified as differentially expressed between MGUS and MM samples (52 down-regulated in MM and 22 up-regulated), indicating that there are fewer differences at the gene expression level between MGUS and MM than between N and MM or N and MGUS (Table 2). Another analysis incorporating an additional 2 MGUS and 15 MM was in keeping with these initial results, and the genes and gene lists discussed were identified in both sets of analyses. The complete lists of genes are available at http://www.egu.leeds.ac.uk/genearray.
Genes differentially expressed in N, MGUS, and MM
. | . | . | Fold change . | . | |
|---|---|---|---|---|---|
| Gene . | Name . | Accession no. . | N vs MGUS . | N vs MM . | |
| FABP4 | Fatty acid binding protein 4, adipocyte | AA128249 | −12.2 | −13.94 | |
| PPBP | Pro-platelet basic protein (chemokine [C-X-C motif] ligand 7) | M54995 | NA | −12.00 | |
| CD5L | CD5 antigen-like (scavenger receptor cysteine rich family) | U82812 | −10.49 | −11.2 | |
| CD163 | CD 163 antigen | Z22971 | −6.51 | −7.81 | |
| HLA-DRA | Major histocompatibility complex, class 2, DR α | J00194 | −6.18 | −9.42 | |
| SEPP1 | Selenoprotein P, plasma, 1 | Z11793 | −5.89 | −9.29 | |
| FNBP1 | Formin-binding protein 1 | AB011126 | −5.5 | −8.48 | |
| SAT | Spermidine/spermine N1-acetyltransferase | M77693 | −4.8 | −11.24 | |
| Cluster Incl* | Unknown | AA203487 | −4.61 | −6.91 | |
| CTSH | Cathepsin H | X16832 | −4.57 | −9.52 | |
| PF4 | Platelet factor 4 (chemokine [C-X-C motif] ligand 4) | M25897 | −4.15 | −15.38 | |
| CLECSF2 | C-type (calcium-dependent, carbohydrate-recognition domain) lectin, superfamily member 2 (activation-induced) | X96719 | −4.47 | −7.07 | |
| HLA-DPA1 | Major histocompatibility complex, class 2, DP α 1 | X00457 | −3.27 | −5.82 | |
| TMSB4X | Thymosin, β4, X chromosome | M17733 | −3.04 | −7.05 | |
| TNFRSF7 | Tumor necrosis factor receptor superfamily, member 7 | M63928 | −2.96 | −7.64 | |
| MARCKS | Myristoylated alanine-rich protein kinase C substrate | D10522 | −1.95 | −4.71 | |
| RING1 | Ring finger protein 1 | AL031228 | 1.4 | 2.92 | |
| NY-REN-24* | NY-REN-24 antigen | AF052087 | NA | 2.18 | |
| NME1 | Nonmetastatic cells 1, protein (NM23A) expressed in | X73066 | 2.4 | 2.18 | |
| GLI3 | GLI-Kruppel family member GLI3 (Greig cephalopolysyndactyly syndrome) | M57609 | 2.41 | 2.14 | |
| WIT1* | Wilms tumor-associated protein | X69950 | 2.53 | 2.42 | |
| RPL14 | Ribosomal protein L 14 | D87735 | NA | 2.56 | |
| PPP1CA | Protein phosphatase 1, catalytic subunit, α isoform | S57501 | 2.53 | 2.61 | |
| Cluster Incl* | Unknown | A1687419 | NA | 2.71 | |
| GNL1 | Guanine nucleotide binding protein-like 1 | L25665 | 4.36 | 2.85 | |
. | . | . | Fold change . | . | |
|---|---|---|---|---|---|
| Gene . | Name . | Accession no. . | N vs MGUS . | N vs MM . | |
| FABP4 | Fatty acid binding protein 4, adipocyte | AA128249 | −12.2 | −13.94 | |
| PPBP | Pro-platelet basic protein (chemokine [C-X-C motif] ligand 7) | M54995 | NA | −12.00 | |
| CD5L | CD5 antigen-like (scavenger receptor cysteine rich family) | U82812 | −10.49 | −11.2 | |
| CD163 | CD 163 antigen | Z22971 | −6.51 | −7.81 | |
| HLA-DRA | Major histocompatibility complex, class 2, DR α | J00194 | −6.18 | −9.42 | |
| SEPP1 | Selenoprotein P, plasma, 1 | Z11793 | −5.89 | −9.29 | |
| FNBP1 | Formin-binding protein 1 | AB011126 | −5.5 | −8.48 | |
| SAT | Spermidine/spermine N1-acetyltransferase | M77693 | −4.8 | −11.24 | |
| Cluster Incl* | Unknown | AA203487 | −4.61 | −6.91 | |
| CTSH | Cathepsin H | X16832 | −4.57 | −9.52 | |
| PF4 | Platelet factor 4 (chemokine [C-X-C motif] ligand 4) | M25897 | −4.15 | −15.38 | |
| CLECSF2 | C-type (calcium-dependent, carbohydrate-recognition domain) lectin, superfamily member 2 (activation-induced) | X96719 | −4.47 | −7.07 | |
| HLA-DPA1 | Major histocompatibility complex, class 2, DP α 1 | X00457 | −3.27 | −5.82 | |
| TMSB4X | Thymosin, β4, X chromosome | M17733 | −3.04 | −7.05 | |
| TNFRSF7 | Tumor necrosis factor receptor superfamily, member 7 | M63928 | −2.96 | −7.64 | |
| MARCKS | Myristoylated alanine-rich protein kinase C substrate | D10522 | −1.95 | −4.71 | |
| RING1 | Ring finger protein 1 | AL031228 | 1.4 | 2.92 | |
| NY-REN-24* | NY-REN-24 antigen | AF052087 | NA | 2.18 | |
| NME1 | Nonmetastatic cells 1, protein (NM23A) expressed in | X73066 | 2.4 | 2.18 | |
| GLI3 | GLI-Kruppel family member GLI3 (Greig cephalopolysyndactyly syndrome) | M57609 | 2.41 | 2.14 | |
| WIT1* | Wilms tumor-associated protein | X69950 | 2.53 | 2.42 | |
| RPL14 | Ribosomal protein L 14 | D87735 | NA | 2.56 | |
| PPP1CA | Protein phosphatase 1, catalytic subunit, α isoform | S57501 | 2.53 | 2.61 | |
| Cluster Incl* | Unknown | A1687419 | NA | 2.71 | |
| GNL1 | Guanine nucleotide binding protein-like 1 | L25665 | 4.36 | 2.85 | |
Values in -fold change are expressed using the N PC group as the baseline. NA indicates not applicable.
Not a human gene nomenclature database symbol.
Genes differentially expressed between MGUS and MM
Gene . | Name . | Accession number . | Fold change . | P . |
|---|---|---|---|---|
| Cell growth and maintenance | ||||
| LILRB1 | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 | AF004230 | −8.26 | .03 |
| DEFA3 | Defensin, α3, neutrophil-specific | L12691 | −2.9 | .04 |
| CTSG | Cathepsin G | M16117 | −2.78 | .03 |
| TNFRSF7 | Tumor necrosis factor receptor superfamily, member 7 | M63928 | −2.58 | .03 |
| GLUL | Glutamate-ammonia ligase (glutamine synthase) | X59834 | −2.47 | .03 |
| MARCKS | Myristoylated alanine-rich protein kinase C substrate | D10522 | −2.41 | .04 |
| TLR1 | Toll-like receptor 1 | AL050262 | −2.09 | .04 |
| S100A8 | S100 calcium binding protein A8 (calgranulin A) | A1126134 | −1.88 | .01 |
| LITAF | Lipopolysaccharide-induced TNF factor | AL120815 | −1.65 | .04 |
| VNN2 | Vanin 2 | D89974 | −1.63 | .04 |
| SLC7A7 | Solute carrier family 7 | AJ130718 | −1.61 | .04 |
| PDE6G | Phosphodiesterase 6G, cGMP-specific, rod, γ | X62025 | 1.32 | .02 |
| MTA1L1 | Metastasis-associated 1-like 1 | AB012922 | 1.37 | .03 |
| UBE2G2 | Ubiquitin-conjugating enzyme E2G 2 | AF032456 | 1.65 | .006 |
| Signal transduction | ||||
| MERTK | c-mer proto-oncogene tyrosine kinase | U08023 | −3.52 | .04 |
| FCER1G | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | M33195 | −2.89 | .01 |
| RNASE6 | Ribonuclease RNase A family k6 | A1142565 | −2.75 | .02 |
| LY96 | Lymphocyte antigen 96 | AB018549 | −2.23 | .04 |
| LASP1 | LIM and SH3 protein 1 | X82456 | −1.91 | .02 |
| ARHGAP1 | Rho GTPase activating protein 1 | U02570 | −1.8 | .03 |
| PFTK1 | PFTAIRE protein kinase 1 | AB020641 | −1.69 | .0009 |
| CAP | Adenylyl cyclase-associated protein | L12168 | −1.67 | .03 |
| DOK1 | Docking protein 1 62 kDa (downstream of tyrosine kinase 1) | U70987 | −1.62 | .05 |
| RPS6KA4 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 4 | AJ010119 | 1.44 | .03 |
| ITGA8 | Integrin α8 | L36531 | 2.14 | .01 |
| Structural proteins | ||||
| PPBP | Pro-platelet basic protein (chemokine [C-X-C motif] ligand 7) | M54995 | −5.54 | .01 |
| ACTN1 | Actinin, α1 | X15804 | −4.83 | .01 |
| MYH9 | Myosin, heavy polypeptide 9, nonmuscle | Z82215 | −2.07 | .02 |
| ADD1 | Adducin 1 (α) | X58141 | −1.92 | .02 |
| VCL | Vinculin | M33308 | −1.83 | .05 |
| SGCE | Sarcoglycan epsilon | AJ000534 | 1.53 | .003 |
| Developmental processes | ||||
| TMSB4X | Thymosin, β4, X chromosome | M17733 | −2.32 | .04 |
| PITX3 | Paired-like homeodomain transcription factor 3 | AF041339 | 1.3 | .003 |
| FAT | FAT tumor-suppressor homolog 1 (Drosophila) | X87241 | 1.55 | .04 |
| MFNG | Manic fringe homolog (Drosphila) | Z93096 | 1.6 | .02 |
| RING1 | Ring finger protein 1 | AL031228 | 2.09 | .01 |
| FRZB | Frizzled-related protein | U91903 | 6.4 | .05 |
| Others | ||||
| NPC2 | Niemann-Pick disease, type C2 | AI525834 | −3.19 | .003 |
| SAT | Spermidine/spermine N1-acetyltransferase | AL050290 | −2.95 | .01 |
| RAGD* | Rag D protein | W27549 | 1.93 | .03 |
| HIST1H2BG | Histone 1, H2bg | AJ223352 | 1.93 | .02 |
Gene . | Name . | Accession number . | Fold change . | P . |
|---|---|---|---|---|
| Cell growth and maintenance | ||||
| LILRB1 | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 | AF004230 | −8.26 | .03 |
| DEFA3 | Defensin, α3, neutrophil-specific | L12691 | −2.9 | .04 |
| CTSG | Cathepsin G | M16117 | −2.78 | .03 |
| TNFRSF7 | Tumor necrosis factor receptor superfamily, member 7 | M63928 | −2.58 | .03 |
| GLUL | Glutamate-ammonia ligase (glutamine synthase) | X59834 | −2.47 | .03 |
| MARCKS | Myristoylated alanine-rich protein kinase C substrate | D10522 | −2.41 | .04 |
| TLR1 | Toll-like receptor 1 | AL050262 | −2.09 | .04 |
| S100A8 | S100 calcium binding protein A8 (calgranulin A) | A1126134 | −1.88 | .01 |
| LITAF | Lipopolysaccharide-induced TNF factor | AL120815 | −1.65 | .04 |
| VNN2 | Vanin 2 | D89974 | −1.63 | .04 |
| SLC7A7 | Solute carrier family 7 | AJ130718 | −1.61 | .04 |
| PDE6G | Phosphodiesterase 6G, cGMP-specific, rod, γ | X62025 | 1.32 | .02 |
| MTA1L1 | Metastasis-associated 1-like 1 | AB012922 | 1.37 | .03 |
| UBE2G2 | Ubiquitin-conjugating enzyme E2G 2 | AF032456 | 1.65 | .006 |
| Signal transduction | ||||
| MERTK | c-mer proto-oncogene tyrosine kinase | U08023 | −3.52 | .04 |
| FCER1G | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | M33195 | −2.89 | .01 |
| RNASE6 | Ribonuclease RNase A family k6 | A1142565 | −2.75 | .02 |
| LY96 | Lymphocyte antigen 96 | AB018549 | −2.23 | .04 |
| LASP1 | LIM and SH3 protein 1 | X82456 | −1.91 | .02 |
| ARHGAP1 | Rho GTPase activating protein 1 | U02570 | −1.8 | .03 |
| PFTK1 | PFTAIRE protein kinase 1 | AB020641 | −1.69 | .0009 |
| CAP | Adenylyl cyclase-associated protein | L12168 | −1.67 | .03 |
| DOK1 | Docking protein 1 62 kDa (downstream of tyrosine kinase 1) | U70987 | −1.62 | .05 |
| RPS6KA4 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 4 | AJ010119 | 1.44 | .03 |
| ITGA8 | Integrin α8 | L36531 | 2.14 | .01 |
| Structural proteins | ||||
| PPBP | Pro-platelet basic protein (chemokine [C-X-C motif] ligand 7) | M54995 | −5.54 | .01 |
| ACTN1 | Actinin, α1 | X15804 | −4.83 | .01 |
| MYH9 | Myosin, heavy polypeptide 9, nonmuscle | Z82215 | −2.07 | .02 |
| ADD1 | Adducin 1 (α) | X58141 | −1.92 | .02 |
| VCL | Vinculin | M33308 | −1.83 | .05 |
| SGCE | Sarcoglycan epsilon | AJ000534 | 1.53 | .003 |
| Developmental processes | ||||
| TMSB4X | Thymosin, β4, X chromosome | M17733 | −2.32 | .04 |
| PITX3 | Paired-like homeodomain transcription factor 3 | AF041339 | 1.3 | .003 |
| FAT | FAT tumor-suppressor homolog 1 (Drosophila) | X87241 | 1.55 | .04 |
| MFNG | Manic fringe homolog (Drosphila) | Z93096 | 1.6 | .02 |
| RING1 | Ring finger protein 1 | AL031228 | 2.09 | .01 |
| FRZB | Frizzled-related protein | U91903 | 6.4 | .05 |
| Others | ||||
| NPC2 | Niemann-Pick disease, type C2 | AI525834 | −3.19 | .003 |
| SAT | Spermidine/spermine N1-acetyltransferase | AL050290 | −2.95 | .01 |
| RAGD* | Rag D protein | W27549 | 1.93 | .03 |
| HIST1H2BG | Histone 1, H2bg | AJ223352 | 1.93 | .02 |
Values in -fold change are expressed using the MGUS group as the baseline.
Not a human gene nomenclature database symbol.
Differentially expressed genes identified by microarray were validated using PCR and flow cytometry
To confirm that the microarray analysis accurately reflected the pattern of changes at the protein level, the data sets generated were validated using flow cytometry comparing the expression of membrane proteins on N, MGUS, and MM PCs. Transcriptional profiles identified down-regulation of genes encoding CD38, CD27, and CCR2 from N through MGUS to MM; these findings were confirmed at the protein level (Figure 3A-C). Validation was extended using RT-PCR for FRZB and SEPP 1, selected because of their large differential expression between N, MGUS, and MM (Figure 3D-E).
Validation of gene array expression levels. Gene expression levels (left panels) were validated (right panels) using flow cytometry (A-C) or RQ-PCR (D-E). (A) CD38 expression is higher in N PCs than in MGUS PCs (P < .001) or MM PCs (P < .001). (B) CD27 expression is higher in N PCs than in MM PCs (P = .007). (C) CCR2 expression is higher in N PCs than in MGUS or MM PCs (P = .03). (D) Frizzled related protein is higher in MM PCs than in N PCs and MGUS PCs. (E) Selenoprotein P is lower in MM PCs than MGUS PCs and N PCs. (F) Gene expression levels demonstrate that XBP1 is lower in MGUS and MM PCs than in N PCS, though the level of expression is still relatively high. (G) RT-PCR for XBP1 and XBP1s demonstrates both forms are present (2 distinct bands, 442p and 416bp) in plasma cells selected from N, MGUS, and MM patients and in MM cell lines. There is a decreased expression of XBP1s in MM PCs and MM cell lines compared with N PCs and MGUS PCs.
Validation of gene array expression levels. Gene expression levels (left panels) were validated (right panels) using flow cytometry (A-C) or RQ-PCR (D-E). (A) CD38 expression is higher in N PCs than in MGUS PCs (P < .001) or MM PCs (P < .001). (B) CD27 expression is higher in N PCs than in MM PCs (P = .007). (C) CCR2 expression is higher in N PCs than in MGUS or MM PCs (P = .03). (D) Frizzled related protein is higher in MM PCs than in N PCs and MGUS PCs. (E) Selenoprotein P is lower in MM PCs than MGUS PCs and N PCs. (F) Gene expression levels demonstrate that XBP1 is lower in MGUS and MM PCs than in N PCS, though the level of expression is still relatively high. (G) RT-PCR for XBP1 and XBP1s demonstrates both forms are present (2 distinct bands, 442p and 416bp) in plasma cells selected from N, MGUS, and MM patients and in MM cell lines. There is a decreased expression of XBP1s in MM PCs and MM cell lines compared with N PCs and MGUS PCs.
Our analysis also identified a number of genes already shown to have altered expression between N and MM plasma cells, including vascular cell adhesion molecule 1 (VCAM-1), stromal cell-derived factor 1 (SDF-1), and platelet factor 4 (PF4),13-15 further confirming the validity of this approach. Importantly, a number of genes identified in this analysis using amplified clinical material have also been identified in other gene array reports examining MM cells or cell lines,13-15 suggesting that amplification using the SMART system combined with Affymetrix arrays is possible on small amounts of clinical material and that consistent results can be obtained.
Functional classes of genes differentially expressed between N, MGUS, and MM
The underlying basis of the transition from N to MGUS and MM may be understood more clearly by looking at the functional classes of the genes altered. For each gene the gene name approved by the HUGO gene nomenclature committee is used where possible, followed by the abbreviation and -fold change in brackets. Within each analysis genes have been grouped for discussion into oncogenes and tumor suppressors, cell-signaling genes, death genes, DNA binding and transcription factors, developmental genes, and genes affecting chromatin structure.
Transition of N to MGUS
When comparing the expression profiles of N and MGUS PCs, only one oncogene was demonstrated to have altered expression, and surprisingly no tumor-suppressor genes were differentially expressed. The oncogene arginine-rich gene mutated in early-stage cancers, ARMET, was down-regulated (-5.40); deletions and mutations having previously been noted in solid tumors.
Dependence on IL-6 signaling and the abrogation of this dependence late in the natural history of the disease by Ras mutations, make changes in RAS and in G-protein signaling important features to document. Despite changes in RAS usually being characteristic of later stages of disease, a number of changes were seen in the N versus MGUS comparison. RAS family members altered included the down-regulation of RAB1A (-2.74), RAP1B (-1.9), RAS suppressor protein (-2.56), and ras homolog gene family member A (ARHA) (-2.32). Other genes altered included seven in absentia homolog 2 (SIAH2) (-2.22), which is a negative regulator of Vav and DCC signal transduction; and regulator of G-protein signaling 1 (RGS1) (-4.35), a negative regulator of G-protein-coupled receptor signaling, which plays a role in B-cell activation and proliferation.
Nuclear factor κB (NFκB) and its regulatory protein, inhibitor (IκB), are known to play central roles in a number of pathways in myeloma, and its downstream effects include modulation of cytokines, chemokines, adhesion molecules, cell proliferation, and survival. Phosphorylation of IκB within the IκB/NFκB complex results in IκB ubiquitination and degradation, allowing NFκB to translocate to the nucleus and mediate signal transduction by binding to the promoters of target genes. An important member of the NFκB pathway, regulating the activation of NFκB by modulating IκB phosphorylation, is peroxiredoxin 4 (PRDX4), which was noted to be down-regulated (-2.25). Nuclear factor κ light-chain polypeptide gene enhancer in B cells inhibitor (NFKBIA), which interacts with NFκB, trapping it in the cytoplasm, was also noted to be down-regulated (-3.38).
A central feature of plasma cell development is transcriptional repression, and we have identified decreased expression of X box binding protein 1 (XBP1) (-1.84) in MGUS PCs compared with N PCs; however, the level of expression is still relatively high (Figure 3F). A recent report identifies a splice variant of XBP1, XBP1s, as important in antibody production and plasma cell differentiation.16 Array-based analysis cannot identify the splice variant; therefore, using RT-PCR, we have demonstrated that both forms (XBP1 and XBP1s) are present in PCs from N, MGUS, and MM, but the expression ratios differ (Figure 3G). In the N and MGUS groups, XBP1s was expressed at similar or increased levels compared with XBP1, whereas in MM PCs it was expressed at a lower level. A number of genes were coregulated with XBP-1, implicating them as possible downstream targets of the transcription factor. These included basic leucine zipper protein (BZW1) (correlation coefficient, R = 0.92), and lamin B receptor (LBR) (R = 0.86), each of which is involved in cell-cycle regulation.
Transcription factors shown to be down-regulated included members of the zinc finger family, which modulate response to growth factor signaling, including zinc finger protein 36-like 1 (ZFP36L1) (-5.64), and ZFP36 (-3.55). A fourth related transcription factor, YY1, located at 14q32, the site of the immunoglobulin H (IgH) locus, was also noted to be down-regulated (-2.35). This is an important gene in the generation of polycomb complexes, and it has recently been identified as important in transforming growth factor-β-induced cell differentiation. Up-regulated genes included paired box gene 4 (PAX4) (+1.25), important in pancreatic islet cell development; forkhead box G1A (FOXG1A) (+2.18), a forkhead transcription factor important in embryonic development; and Pou domain class 2 (POU2F2) (+1.50), a transcription factor that activates immunoglobulin gene expression. The binding factor ring finger 1 (RING1), a transcription repressor involved in the poly-comb complex, was also up-regulated (+1.4).
A consistent feature of malignant transformation is deranged cellular programming and aberrant expression of genes important in patterning and development. Changes in the expression of members of the hedgehog pathway were observed. Sonic hedgehog (SHH) expression was up-regulated (+1.82), as was its downstream target GLI-Kruppel family member 3 (GLI3) (+2.41). Interestingly patched-related protein translocated in renal cancer (TRC8), a protein with homology to patched protein in this pathway, was down-regulated in MGUS PCs compared with N PCs (-2.12). Von Hippel-Lindau syndrome gene (VHL) and COP9 homology gene, proteins that physically interact with TRC8, were also down-regulated (-2.50 and -2.41, respectively).
Given that the normal versus malignant comparison examines cells with different transcriptional status, it is not surprising that a number of structural nuclear proteins and genes responsible for acetylation/methylation of DNA are altered. These genes include SMARCE1 (-3.15), which is part of the chromatin remodeling complex SWI/SNF; sin-3-associated peptide (SAP18) (-2.81), a component of the histone deacetylase complex, which enhances sin-3-mediated transcriptional repression; and histone deacetylase 2 (HDAC2) (-2.82), a regulator of chromatin structure during transcription, which associates with YY1.
Transition of MGUS to MM
The number of genes differentially expressed between MGUS and MM was much smaller (Table 2). Many showed a progressively different expression from N to MGUS to MM, including spermidine/spermine N1-acetyltransferase (SAT) (-2.95), an enzyme involved in intracellular transport; the membrane protein tumor necrosis factor receptor superfamily member 7 (TNFRSF7/CD27) (-2.58); and RING1 (+2.09). Other potentially important genes included those involved in the developmental pathways. FRZB, a negative regulator of the WNT pathway, was the most up-regulated gene (+6.4). Manic fringe homolog (MFNG) was also noted to be up-regulated (+1.6), which plays a central role in notch signaling. A number of genes involved in the cytoskeleton were also differentially regulated. These included LIM and SH3 protein 1 (LASP1) (-1.91), and actinin α1 (ACTN1) (-4.83), which are involved in actin binding; myosin heavy polypeptide 9-nonmuscle (MYH9) (-2.07), involved in shape change function; and thymosin β4 X chromosome (TMSB4X) (-2.32), which plays an important organizational role in the cytoskeleton by inhibiting actin polymerization.
Transition of N to MM
A number of genes were differentially expressed between N and MM that were not highlighted as altered in the MGUS set. This may indicate a gradual change in gene expression through the disease phases that fails to reach statistical significance between sequential stages but does reach significance overall. Another 26 oncogenes showed altered expression, all but 4 of which were decreased in MM cells compared with normal PCs. These genes included lymphoid nuclear protein related to AF4 (LAF4) (+1.78), a transcriptional repressor in lymphoid development, which is related to MLLT2; v-myb myeloblastosis viral oncogene homolog (MYB) (+1.97), an important transcriptional activator in proliferation and differentiation control, and BCL2 (+1.51). Seven tumor-suppressor genes were down-regulated, including CDK2-associated protein 1 (CDK2AP1) (-4.69), a negative regulator of CDK activity; transforming growth factor-β-induced gene product (TGFBI) (-7.55); and disabled homolog 2 (DAB2) (-6.5). The tumor-suppressor gene retinoblastoma 1 (RB1), which is located on chromosome 13q, was also down-regulated (-4.62), and recent FISH studies have noted this area to be deleted in a significant proportion of MGUS and MM cases.7,8 The down-regulation of the tumor-suppressor gene, jumping translocation breakpoint (JTB) (-1.92), located at the site of the jumping translocation on chromosome 1, is of particular interest; previous reports using CGH and SKY have identified this region as a hot spot.17,18
A comprehensive screen of genes known to be involved in the early stages of B-cell development identified only a few genes that were differentially expressed between normal and malignant plasma cells, consistent with the transforming event being late in B-cell development. Genes included CD79A (-3.59); the major histocompatibility complex (MHC) class II DR (-9.42) and class II DPα1 (-5.82); B-cell linker (BLNK) (-3.34), a critical component of the BCR transduction complex; and signaling lymphocytic activation molecule (SLAM) (-2.13), which is involved in bidirectional T- to B-cell interaction.
For an indolent tumor such as multiple myeloma that only has a low growth fraction, alterations in apoptotic genes are a potentially important mechanism of tumorigenesis. However, changes in only a limited number of apoptosis-related genes were identified, indicating that plasma cells may normally express genes conferring resistance to apoptosis. The genes identified include forkhead box O3A (FOXO3A) (-1.68), a transcription factor that triggers apoptosis by inducing genes critical in cell death; beclin1 (BECN1) (-2.00), which interacts with BCL2; thioredoxin-like (TXNL) (-2.36), a redox regulator implicated in apoptosis inhibition; and Tax 1 binding protein (TAX1BP1) (-2.20), which interacts with a zinc finger protein important in apoptosis.
Other transcription factor genes were identified, including ribosomal protein L14 (RPL14) (+2.56), a component of the 60S subunit, and SRY box 22 (SOX22), which has a role in differentiation (+1.46). Other important binding factors included ring fingers 4 and 11, which were both down-regulated (-1.9 and -3.49, respectively). In keeping with the up-regulation of the developmental gene FRZB in the MGUS versus MM comparison, important downstream targets of the WNT pathway were noted to be down-regulated, including Ras homolog gene family member A (ARHA) (-3.53) and β catenin (CTNNB1) (-2.57). Figure 4 combines these data into a transcriptomic model of the multistep pathogenesis of myeloma.
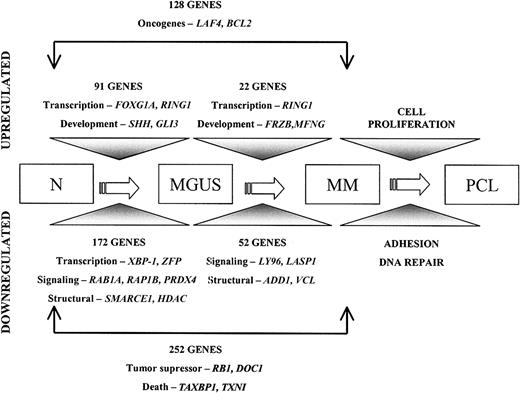
A diagrammatic representation of the multistep pathogenesis of MM. This transcriptomic model shows genes differentially expressed between plasma cells from healthy donors and individuals with MGUS or MM.
A diagrammatic representation of the multistep pathogenesis of MM. This transcriptomic model shows genes differentially expressed between plasma cells from healthy donors and individuals with MGUS or MM.
Discussion
One of the important uses of microarray data in multiple myeloma is to define global patterns of expression relevant to the biology of normal and malignant plasma cells that may be applied in the clinical setting. This approach has been successfully used in non-Hodgkin lymphoma to define novel subtypes of diffuse large B-cell lymphoma19,20 and to relate these to normal B-cell populations at different stages of differentiation. Taking a similar approach in myeloma is difficult because the normal comparator group is not yet fully defined, and isolating cells representing the normal stages of plasma cell development is difficult. Published data in myeloma have addressed this issue in 2 broad ways. One approach is to differentiate normal peripheral blood lymphocytes into plasma cells in an artificial system.21 This has the advantage of being a relatively clean system capable of defining changes in gene expression; however, its relationship to myeloma is less clear. An alternative approach is to isolate normal B cells and plasma cells from tonsil and bone marrow and to compare these with PCs isolated from MGUS and myeloma cases and with cell lines.13 The latter approach is directly relevant to malignancies of plasma cells, but it is difficult to understand how these populations relate to each other. Because of this uncertainty, we have restricted our analysis to a comparison of normal bone marrow plasma cells and bone marrow plasma cells from individuals with MGUS and patients with myeloma. The relationship of the normal CD138+ plasma cell to myeloma is unclear, but it does serve to anchor the changes described in malignant plasma cells to a defined stage of B-cell differentiation. There is a strong argument in future experiments to use cell sorting to define more stages within the differentiation pathway of normal plasma cells.
In terms of global gene expression patterns, the major feature separating normal and malignant plasma cells is the decreased level of expression of a large number of genes. In contrast the list of up-regulated genes is more limited. Transcription factors are key genes in regulating gene expression patterns, and we have identified a number of such genes that may be important in myeloma. XBP1 was decreased in expression in the CD138+ malignant plasma cells compared with their normal counterparts (CD138+ normal PCs), which is consistent with a number of other reports.13 Recent reports have shown that XBP1, along with BLIMP1 (PRDFI-BF1), are important transcription factors required for the terminal differentiation of B lymphocytes to plasma cells.22,23 More recently it has been reported that a splice variant of XBP1 (XBP1s) plays a crucial role in normal plasma cell differentiation.16 Using RT-PCR we looked at the expression of XBP1 and XBP1s and found that though both forms were expressed in normal and malignant PCs, the relative levels of XBP1s compared to XBP1 were decreased in myeloma compared with normal PCs. In normal PC development XBP1s is important in IL-6 up-regulation and secretion,16 and previous experiments in myeloma have shown that XBP1 and IL-6 are dependent on each other.24,25 However, the role of XBP1s in this experimental system has not been examined. Ongoing experiments are examining the role of XBP1/XBP1s within PCs and bone marrow stromal cells. A bioinformatic approach to identify other molecules important in this process was used to determine genes that co-regulate with XBP1. We identified a number of transcription factors fulfilling these criteria (ZFP36, ZFP36L1, and ZFP36L2), all of which were expressed at lower levels. The zinc finger proteins are a family of transcription factors important in governing the response to growth factor signaling. Interestingly, ZFP36L1, a member of the Kruppel C2H2 zinc finger family, is located at the site of the IgH locus (14q22-24), which is commonly translocated in myeloma.
Patterning and developmental genes are important in determining the differentiated architecture of normal tissue and are tightly controlled during development. We found a number of abnormalities in the WNT and hedgehog pathways, suggesting that they may be important in late normal B-cell development and myeloma pathogenesis. This finding is consistent with the description of a functional WNT pathway in myeloma cell lines.26 The relevance of these findings has been confirmed by other array studies, which have also identified changes in the expression of FRZB.13,21 In our analysis FRZB is the most up-regulated gene in the transition of MGUS to MM. The normal function of this secreted protein is to act as a negative regulator of the WNT pathway; therefore, in myeloma it may have either an autocrine or a paracrine effect by inhibiting WNT signaling in PCs or in the microenvironment. In particular, inhibiting WNT signaling in osteoblasts results in decreased bone formation.27 Hence, FRZB may contribute to myeloma bone disease. In our data set we found overexpression of FRZB in approximately 45% of myeloma cases and overexpression of Dickkopf1 (DKK1), another secreted inhibitor of the WNT pathway.
The sonic pathway is important in normal stem cell and thymocyte development and has been implicated in normal development and in tumor formation. Overexpression, mutation, and translocation of patched protein (PTC), patched-related protein (TRC8), and GLI-Kruppel family member 3 (GLI3) are associated with basal cell carcinomas, gliomas, and hereditary renal cell carcinoma. We noted an overexpression of SHH and GLI3 and a decrease in expression of TRC8 between N, MGUS, and MM PCs. Furthermore many of the downstream targets of the sonic pathway include pathways previously demonstrated as important in MM pathogenesis, including WNT and TGFβ, further implicating this pathway in MM biology.
As in other hematologic malignancies, the use of gene expression analysis offers the opportunity to investigate disease biology by comparing malignant cells with their normal developmental counterparts. We have anchored our analysis to normal plasma cells and have demonstrated that most changes occur between N and MGUS plasma cells, with only a relatively small list of differentially expressed genes distinguishing MGUS from MM. Further investigation of this list is warranted because it potentially contains the key genes important in the transformation process.
Prepublished online as Blood First Edition Paper, August 28, 2003; DOI 10.1182/blood-2003-01-0016.
Supported by the Department of Health, Leukaemia Research Fund UK (F.E.D., G.J.M.); British Society of Haematology (A.C.R., F.E.D.); Multiple Myeloma Research Foundation (D.C., T.H., N.M., C.M.); Doris Duke Distinguished Clinical Research Scientist Award (K.C.A.); VA Merit Review and Leukemia and Lymphoma Society Scholar in Translational Research Award (N.C.M.); National Institutes of Health grants 50947 (K.C.A.), CA78373 (K.C.A.), HG02341 (C.L., W.H.W.), and CA96470 (W.H.W.); The Myeloma Research Fund; and The Cure Myeloma Fund.
F.E.D. and A.M.D. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


![Figure 2. Gene array analysis demonstrates the differences between normal and malignant PCs. (A) “Filter gene” function within dChip identified 125 genes with a large variation across samples (filtering criteria: [1.10 < SD/mean between < 10.00] P call % in the array used, 20% or greater). Unsupervised clustering using this list separates the samples into 2 trees: normal samples in one tree versus MGUS and MM samples in the other. The second tree further separates into 2 branches, one containing mainly MM samples and the other containing mainly MGUS samples. (B) To identify specific genes differentially expressed between N and MGUS, arrays were grouped according to sample type, and their mean expression levels were compared using the dChip compare samples facility (criteria, lower bound -fold change [E/B > 1.1] or [B/E > 1.3], mean difference [E - B > 50] or [B - E > 150], t test P < .05). Two hundred sixty-three genes were identified, most of which were down-regulated. (C) Using this gene list to hierarchically cluster all the samples defines 2 branches: one contains the N samples, and the other contains MGUS and MM samples.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/13/10.1182_blood-2003-01-0016/6/m_h82435332002.jpeg?Expires=1765901338&Signature=A9BldrajppjA3kLFw5qx72b5m8rfHP3rkd-Tv1sTdcqq-mVAOMM0WHqnAF1rABa~8FfRlIB0Cbl94qoSxROP4UPuiwnzn5gVpb3PfiPQsgRJhmgaah2R0cL3-FQiYJaE4T7~TFYkddJuzdi5ezK3wmbrrJ7q-jsvtl-NLZNMbKT3MI3J3f1cknzdl2F2SWxIhnvblvTSP~pBooreW0zxeQecyvsCi2Z~ClZopfxm4IDM4qP0xvOeJCI7OjIaKH6k2TkKQX8DFsMwfy4HnKdGu9CsFIYJij-zhZnzSVHTGTbepuaR9WAAPQDRpHCL0L50qEM0JcKOqGHEpt19Ivr~7g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal