Abstract
Mycosis fungoides (MF) is the most frequent type of cutaneous T-cell lymphoma, whose diagnosis and study is hampered by its morphologic similarity to inflammatory dermatoses (ID) and the low proportion of tumoral cells, which often account for only 5% to 10% of the total tissue cells. cDNA microarray studies using the CNIO OncoChip of 29 MF and 11 ID cases revealed a signature of 27 genes implicated in the tumorigenesis of MF, including tumor necrosis factor receptor (TNFR)–dependent apoptosis regulators, STAT4, CD40L, and other oncogenes and apoptosis inhibitors. Subsequently a 6-gene prediction model was constructed that is capable of distinguishing MF and ID cases with unprecedented accuracy. This model correctly predicted the class of 97% of cases in a blind test validation using 24 MF patients with low clinical stages. Unsupervised hierarchic clustering has revealed 2 major subclasses of MF, one of which tends to include more aggressive-type MF cases including tumoral MF forms. Furthermore, signatures associated with abnormal immunophenotype (11 genes) and tumor stage disease (5 genes) were identified.
Introduction
Mycosis fungoides (MF) is a peripheral cutaneous T-cell lymphoma (CTCL) characterized by the infiltration and accumulation of tumoral T cells in the skin (epidermal and dermal infiltration).1 It is the most frequent type of CTCL, representing more than half of all lymphomas originating in the skin.2
Clinical and pathologic diagnosis of MF has proved very difficult since it bears many resemblances to common inflammatory dermatoses (ID) such as interface and spongiotic dermatitis, conditions involving infiltration and accumulation of benign T cells in the skin. The study of MF is further compounded by the small number of tumoral cells, which may be present in the skin, with tumoral T-cell infiltrate often accounting for only 5% to 10% of the total tissue cells. Most CTCLs have the phenotype of T-helper memory lymphocytes (CD3+, CD4+) with only a minority of cases showing phenotypes such as CD4- or CD8+, with CD8+ cases possibly representing a subset of aggressive cases.3 Additionally, loss of CD7 expression is considered a distinguishing characteristic of MF.4
The etiology of MF remains largely unknown, although some triggering factors such as chronic antigen stimulation5 or human T-cell lymphotropic virus type 1 or other viral infections have been proposed. However, some of these findings have been somewhat contentious.6,7 Molecular studies of MF have revealed some interesting facts, although the studies are confined to small groups of genes and proteins. Defective FAS signaling has been suggested as a possible causative agent in MF pathogenesis due to defects in apoptosis signaling in skin-homing T cells.8 Reduction of FAS expression has been documented in peripheral blood CD4+ T lymphocytes in CTCL.9 This decreased FAS expression may be the result of FAS mutations, which are infrequent but are present in some cases.8 Defective FAS signaling may also be supplemented by mutations or defects in caspases or BCL2 family members. However studies of BCL210 and BAX8 do not favor a role of alterations in these genes in MF pathogenesis. Similarly molecular analysis of P53, the most frequently mutated gene in human malignancies, has shown that mutations are found only at later stages of disease and are unlikely to play a role in the early pathogenesis of MF.8 Studies of p16 have revealed reduced p16 expression in progressing MF lesions,11,12 and some studies have also revealed silencing of p15 gene expression associated with MF and Sezary syndrome.13 Additional studies have implicated signal transducer and activator of transcription 1 (STAT1),14 STAT3,15 CD30,16 CD40/CD40L,17 interleukin-15 (IL15),18 IL16,18 chemokine receptors,19 and telomere shortening and increased telomerase activity20 in pathogenesis and progression of MF. Finally, in vitro studies of MF have shown that nuclear factor κB (NF-κB) may play a role in the pathogenesis of MF.21
Here we have focused on the mechanisms of tumorigenesis of MF and MF heterogeneity using cDNA microarray analysis. The resulting data allow the identification of an MF signature, which can distinguish MF and ID cases using a prediction model, validated in an external blind patient set. cDNA microarrays are a powerful technique, which allow the analysis of the expression of thousands of genes simultaneously. These types of studies have revolutionized the study of cancer allowing finer tumor classification,22 prediction of treatment response,23,24 and prediction of overall survival.25 This type of analysis has been previously used in the in vitro study of MF cells where it was possible to identify genes with a possible role in the resistance to interferon alpha in patient samples.24 Here these cDNA microarrays have been used for examining the expression profile of a total of 53 MF samples (29-patient study group and an additional 24-patient validation group) and 11 ID samples, allowing the identification of 27 genes implicated in the tumorigenesis of MF, many of which are implicated directly in antiapoptotic signaling by tumor necrosis factor (TNF). Subsequently, a 6-gene prediction model was identified and validated for both this study group and an external blind set of 24 MF patients. Hierarchic clustering revealed the presence of 2 main MF groups, whose characteristics have been investigated here. cDNA microarray analysis also allowed the identification of molecular signatures associated with abnormal immunophenotype (11 genes) and tumor stage disease (5 genes).
Patients, materials, and methods
Case selection
A total of 11 ID samples, 29 MF samples, and 6 normal nonphotoexposed control skin samples in addition to a set of 24 MF patients for blind testing were selected from the medical records of hospitals included in the Spanish Tumor Bank Network. All tissue samples, both paraffin embedded and frozen samples, were collected through the protocols of the CNIO tumor bank. Informed consent was obtained from all patients enrolled in the study under the supervision of the local ethics committees. The MF samples represented consecutive biopsies, randomly chosen, from the contributing hospitals. All samples were centrally reviewed by a panel of pathologists (A.M.D., J.L.R.P., J.F.G., J. Fraga and M.A.P.), and diagnosed using uniform criteria based on the clinical, histologic, immunophenotypical, and molecular characteristics generally accepted and previously defined,26 with the aid of hematoxylin/eosin (H/E), CD3, CD4, and CD8 staining, and T-cell receptor (TCR) gamma gene rearrangement analysis by polymerase chain reaction (PCR). The MF samples came from initial diagnostic samples from patients in various stages of the disease, including plaque and tumor stages, while external validation group MF patients included in this study were enrolled in a randomized clinical trial and were all stage Ia, Ib, or IIa patients. For blind set patients only frozen tissue was supplied for this study. The 11 ID samples used represented cases of interface and spongiotic dermatitis, including rosacea, lupus erythematosus, dermatitis herpetiformis, seborrheic dermatitis, and spongiotic dermatitis. MF patients were treated with curative purposes, including a combination of PUVA (combination of psoralen and ultraviolet A [UVA] irradiation), alpha-interferon and standard chemotherapy. Responders are defined as patients who achieve complete remission at any stage during the treatment under any treatment regimen.
RNA isolation and cDNA microarray target preparation
Total RNA extraction was carried out using the Trizol reagent (Life Technologies, Grand Island, NY), and RNA was further purified using the RNeasy kit (Qiagen, Valencia, CA) and digested with RNase-free DNase I following the manufacturer's instructions. Target RNA (1-3 μg) was amplified using a T-7–based in vitro transcription system as described previously.24 aRNA (5 μg) from each sample was directly labeled with cyanine 5 (Cy5)–conjugated deoxyuracil triphosphate (dUTP), while 5 μg aRNA from the Universal Human Reference RNA (Stratagene, La Jolla, CA) was labeled with cyanine 3 (Cy3)–conjugated dUTP as reference. For all microarray studies the CNIO OncoChip was used and hybridizations were performed as previously described.24 Scanning was performed using a Scanarray 5000 XL (GSI Lumonics, Kanata, ON, Canada) and images were analyzed with the GenePix 4.0 Pro Software (Axon Instruments, Union City, CA).
Data analysis and normalization
Fluorescence intensity measurements were subjected to automatic background subtraction and the Cy3/Cy5 ratio values were normalized to the median ratio value of all spots in the array. The sum of the median background for each channel was calculated, and spots with total intensity values below the calculated sum of median backgrounds were discarded. All ratio values were log transformed (base 2). Inconsistent duplicates were discarded, all consistent duplicate spots and genes were averaged, and genes with less than 80% of the data available were excluded from further analysis.27 In the case of comparison of immunohistochemical MF subgroups, only genes with 100% of the data available were used.
In order to minimize the background noise caused by the presence of normal cells in the tissue, 2 methods were used: median centering normalization of the genes in all MF and ID cases and normalization of MF and ID samples against a pool of normal skin samples. The latter method was found to be more effective in separation of MF and ID cases using clustering analysis, and therefore each sample in the study was normalized against a collection of normal skin samples. For this purpose, 6 nonphotoexposed normal skin samples were chosen, and an average for each gene was calculated for those genes for which data were available for at least 4 of the samples. This average was used to normalize all ID and MF samples. All raw data and normalized data are included in the supplementary information (http://bioinfo.cnio.es/data/mycosis_fungoides).
Clustering and statistical analysis
For clustering analysis the Self-Organizing Tree Algorithm (SOTA) clustering program28 (http://gepas.bioinfo.cnio.es/cgi-bin/sotarray) was used and trees were viewed using the TreeView program.
Gene biologic functions were assigned by using the GENECARDS database29 (http://bioinformatics.weizmann.ac.il/cards).
To identify the genes important in distinguishing subgroups within our patient set we used a t statistic (Welch's version that does not require equal variances30 ), and unadjusted P values were obtained from permutation tests. Adjusted P values were obtained using Benjamini and Hochberg's procedure for the control of the false discovery rate.31 Genes with unadjusted P values less than .001 and adjusted P values less than .1 were deemed to be significant for group separation. These procedures are implemented in http://bioinfo.cnio.es/cgi-bin/tools/multest/multest.cgi.
In order to validate the 27-gene MF prediction model, gene clustering was carried out using the self-organizing map (SOM) algorithm32 defining a maximum of 7 gene clusters. An average for each gene cluster was calculated, and this average was used for validation using discriminant analysis in the study group and blind set of patients, with the software SPSS Base version 10.0 (SPSS, Chicago, IL). Additionally, in order to identify a smaller subset of class predictor genes, the gene with the lowest P value was selected from each SOM cluster. Again, these genes were validated by discriminant analysis in both the study set and the blind patient set.
To compare histologic and clinical characteristics between the 2 MF clusters identified, the Fisher exact test was used.
Immunohistochemistry
Techniques for immunohistochemistry have been previously described.24 H/E staining of all test group samples was examined to identify cases with tumor-type disease compared with plaque-type disease and samples containing large cells (Figure 1). Immunohistochemistry and histologic assays were performed on the paraffin-embedded tissue samples from the test group using commercially available monoclonal CD3, CD8, and Ki67 antibodies (DakoCytomation, Glostrup, Denmark); CD4 antibody (Master Diagnóstica, Granada, Spain); CD7 and CD30 antibodies (Novocastra Laboratories, Newcastle upon Tyne, United Kingdom); and STAT3 and STAT1 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA; Figure 1).
Relevant histologic and immunophenotypical characteristics were analyzed. Plaque versus tumor stage (H/E stain; top left and right panels, respectively; original magnifications, × 50, × 100 [inset]); high proliferative index vs low proliferative index (Ki67 low, Ki67 high, middle panels; original magnification, × 100); and common phenotype (CD4+, CD8-, middle panels; original magnification, × 200) vs aberrant phenotype (CD4- and/or CD8+, bottom panels; original magnification, × 200).
Relevant histologic and immunophenotypical characteristics were analyzed. Plaque versus tumor stage (H/E stain; top left and right panels, respectively; original magnifications, × 50, × 100 [inset]); high proliferative index vs low proliferative index (Ki67 low, Ki67 high, middle panels; original magnification, × 100); and common phenotype (CD4+, CD8-, middle panels; original magnification, × 200) vs aberrant phenotype (CD4- and/or CD8+, bottom panels; original magnification, × 200).
Common phenotype was defined as CD3+/CD4+/CD8-/CD7-, while any other phenotype was considered aberrant. High proliferative index was defined as samples displaying high or intermediate levels of Ki67 (more than 30% of positive tumoral cells), while low proliferative index cases were those with low Ki67 expression (Figure 1).
Results
Data normalization
Preliminary studies were carried out using 6 ID and 8 MF samples. Hierarchic clustering of all MF and ID cases did not separate cases clearly into 2 groups (Figure 2A). Taking into account that in some MF and ID cases, T-cell infiltration accounts for less than 10% of the total tissue, we attempted to eliminate the signal coming from normal surrounding tissue in both sample types. We used 2 methods to reduce background: (1) median centering normalization of each gene in MF and ID samples and (2) normalization of each sample against a pool of normal skin samples. Optimal results were achieved by normalizing each MF and ID sample against a pool of 3 normal nonphotoexposed skin samples (Figure 2B). Therefore, this method of normalization against normal skin samples is a valid method for enriching the data obtained from microarray data in these types of samples where the number of tumoral cells is low, allowing better clustering and classification of these tumors. For the remainder of the study this type of normalization was used for all cases of MF and ID using a total of 6 normal skin samples for normalization.
Unsupervised clustering of raw and normalized MF and ID data. (A) Using raw data from 8 MF patients and 6 ID patients, group separation was not possible. (B) Normalization of all samples against normal skin allowed the separation of MF from ID cases.
Unsupervised clustering of raw and normalized MF and ID data. (A) Using raw data from 8 MF patients and 6 ID patients, group separation was not possible. (B) Normalization of all samples against normal skin allowed the separation of MF from ID cases.
MF signature: differential expression with inflammatory skin conditions
To identify genes implicated in MF tumorigenesis, a total of 29 samples from MF patients and 11 ID samples were used. Data from microarray studies of each of these cases were normalized against the average signal from 6 normal skin samples. Using a Student t test, with an adjusted P value to control the false discovery rate, 27 genes significant in the separation of MF from ID samples were identified (Table 1). The expression pattern of these 27 genes is notably different between MF and ID cases (Figure 3). These genes are involved in a large variety of functions, such as cell cycle control, apoptosis, and signal transduction. However, the most striking finding is the up-regulation of almost 10 genes implicated directly in TNF signaling, including genes such as TRAF1, BIRC3, and TNFSF5.
A total of 27 genes significant in separating MF from ID cases
Gene symbol . | P, unadjusted . | P, adjusted . | Function . |
|---|---|---|---|
| FJX1 | .00002000 | .0210896 | Differentiation and development |
| Hs. 127160 | .00002000 | .0210896 | ESTs |
| STAT4 | .00002000 | .0210896 | Signal transduction, T-cell differentiation/proliferation |
| SYNE-1B | .00004000 | .0210896 | Signal transduction |
| CDC16 | .00004000 | .0210896 | Cell cycle control |
| BCL7A | .00004000 | .0210896 | Oncogenesis, actin binding |
| TRAF1 | .00006000 | .0210896 | TNF signaling, apoptosis regulator |
| BIRC3 | .00006000 | .0210896 | Inhibition of apoptosis |
| PRO0823 | .00006000 | .0210896 | Hypothetical protein PRO0823, function unknown |
| WBSCR14 | .00006000 | .0210896 | Role in cell differentiation and/or proliferation |
| ITGAX | .00010000 | .0319539 | Integrin, cell adhesion |
| MGC5363 | .00014000 | .0395430 | Hypothetical protein MGC5363, function unknown |
| LYN | .00016000 | .0395430 | Signal transduction, oncogene |
| EB12 | .00016000 | .0395430 | EBV induced, receptor signaling, immune response |
| BARX2 | .00018000 | .0395430 | Differentiation and development, cell adhesion |
| FBP17 | .00018000 | .0395430 | Forming binding |
| RASSF2 | .00022000 | .0454873 | Oncogene |
| WASPIP | .00026000 | .0507712 | Cytoskeleton organization, signal transduction |
| LCP2 | .00031999 | .0562389 | T-cell signaling |
| PCTAIRE2BP | .00031999 | .0562389 | Cell cycle control |
| HCK | .00033999 | .0569084 | Signal transduction |
| DEDD | .00037999 | .0607124 | Apoptosis |
| ACTN1 | .00039999 | .0611292 | Actinin, cytoskeleton organization |
| SELP | .00043999 | .0644404 | Selectin, cell adhesion |
| CD5 | .00047999 | .0674867 | T-cell marker, T-cell proliferation |
| UBE2L3 | .00061999 | .0838176 | Ubiquitination |
| TNFRSF5 | .00069999 | .0911278 | TNF family, T-cell activation, apoptosis inhibitor |
Gene symbol . | P, unadjusted . | P, adjusted . | Function . |
|---|---|---|---|
| FJX1 | .00002000 | .0210896 | Differentiation and development |
| Hs. 127160 | .00002000 | .0210896 | ESTs |
| STAT4 | .00002000 | .0210896 | Signal transduction, T-cell differentiation/proliferation |
| SYNE-1B | .00004000 | .0210896 | Signal transduction |
| CDC16 | .00004000 | .0210896 | Cell cycle control |
| BCL7A | .00004000 | .0210896 | Oncogenesis, actin binding |
| TRAF1 | .00006000 | .0210896 | TNF signaling, apoptosis regulator |
| BIRC3 | .00006000 | .0210896 | Inhibition of apoptosis |
| PRO0823 | .00006000 | .0210896 | Hypothetical protein PRO0823, function unknown |
| WBSCR14 | .00006000 | .0210896 | Role in cell differentiation and/or proliferation |
| ITGAX | .00010000 | .0319539 | Integrin, cell adhesion |
| MGC5363 | .00014000 | .0395430 | Hypothetical protein MGC5363, function unknown |
| LYN | .00016000 | .0395430 | Signal transduction, oncogene |
| EB12 | .00016000 | .0395430 | EBV induced, receptor signaling, immune response |
| BARX2 | .00018000 | .0395430 | Differentiation and development, cell adhesion |
| FBP17 | .00018000 | .0395430 | Forming binding |
| RASSF2 | .00022000 | .0454873 | Oncogene |
| WASPIP | .00026000 | .0507712 | Cytoskeleton organization, signal transduction |
| LCP2 | .00031999 | .0562389 | T-cell signaling |
| PCTAIRE2BP | .00031999 | .0562389 | Cell cycle control |
| HCK | .00033999 | .0569084 | Signal transduction |
| DEDD | .00037999 | .0607124 | Apoptosis |
| ACTN1 | .00039999 | .0611292 | Actinin, cytoskeleton organization |
| SELP | .00043999 | .0644404 | Selectin, cell adhesion |
| CD5 | .00047999 | .0674867 | T-cell marker, T-cell proliferation |
| UBE2L3 | .00061999 | .0838176 | Ubiquitination |
| TNFRSF5 | .00069999 | .0911278 | TNF family, T-cell activation, apoptosis inhibitor |
Using a t statistic and an adjusted P value, 27 genes significant in the separation of MF from ID samples were identified (29 MF samples, 11 ID samples).
Using a t statistic and an adjusted P value, 27 genes significant in the separation of MF from ID samples were identified (29 MF samples, 11 ID samples).
MF prediction model construction and validation by blind testing
Clustering, using the SOM algorithm, of the 27 genes significant in separating MF and ID cases revealed 6 gene clusters (supplementary information: http://bioinfo.cnio.es/data/mycosis_fungoides). An average was calculated for each cluster, and using these averages the 27-gene model was validated by discriminant analysis, both in the original sample and in a blind set of 24 MF samples, yielding a success rate of 100% in both cases (Table 2). In order to identify a smaller subset of class predictor genes, the gene with the lowest P value (Table 1) was selected from each of the 6 clusters generated by the SOM algorithm. The final class prediction model of 6 genes (Table 3) was validated by discriminant analysis using only the samples for which all data were available for all 6 genes without imputation of missing values. This class prediction model classifies all cases with 97.3% accuracy in the original sample set and with 97.0% accuracy in the blind set (Table 4).
Discriminant analysis of averaged 27-gene MF prediction model in study group of 29 MF and 11 ID cases and in blind test group of 24 MF cases
Class . | Predicted group membership . | . | Total . | |
|---|---|---|---|---|
. | MF . | ID . | . | |
| Original study group, count (%) | ||||
| MF | 29 (100) | 0 (0) | 29 (100) | |
| ID | 0 (0) | 11 (100) | 11 (100) | |
| Blind set, count (%) | ||||
| MF | 24 (100) | 0 (0) | 24 (100) | |
| ID | 0 (0) | 11 (100) | 11 (100) | |
Class . | Predicted group membership . | . | Total . | |
|---|---|---|---|---|
. | MF . | ID . | . | |
| Original study group, count (%) | ||||
| MF | 29 (100) | 0 (0) | 29 (100) | |
| ID | 0 (0) | 11 (100) | 11 (100) | |
| Blind set, count (%) | ||||
| MF | 24 (100) | 0 (0) | 24 (100) | |
| ID | 0 (0) | 11 (100) | 11 (100) | |
100% of original study group cases correctly classified
100% of blind set cases correctly classified
A 6-gene MF prediction model
Gene symbol . | P, unadjusted . | P, adjusted . | Function . |
|---|---|---|---|
| FJX1 | .0000200 | .0210896 | Differentiation and development |
| Hs. 127160 | .0000200 | .0210896 | EST, function unknown |
| STAT4 | .0000200 | .0210896 | Signal transduction, T-cell differentiation/proliferation |
| SYNE-1B | .0000400 | .0210896 | Signal transduction |
| TRAF1 | .0000600 | .0210896 | Apoptosis regulator, TNF signaling |
| BIRC3 | .0000600 | .0210896 | Inhibition of apoptosis |
Gene symbol . | P, unadjusted . | P, adjusted . | Function . |
|---|---|---|---|
| FJX1 | .0000200 | .0210896 | Differentiation and development |
| Hs. 127160 | .0000200 | .0210896 | EST, function unknown |
| STAT4 | .0000200 | .0210896 | Signal transduction, T-cell differentiation/proliferation |
| SYNE-1B | .0000400 | .0210896 | Signal transduction |
| TRAF1 | .0000600 | .0210896 | Apoptosis regulator, TNF signaling |
| BIRC3 | .0000600 | .0210896 | Inhibition of apoptosis |
Discriminant analysis of 6-gene MF prediction model in study group of 29 MF and 11 ID cases and in blind test group of 24 MF cases, in which data were available for all 6 genes in the model
. | Predicted group membership . | . | . | |
|---|---|---|---|---|
| Class . | MF . | ID . | Total . | |
| Original study group, count (%) | ||||
| MF | 27 (100) | 0 (0) | 27 (100) | |
| ID | 1 (10) | 9 (90) | 10 (100) | |
| Blind set, count (%) | ||||
| MF | 22 (95.7) | 1 (4.3) | 23 (100) | |
| ID | 0 (0) | 10 (100) | 10 (100) | |
. | Predicted group membership . | . | . | |
|---|---|---|---|---|
| Class . | MF . | ID . | Total . | |
| Original study group, count (%) | ||||
| MF | 27 (100) | 0 (0) | 27 (100) | |
| ID | 1 (10) | 9 (90) | 10 (100) | |
| Blind set, count (%) | ||||
| MF | 22 (95.7) | 1 (4.3) | 23 (100) | |
| ID | 0 (0) | 10 (100) | 10 (100) | |
97.3% of original study group cases correctly classified
97.0% of blind set cases correctly classified
Identification of MF subgroups
In order to identify subgroups of MF patients, unsupervised hierarchic cluster analysis was performed on all 53 MF samples using data from all genes (3393) for which at least 80% of the data were available, and 2 main subgroups or clusters of MF were identified (Figure 4A). At the same time, the heterogeneity of the MF samples was analyzed in terms of clinical (treatment responders vs nonresponders), histologic (plaque vs tumor stage; large cell component), or immunophenotypical (common vs aberrant phenotype, high proliferative index vs low proliferative index, and immunostaining for STAT1, STAT3, or CD30 antibodies) characteristics of the patients. The Fisher exact test was used to find significant differences between the 2 clusters. Some characteristics associated with more aggressive phenotypes have a tendency to be found in cluster 2. All tumor-stage disease cases (100%, P = .061), along with cases expressing activated nuclear STAT3, an oncogene that has been demonstrated to play a role in apoptosis escape in MF cell lines,15 are overrepresented in cluster 2 (47% compared with 17%, P = .069). Other characteristics associated with aggressive disease (large cell component, high proliferative index, and high-stage disease) are more predominant in cluster 2, although these differences do not reach significance.
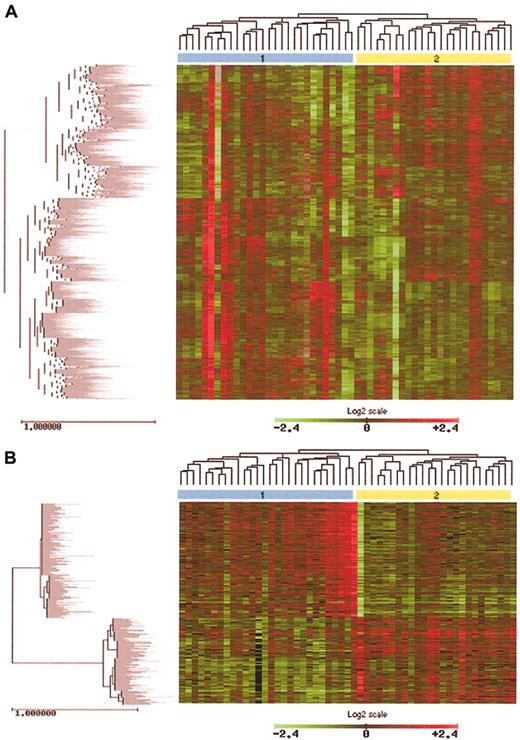
Identification of MF subgroups using hierarchic clustering methods. (A) Unsupervised clustering analysis of approximately 3400 genes in 53 MF cases revealed 2 main MF clusters. Cluster 2 tends to represent more patients with tumor stage disease and nuclear STAT3 expression, characteristics associated with aggressive disease. (B) Using a T statistic and an adjusted P value, genes significant in the differentiation of MF cluster 1 and MF cluster 2 were identified. After removal of ESTs and hypothetical genes, a total of 334 named genes were found. The expression pattern of these 334 genes is notably different between MF cluster 1 and cluster 2, as shown by unsupervised hierarchic clustering.
Identification of MF subgroups using hierarchic clustering methods. (A) Unsupervised clustering analysis of approximately 3400 genes in 53 MF cases revealed 2 main MF clusters. Cluster 2 tends to represent more patients with tumor stage disease and nuclear STAT3 expression, characteristics associated with aggressive disease. (B) Using a T statistic and an adjusted P value, genes significant in the differentiation of MF cluster 1 and MF cluster 2 were identified. After removal of ESTs and hypothetical genes, a total of 334 named genes were found. The expression pattern of these 334 genes is notably different between MF cluster 1 and cluster 2, as shown by unsupervised hierarchic clustering.
To identify the biologic factors responsible for the clustering observed within the MF cases, a Student t test comparing MF clusters 1 and 2 was performed. A total of 583 genes whose expression was significantly different in distinguishing between these MF clusters were identified. To simplify the analysis, all expressed sequence tags (ESTs) and hypothetical genes were excluded from further analysis, leaving a total of 334 named genes implicated in the differentiation of MF cluster 1 and cluster 2 (Figure 4B). (Supplementary information: http://bioinfo.cnio.es/data/mycosis_fungoides.) Cluster 2, which tends to include the more aggressive cases, shows a more pronounced up-regulation of many genes implicated in TNF pathway signaling, and parallel up-regulation of oncogenes, positive cell-cycle regulators, and antiapoptotic genes. Cluster 1 cases, which tend to be less aggressive in terms of STAT3 expression and disease type (plaque vs tumor), show increased expression of a variety of tumor suppressor genes. Increased expression of growth factors, interleukins, oncogenes, and positive cell-cycle regulators is also observed.
Genes implicated in separation of MF immunohistochemical/histologic subgroups
The gene expression pattern between aberrant and common phenotype cases and tumor stage and plaque stage disease were compared using Student t tests.
A total of 11 genes significant in the separation of common phenotype and aberrant phenotype cases were identified (Figure 5A), 10 of which were up-regulated in aberrant phenotype cases. Some of the up-regulated genes are of unknown function, but there are also genes implicated in oncogenesis, signal transduction, and protein ubiquitination.
Genes implicated in separation of MF immunohistochemical/histologic subgroups. (A) A total of 11 genes that are significant in the separation of common phenotype and aberrant phenotype cases were identified, 10 of which are up-regulated in aberrant phenotype cases, with one EST (Hs.199961) down-regulated in aberrant phenotype cases. (B) A total of 5 genes significant in the separation of tumor stage disease from plaque stage disease were identified, all 5 of which are down-regulated in cases with tumor stage disease.
Genes implicated in separation of MF immunohistochemical/histologic subgroups. (A) A total of 11 genes that are significant in the separation of common phenotype and aberrant phenotype cases were identified, 10 of which are up-regulated in aberrant phenotype cases, with one EST (Hs.199961) down-regulated in aberrant phenotype cases. (B) A total of 5 genes significant in the separation of tumor stage disease from plaque stage disease were identified, all 5 of which are down-regulated in cases with tumor stage disease.
A total of 5 genes significant in the separation of tumor stage disease from plaque stage disease were identified (Figure 5B), all 5 of which are down-regulated in cases with tumor stage disease. These genes were implicated in inhibition of NFκB activation, signal transduction, transcription regulation, and stress response.
Discussion
Data normalization
In order to better understand MF tumorigenesis, samples from MF and ID patients have been analyzed using the cDNA microarray CNIO OncoChip, which has been specifically designed for the study of cancer.24 The raw data from each sample were normalized to an average of the signal from a pool of normal nonphotoexposed skin samples in an effort to reduce the signal coming from surrounding normal tissues. This step has allowed the reduction of background noise in these samples, in which tumoral cells account for less than 10% of the total cells, and subsequent clustering of samples into the correct subgroups (Figure 2B). Without this normalization, separation of the samples by hierarchic clustering methods was not possible (Figure 2A).
TNF antiapoptotic pathway activation in the tumorigenesis of MF
Using the normalized microarray data from 29 MF patients and 11 ID patients, 27 genes were identified whose expression is significantly different between MF and ID cases (Table 1; Figure 3). Of the 27 genes identified, 20 were up-regulated in the MF cases, while the remaining 7 genes were down-regulated in the MF cases.
Most interesting is the observation of the up-regulation of almost 10 genes directly implicated in the regulation of TNF signaling including BIRC3, a member of the inhibitor of apoptosis (IAP) gene family, transcriptionally regulated by NF-κB.33 The genes of this family are characterized by the presence of IAP repeat or baculoviral IAP repeat domain (BIR) regions,34 essential for their antiapoptotic effects.35 BIRC3 also known as cellular IAP2 (c-IAP2), possesses an antiapoptotic BIR domain,35 a caspase binding caspase recruitment domain (CARD), and a finger (RING) domain for degradation of target proteins36 such as caspase-3 and caspase-737 (Figure 6).
TNF signaling pathway deregulation in MF tumorigenesis. A combination of antiapoptotic signaling by TNFR1, propelled by up-regulation of TRAF1 and BIRC/IAP proteins and inhibition of proapoptotic TNFR1 signaling by BIRC1/BIRC3 caspase inactivation. TNFR2 antiapoptotic signaling is active due to the overexpression of TRAF1 and BIRC3. Meanwhile, CD40L/TNFSF5 overexpression may induce T-cell proliferation via CD40 receptor and TRAF1. Overexpression of IL2R activates Jak2 and STAT4 and subsequently induces expression of oncogenes c-MYC, LYN, and HCK. Oncogene LYN and HCK participate in a feedback loop of TNF antiapoptotic TNF signaling by producing endogenous TNF, thereby autostimulating TNFR1 and TNFR2 antiapoptotic pathways. Genes up-regulated in MF cases are indicated by red text label, whereas green text labels indicate down-regulated genes.
TNF signaling pathway deregulation in MF tumorigenesis. A combination of antiapoptotic signaling by TNFR1, propelled by up-regulation of TRAF1 and BIRC/IAP proteins and inhibition of proapoptotic TNFR1 signaling by BIRC1/BIRC3 caspase inactivation. TNFR2 antiapoptotic signaling is active due to the overexpression of TRAF1 and BIRC3. Meanwhile, CD40L/TNFSF5 overexpression may induce T-cell proliferation via CD40 receptor and TRAF1. Overexpression of IL2R activates Jak2 and STAT4 and subsequently induces expression of oncogenes c-MYC, LYN, and HCK. Oncogene LYN and HCK participate in a feedback loop of TNF antiapoptotic TNF signaling by producing endogenous TNF, thereby autostimulating TNFR1 and TNFR2 antiapoptotic pathways. Genes up-regulated in MF cases are indicated by red text label, whereas green text labels indicate down-regulated genes.
BIRC1, also known as neuronal apoptosis inhibitor protein (NAIP), which binds to the cytoplasmic region of TNFR2, the type 2 tumor necrosis family receptor,38 was also found to be up-regulated in MF cases (Figure 6). However, the significance of this up-regulation was just outside the adjusted P value threshold set (P = .00088, adjusted P = .100698). Although BIRC1 lacks a CARD domain and a RING domain, it can inhibit caspase-3 and caspase-7 via its BIR domains alone39 (Figure 6).
TRAF1 has been identified here as being up-regulated in MF cases, extending previous observations concerning TNFR-associated factor 1 (TRAF1) relevance in B-cell lymphomas.40 TRAF1 is inducible by NF-κB41 and interacts with TNFR242 and several TNF family members such as CD4043,44 (Figure 6). TNFR2, a receptor of the TNF receptor superfamily lacking an intracellular death domain, mediates survival signaling by recruiting TRAF1, TRAF2,38,42 and inhibitor of apoptosis proteins such as BIRC3. TNFR2 activates NF-κB and Jun kinase (JNK) survival pathways,38,42,45,46 and the IAP/BIRC protein component of the complex inhibits apoptotic signaling (Figure 6).
The TNF receptor 1, TNFR1, possesses an intracellular death domain and induces apoptosis by recruitment of TNF receptor–associated via death domain (TRADD) and subsequent activation of caspase-8 through the adapter protein Fas-associated via death domain (FADD)46,47 (Figure 6). However, TRAF2 binds with high affinity to TRADD,48 and subsequent recruitment of RIP1 (RALBP1), TRAF1, and IAP/BIRC protein family members leads to suppression of apoptotic signaling from this receptor complex33,48 (Figure 6).
TNFSF5 (CD40L or TNF-related activation protein [TRAP]) is a TNF family member implicated in prevention of apoptosis and enhancement of cell proliferation.49,50 Expression of CD40L has previously been reported in cases of MF where it was suggested that CD40L might play a role in a paracrine loop important for preventing apoptosis and/or positively regulating growth.17 It has also been suggested that CD40L expression may be important for homing of neoplastic T cells to the skin17 (Figure 6).
Hemopoietic cell kinase (HCK) and oncogene v-yes-1 Yamaguchi sarcoma viral-related oncogene homolog (LYN) are tyrosine kinases up-regulated in MF cases. HCK is responsible for production of TNF in murine cells,51 and both HCK and LYN have been implicated in TNF production in human monocytes.52 In this study, in addition to HCK and LYN up-regulation in MF cases, TNF was overexpressed, although significance was just outside the stringent range set here. Additionally, HCK53 and LYN54 and the oncogene c-MYC55 are induced by IL2 signaling by Janus kinase 2 (JAK2) and STAT4 activation. In this study STAT4 is overexpressed in MF cases, as is the IL2 receptor (IL2R), but with a lower significance. According to the findings of this study, it is possible that in MF cases, IL2-induced up-regulation of HCK and LYN leads to production of TNF and subsequent antiapoptotic signaling via TNFR1 and TNRF2 (Figure 6).
UBE2L3, a ubiquitin pathway protein also known as UbcH7, is down-regulated in this MF series. This protein is involved in the ubiquitination and degradation of the oncogenes MYC56 and c-FOS.57 Therefore, UBE2L3 down-regulation in MF cases may assist MF tumorigenesis (Figure 6).
Since only genes with a maximum of 20% missing values could be included in this study, many genes with a possible role in this pathway were excluded from t test analysis. By averaging the gene expression patterns for the MF and ID subgroups for genes with no more than 50% missing values, some supporting evidence for this model is provided. Briefly, NFκB is up-regulated in MF cases compared with ID cases (1.54-fold up-regulation of NFKB1), while IL2 shows a 1.86-fold increased expression in MF cases (Figure 6). Inhibitor of NFκB (IκB) kinase, responsible for the phosphorylation of IκB and therefore the activation of NFκB, also showed a tendency to be up-regulated in MF cases. Finally, we found that in MF cases there is also a very slight increase in the expression of genes involved in JNK and p38 proliferation and inflammation pathways (JunB, JunD, mitogen-activated protein kinase 14 [MAPK14; p38], activating transcription factor 3 [ATF3], and TRAF2) and NFκB activation pathways (heat shock protein 90 [HSP90] family members, CDC37 and TRAF2).
In conclusion, MF tumorigenesis is associated with changes in the regulation of a combination of antiapoptotic signaling through TNFR1 (induced by up-regulation of TRAF1 and BIRC/IAP expression) and inhibition of the proapoptotic pathways of TNFR1 (by BIRC1/BIRC3 caspase inhibition). Additionally, antiapoptotic signaling via TNFR2 is active as signaled by the overexpression of TRAF1 and BIRC3. CD40L/TNFSF5 overexpression contributes to MF tumorigenesis by inducing T-cell proliferation via CD40 receptor and TRAF1. Interleukin-2 may also play a critical role in MF tumorigenesis through its receptor by activating JAK2 and STAT4 and subsequently inducing expression of oncogenes c-MYC, LYN, and HCK. Finally, LYN and HCK participate in an autocrine loop by participating in endogenous TNF production and thereby autostimulating TNFR1 and TNFR2 antiapoptotic pathways and creating a feedback loop of TNF survival pathway activation. TNF apoptotic pathway silencing may be due to BIRC1/BIRC3 overexpression and caspase inhibition.
MF class-prediction model
The prediction model using the average signal from the clusters of all 27 genes can correctly assign class in 100% of both the original MF series and the blind set of 24 MF patients. Subsequently, the gene with the lowest P value from each cluster was chosen, and this 6-gene prediction model correctly classified 97.3% of the original series and 97.0% of the MF blind set. Further weight to the validation of this prediction model is provided by the exclusive presence in the blind test of cases with early stages of MF, whose histology more closely mimics that of ID cases.
Subclasses of MF cases based on expression patterns
Hierarchic clustering of approximately 3400 genes in all 53 MF samples revealed 2 main MF clusters (Figure 4). Cluster 2 tends to include a higher proportion of cases with tumor-stage disease and nuclear STAT3, both of which are indicative of a more aggressive phenotype.
After exclusion of ESTs and hypothetical genes, a total of 334 genes whose expression was significantly different in distinguishing between these MF clusters were identified (Figure 4B). Cluster 2 shows a stronger up-regulation of many genes implicated in TNF pathway signaling. Members of the activator protein 1 (AP1) transcription complex involved in JNK/p38 pathway activation such as ATF and FOS46 are also highly expressed in cluster 2 cases along with members of the TNF receptor family such as TNFSF13 (APRIL), which contributes to tumor cell growth.58 Finally, members of the BIRC family of apoptosis inhibitors such as BIRC2 are up-regulated in cluster 2 in addition to IL2RB, FOS, and MYC, which play an important role in TNF signaling (Figure 6). Parallel to up-regulation of TNF signaling genes, up-regulation of oncogenes, such as members of the RAS family of oncogenes, MYC, FOS, VAV3, WNT, and PIM1, is observed and is accompanied by up-regulation of positive cell-cycle regulators such as cyclin D2.
Cluster 1 cases show higher expression of a number of genes with tumor-suppressing properties. These include HIC1, LGI1, DLEU1, ST14, MSH5, and PTEN, whose alteration has been described in lymphoid malignancies.59 However, increased expression of numerous oncogenes and positive cell-cycle regulators is also observed in this cluster 1, including growth factors such as those of the fibroblast growth factor family (FGFR1 and FGF5), epidermal growth factor family (GRB7, EGFL3, and ERBB3), platelet derived growth factor family (PDGFA), vascular endothelial growth factor family (ANGPTL3), and the transforming growth factor family (BMP5, TAB1, TGFBR3, MADHIP, and GDF1). In parallel, higher expression of oncogenes (RAB1, RAB14, RABL2B, TIM) and interleukins (IL15RA, IL7, IL1RAPL1, and IL11) is observed accompanied by up-regulation of positive cell-cycle regulators such as cyclin A1, cyclin-dependent kinase family genes (CDKL2 and CDKL3), and cell division cycle genes (CDC27 and CDC20).
Genes associated with aberrant phenotype expression in MF cases
The common phenotype (CD3+/CD4+/CD8-/CD7-) in MF is generally considered to be less aggressive than phenotypes such as CD8+ (Berti et al3 ) and/or CD7+ (Berti et al3 and Murphy et al4 ) and/or CD4-.3,60 Corroborating evidence has been found for this in the current study with the up-regulation of oncogenes (CSF1R, LHFPL1), cell-cycle control genes, and transcription factors in cases with aberrant phenotypes (Figure 5A). CSF1R is the receptor for colony-stimulating factor 1 and product of the FMS proto-oncogene, and its up-regulation has previously been described in hematopoietic malignancies.61,62
Genes associated with tumor stage MF disease
A total of 5 genes differentially expressed between tumor and plaque stage disease were identified (Figure 5B), all of which are up-regulated in cases with tumor stage disease, including IKBKAP, a gene whose overexpression has been shown to block TNF-induced NFκB activation.63 However, its role under normal biologic circumstances is unclear.
In conclusion, this study has shown that while tumoral cells account for less than 10% of the total tissue in MF, by using microarray studies and normalization against normal skin samples it is possible to detect a signature that distinguishes this type of tumor from common inflammatory skin conditions. This MF signature includes deregulation of TNF signaling in addition to an autocrine TNF feedback loop promoting antiapoptotic signaling. From this MF signature it was possible to identify a 6-gene MF prediction model that allows 97.3% accurate distinction from ID cases in the study group, and 97.0% accurate classification in a randomly chosen external blind set of 24 early stage MF patients. These microarray studies allowed the identification of 2 MF subgroups, one of which tends to include mainly the aggressive MF cases and shows increased expression of TNF pathway genes and oncogenes.
Appendix: members of the Cooperative Cutaneous Lymphoma Group of Madrid
Pablo Ortiz, F. Vanaclocha, Dermatology Department, Hospital 12 de Octubre, Madrid; M. García Rodríguez, Dermatology Department, Hospital Principe de Asturias; Alcalá de Henares, J. Fernández Herrera, Dermatology Department, Hospital de la Princesa, Madrid; I. Mora, Dermatology Department, Hospital Clínico San Carlos, Madrid; C. García, Dermatology Department, Hospital Virgen de la Salud, Toledo; S. Vidal, Dermatology Department, Hospital Militar Gomez Ulla, Madrid; J. Fraga, Pathology Department, Hospital de La Princesa, Madrid; and L. Requena, Dermatology Department, Clínica de la Concepción, Madrid, Spain.
Prepublished online as Blood First Edition Paper, April 10, 2003; DOI 10.1182/blood-2002-11-3574.
Supported by grants from the Ministerio de Ciencia y Tecnología (1FD97-0431, BIO2000-0275-C02/01 and /02, and SAF2001-0060) Spain. L.T. is supported by a grant from the Centro Nacional de Investigaciones Oncológicas (CNIO) and the Department of Hematology/Institute for Molecular Medicine HEA PRTL grant.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are indebted to I. Fernandez and M. Lopez for their help with extraction of RNA from frozen tissue and subsequent amplification and to members of the microarray analysis department for help with target preparation and hybridization. We thank Ramon Diaz and Javier Herrero, both from the Bioinformatics unit of the CNIO, for their help and advice about statistical and clustering methods, respectively. Our thanks are also extended to the immunohistochemistry unit of the CNIO for assistance afforded for immunohistochemical assays.
For a list of members of the Cooperative Cutaneous Lymphoma Group of Madrid, see “Appendix.”

![Figure 1. Relevant histologic and immunophenotypical characteristics were analyzed. Plaque versus tumor stage (H/E stain; top left and right panels, respectively; original magnifications, × 50, × 100 [inset]); high proliferative index vs low proliferative index (Ki67 low, Ki67 high, middle panels; original magnification, × 100); and common phenotype (CD4+, CD8-, middle panels; original magnification, × 200) vs aberrant phenotype (CD4- and/or CD8+, bottom panels; original magnification, × 200).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/3/10.1182_blood-2002-11-3574/6/m_h81534709001.jpeg?Expires=1769096693&Signature=CjsORDMvN9xcjLBSEu~vISpxfd0vRxTYTeh2n0H~y1tuTd1IVTlV70yiMNLCZ2ajCF5pDjIBMZq8xyE6V~9wdNDZiAu9ueRfqMnxstYsxsS3JcndejxplWijFhmE6Se46A5xvKaxd-tQezp61xajRrPH7CM08gAusZ5Un9o8Nu6iLITD7wKCbas8sACztmeSgMSKfqwAfwt2jMAOUzM8xL2R2pT9oqSGopukcEIJDco5yqePCtvFROyXlPM2Zj5LOK7DQ-CjI8qHxjgGLu2RwRKB6sxgZCOFN4nkhNZjLrrrV1ltYPgEYf3QrM~m-3nL3n4xqAIdT9Qu6p3yXuzVgQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal