Abstract
We recently reported that human recombinant melanotransferrin (p97) presents a high transport rate across the blood-brain barrier that might involve the low-density lipoprotein receptor–related protein (LRP). We now report new interactions between p97 and another LRP ligand, the urokinase plasminogen activator (uPA) complex. By using biospecific interaction analysis, both pro-uPA and plasminogen are shown to interact with immobilized p97. Moreover, the activation of plasminogen by pro-uPA is increased by soluble p97. Because the uPA system plays a crucial role in cell migration, both in cancer and in angiogenesis, we also measured the impact of both endogenous membrane-bound and exogenous p97 on cell migration. The monoclonal antibody L235 (which recognizes a conformational epitope on p97) inhibited the migration of human microvascular endothelial cells (HMECs-1) and of human melanoma SK-MEL-28 cells, indicating that endogenous membrane-bound p97 could be associated with this process. In addition, low concentrations of exogenous p97 (10 and 100 nM) inhibited HMEC-1 and SK-MEL28 cell migration by more than 50%. These results indicate that membrane-bound and soluble p97 affect the migration capacity of endothelial and melanoma cells and suggest that p97 could be involved in the regulation of plasminogen activation by interacting with pro-uPA and plasminogen.
Introduction
Melanotransferrin (p97) possesses a high level of homology (37%-39%) with human serum transferrin, human lactoferrin, and chicken transferrin.1,2 It is a glycosylated protein that reversibly binds iron and was first found at high levels in malignant melanoma cells.1,3 Two forms of p97 have been reported, one of which is bound to cell membranes by a glycosylphosphatidylinositol anchor while the other form is both soluble and actively secreted.4-6 The exact physiologic role of either membrane-bound p97 or secreted p97 is largely unexplored.7
In the early 1980s, p97 was found to be expressed in much larger amounts in neoplastic cells and fetal tissues than in normal tissues.3,8,9 More recently, it was reported that p97 mRNA is widespread in normal human tissues.10 p97 is also expressed in reactive microglia associated with amyloid plaques in Alzheimer disease.11 Normal serum contains very low levels of p97,3 which were reported to increase by 5- to 6-fold in patients with Alzheimer disease.12
We previously demonstrated that recombinant human melanotransferrin (p97) is transported at high rate into the brain, using both an in vitro model of the blood-brain barrier (BBB) and in situ mouse brain perfusion.13 We also showed that p97 transcytosis might involve the low-density liproprotein receptor–related protein (LRP). This receptor is also known to mediate the internalization of the urokinase:plasminogen activator inhibitor:urokinase receptor complex (uPA:PAI-1:uPAR). Briefly, single-chain proenzyme-uPA is activated upon binding to its cell surface receptor uPAR,14 which is a glycosylphosphatidylinositol (GPI)–anchored membrane protein.15,16 After its activation, uPA (which catalyzes the conversion of plasminogen to plasmin) is quickly inhibited by the plasminogen activator inhibitor type-1 (PAI-1). The inactive uPA:PAI-1 complex binds to uPAR and then is rapidly internalized by LRP.17,18 The uPA:PAI-1 complex is degraded in lysosomes, whereas the uPAR is recycled at the cell surface.19 Other LRP ligands include pro-uPA, PAI-1, receptor-associated protein (RAP), and a diverse spectrum of structurally unrelated proteins.20
Because uPA plays a crucial role in enhancing cell migration and invasion during embryogenesis, wound healing, and metastasis,21-24 and since p97 and the uPA/uPAR complex may share the same pathway for endocytosis, we investigated potential interactions between p97 and components of the uPA/uPAR complex. We used the BIAcore apparatus (BIAcore, Piscataway, NJ) for examining potential protein-protein interactions in real time. We demonstrate here that pro-uPA and plasminogen interact in vitro with immobilized p97. In addition, we report that p97 stimulates the activation of plasminogen by pro-uPA. We also show that either an antibody directed against endogenous p97 or application of low concentrations of exogenous p97 inhibited cell migration of human endothelial and melanoma cells. These data suggest that the balance between membrane-bound p97 and soluble p97 could be important in cell migration, which is crucial for angiogenesis and tumor growth.
Materials and methods
Materials
Soluble human recombinant p97, which is produced by introducing a stop codon following the glycine residue at position 711, and monoclonal antibodies (mAbs) directed against p97 were kindly provided by Biomarin Pharmaceutical (Novato, CA). Tissue plasminogen activator (tPA), PAI-1, and plasmin were from Calbiochem (La Jolla, CA). Pro-uPA and plasminogen were from American Diagnostica (Greenwich, CT). Angiostatin was purchased from Angiogenesis Laboratories (Tucson, AZ), and uPA was from Roche Biochemicals (Laval, QC, Canada). CM5 sensor chips were from BIAcore (Piscataway, NJ). The plasmin substrate (d-Val-Leu-Lys-P-nitraniline, or VLK-pNA) and other biochemical reagents were from Sigma (Oakville, ON).
Blood-brain barrier model and transcytosis experiments
The in vitro model of the blood-brain barrier (BBB) was established by using a coculture of bovine brain capillary endothelial cells (BBCECs) and newborn rat astrocytes as previously described.25 p97 was radioiodinated with standard procedures using an iodo-beads kit and D-Salt Dextran desalting columns from Pierce (Rockford, IL), as previously described.13 Transcytosis experiments were performed as follows: one insert covered with BBCECs was set into a 6-well microplate with 2 mL Ringer–HEPES (N-2-hydroxyethylpiperazine-N-2-ethanesulfonic acid) and was preincubated for 2 hours at 37°C. [125I]-p97 (0.5-1.5 μCi [18-55 kBq] per assay), at a final concentration of 25 nM, was then added to the upper side of the insert. At various times, the insert was sequentially transferred into a fresh well to avoid possible reendocytosis of p97 by the abluminal side of the BBCECs. At the end of the experiment, [125I]-p97 was assayed in 500 μL of the lower chamber of each well following trichloroacetic acid (TCA) precipitation.
Cell culture
Cells were cultured under 5% CO2/95% air atmosphere. Human microvascular endothelial cells (HMECs-1) were from the Centers for Disease Control and Prevention (Atlanta, GA) and were cultured in MCDB-131 medium (Sigma) supplemented with 10 mM l-glutamine, 10 ng/mL epidermal growth factor (EGF), 1 μg/mL hydrocortisone, and 10% inactivated fetal bovine serum (FBS). Human umbilical vein endothelial cells (HUVECs) and SK-MEL-28 cells were obtained from American Type Culture Collection (Manassas, VA). HUVECs were cultured in an EGM-2 bullet kit from BioWhittaker (Walkersville, MD). Melanoma SK-MEL-28 cells were grown in modified Eagle medium (MEM) supplemented with 1 mM Na-pyruvate, 100 U/mL penicillin-streptomycin, 1.5 g/L Nabicarbonate, and 10% FBS.
BIAcore analysis
p97, PAI-1, and plasminogen were covalently coupled to a CM5 sensor chip via primary amine groups using the N-hydroxysuccinimide (NHS)/N-ethyl-N′-(dimethylaminopropyl)carbodiimide (EDC) coupling agents as previously described.26 Briefly, the carboxymethylated dextran was first activated with 50 μL NHS/EDC (50 mM/200 mM) at a flow rate of 5 μL/min. p97, PAI-1, or plasminogen (5 μg) in 20 mM acetate buffer, pH 4.0, was then injected and the unreacted NHS esters were deactivated with 35 μL 1 M ethanolamine hydrochloride, pH 8.5. Approximately 8000 to 10 000 relative units of p97, PAI-1, or plasminogen were immobilized on the sensor chip surface. Ringer solution or a 50 mM Tris (tris(hydroxymethyl)aminomethane)/HCl buffer (pH 7.5) containing 150 mM NaCl and 50 mM CaCl2 was used as the eluent buffer. Proteins were diluted in the corresponding eluent buffer and injected onto the sensor chip surface. Protein interactions were analyzed using both the Langmuir binding model, which is the simplest model for 1:1 interaction between analyte and immobilized ligand, and a 2-state conformational change model that describes a 1:1 binding of analyte to immobilized ligand followed by a conformational change.
Enzymatic assay and cell treatment with soluble p97
The enzymatic activity of pro-uPA was measured using a colorimetric assay. The reaction was performed in a final volume of 200 μL in an incubation medium consisting of 50 mM Tris/HCl buffer (pH 7.5), 150 mM NaCl, and 50 mM CaCl2. This incubation medium also contained 15 μg/mL VLK-pNA with or without plasminogen. Enzymatic activity was assessed in the absence or presence of p97. The reaction was started by the addition of pro-uPA. In this assay, the cleavage of VLK-pNA results in a P-nitraniline molecule that absorbs at 405 nm. The reaction product was monitored at 405 nm using a Microplate Thermomax Autoreader (Molecular Devices, Sunnyvale, CA).
HMECs-1 were grown to 85% confluency in 6-well plates and were incubated for 18 hours under 5% CO2/95% air atmosphere in cell culture medium with or without p97 (100 nM). Endothelial cells were washed twice with Ringer solution and mechanically scraped from the wells. Cells were counted and frozen at –80°C until used. A volume corresponding to 100 000 cells was incubated in the plasmin assay as above and plasmin activity was monitored at 405 nm for 60 minutes. HMECs-1 were also individualized by phosphate-buffered saline (PBS) citrate solution (138 mM NaCl, 2.7 mM KCl, 1.47 mM KH2PO4, 8.1 mM Na2HPO4–7H2O, 15 mM Na citrate pH 6.8) for 15 minutes. Cells were washed twice in Ringer-HEPES solution (150 nM NaCl, 5.2 mM KCl, 2.2 mM CaCl2, 0.2 mM MgCl2–6H2O, 6 mM NaHCO3, 5 mM HEPES, 2.8 mM glucose, pH 7.4) and counted. A volume corresponding to 100 000 cells was incubated in the plasmin assay with mAb L235 (325 nM) or immunoglobulin G (IgG) control. Plasmin activity was monitored at 405 nm for 480 minutes.
Cell migration assay
HMEC-1, HUVEC, and SK-MEL-28 cell migration was performed using Transwell filters (Costar, Corning, NY; 8 μm pore size) precoated with 0.15% gelatin for 2 hours at 37°C. The Transwell filters were assembled in 24-well plates (Falcon 3097, Fischer Scientific, Montreal, QC, Canada) and the lower chambers filled with 500 μL cell culture medium. To study the effect of p97, mAb L235, or mouse IgG on cell migration, HMEC-1, HUVEC, and SK-MEL-28 cells were harvested by trypsinization and centrifuged. Approximately 10 000 cells were resuspended in 100 μL fresh Dulbecco MEM (DMEM) with or without p97, mAb L235, or mouse IgG and added into the upper chamber of each transwell (lower chamber of the transwell also contained p97, mAb L235, or nonspecific mouse IgG). The plates were than placed at 37°C in 5% CO2/95% air for 18 hours. Cells that had migrated to the lower surface of the filters were fixed with 3.7% formaldehyde in PBS, stained with 0.1% crystal violet/20% MeOH, and counted (4 fields per filter). Photomicrographs at 100 × magnification were taken using a digital Nikon Coolpix 5000 camera (Nikon Canada, Mississauga, ON, Canada) attached to a Nikon TMS-F microscope (Nikon Canada).
Data analysis
Statistical analyses were made with the Student paired t test using GraphPad Prism (San Diego, CA). Significant difference was accepted for P values less than .05.
Results
Transcytosis of p97 through BBCEC monolayers
We first evaluated the transcytosis of p97 across an in vitro model of the BBB at 37°C (Figure 1A). A significant (> 50%) reduction in the transport of [125I]-p97 (25 nM) from the apical (blood) side to the basolateral (brain) side of BBCEC monolayers was observed in the presence of 640 nM RAP. Transcytosis of [125I]-p97 was unaffected by a 200-fold molar excess of bovine serum albumin (BSA). The permeability coefficent for sucrose is similar in the absence or presence of RAP, indicating that the integrity of the BBCEC monolayers was unaffected by this protein (data not shown). The results with RAP also suggest that LRP might be involved in p97 transcytosis, since it has been reported to be an LRP ligand, whereas BSA was shown to bind to megalin, another member of the low-density lipoprotein (LDL) receptor family, probably via cubilin.27 To determine whether protein interaction could occur between p97 and RAP, leading to a reduction in p97 transcytosis, protein interactions were investigated by using biologic interaction analysis in real time (Figure 1B). For this analytical approach, p97 was first immobilized on the surface of a sensor chip. Using standard NHS/EDC coupling procedures, about 8 to 10 ng/mm2 of p97 were immobilized. RAP or BSA (0.05 μg/μL) was then injected over immobilized p97. No interactions could be observed between these proteins and p97, suggesting that the inhibition of [125I]-p97 transcytosis is not related to protein interactions between p97 and RAP.
Transcytosis of p97 across BBCEC monolayers. (A) Transcytosis experiments were performed at 37°C for 2 hours. [125I]-p97 (25 nM) was added to the upper side of the cell-covered filter in the absence or presence of RAP (650 nM) or BSA (5 μM). At the end of the experiment, radiolabeled proteins were measured in the lower chamber of each well by TCA precipitation. Results represent means ± SE (n = 6). (B) p97 was immobilized on a sensor chip surface (CM5) as described in “Materials and methods.” p97, RAP, and BSA (5 μg/100 μL) were injected over immobilized p97. One representative experiment is shown (n = 3).
Transcytosis of p97 across BBCEC monolayers. (A) Transcytosis experiments were performed at 37°C for 2 hours. [125I]-p97 (25 nM) was added to the upper side of the cell-covered filter in the absence or presence of RAP (650 nM) or BSA (5 μM). At the end of the experiment, radiolabeled proteins were measured in the lower chamber of each well by TCA precipitation. Results represent means ± SE (n = 6). (B) p97 was immobilized on a sensor chip surface (CM5) as described in “Materials and methods.” p97, RAP, and BSA (5 μg/100 μL) were injected over immobilized p97. One representative experiment is shown (n = 3).
Pro-uPA and p97 interaction
To evaluate the impact of immobilization procedures on the structural integrity of p97, different mAbs directed against various conformational epitopes of p97 were injected over p97 (Figure 2). The surface plasmon resonance (SPR) signal generated by the interaction between p97 and various mAbs varied from 250 relative units (RU) to 2500 RU. These data show that the mAbs could still recognize p97, indicating that the protein is intact following its immobilization on the sensor chip surface. Table 1 shows the kinetic parameters estimated by the BIAevaluation software (BIAcore) for antibody interactions with p97. From these values, the affinity constant (KA = ka/Kd) of these mAbs for immobilized p97 ranged from 0.08 to 2.7 nM–1, and the relative affinities are HybE < L235 < 9B6 < 2C7, HybC < HybF.
Biospecific interaction analysis in real time between p97 and various anti-p97 mAbs. p97 was immobilized on a sensor chip (CM5) using standard coupling procedures incorporating NHS, EDC, and ethanolamine. Different mAbs directed against p97 (HybC, HybE, HybF, L235, 2C7, 9B6), diluted to 0.05 μg/μL in Ringer-HEPES, were injected into the BIAcore at a flow rate of 5 μL/min. The surface plasmon resonance response obtained for these mAbs was plotted (in relative units [RU]) as a function of time. After each injection, immobilized p97 was regenerated with 0.2M glycine at pH 2 for 2 minutes followed by a 2-minute injection of ferric ammonium citrate (1 mM) (n = 4).
Biospecific interaction analysis in real time between p97 and various anti-p97 mAbs. p97 was immobilized on a sensor chip (CM5) using standard coupling procedures incorporating NHS, EDC, and ethanolamine. Different mAbs directed against p97 (HybC, HybE, HybF, L235, 2C7, 9B6), diluted to 0.05 μg/μL in Ringer-HEPES, were injected into the BIAcore at a flow rate of 5 μL/min. The surface plasmon resonance response obtained for these mAbs was plotted (in relative units [RU]) as a function of time. After each injection, immobilized p97 was regenerated with 0.2M glycine at pH 2 for 2 minutes followed by a 2-minute injection of ferric ammonium citrate (1 mM) (n = 4).
Kinetics of interaction between immobilized p97 and mAbs
Antibody . | ΔRU . | Ka, M-1 s-1 . | Kd, s-1 . | KA = Ka/Kd, M-1 . | KD = Kd/Ka, M . |
|---|---|---|---|---|---|
| L235 | 1055 ± 82 | 4.4 × 104 | 5.3 × 10-5 | 0.9 × 109 | 0.1 × 10-10 |
| HybC | 1509 ± 184 | 7.2 × 104 | 4.5 × 10-5 | 1.6 × 109 | 6.4 × 10-10 |
| HybE | 232 ± 52 | 0.9 × 104 | 9.8 × 10-5 | 0.08 × 109 | 0.01 × 10-10 |
| HybF | 2199 ± 150 | 8.0 × 104 | 3.0 × 10-5 | 2.7 × 109 | 3.8 × 10-10 |
| 9B6 | 2440 ± 112 | 1.2 × 104 | 9.1 × 10-5 | 1.3 × 109 | 7.9 × 10-10 |
| 2C7 | 2290 ± 87 | 5.9 × 104 | 3.8 × 10-5 | 1.6 × 109 | 6.5 × 10-10 |
Antibody . | ΔRU . | Ka, M-1 s-1 . | Kd, s-1 . | KA = Ka/Kd, M-1 . | KD = Kd/Ka, M . |
|---|---|---|---|---|---|
| L235 | 1055 ± 82 | 4.4 × 104 | 5.3 × 10-5 | 0.9 × 109 | 0.1 × 10-10 |
| HybC | 1509 ± 184 | 7.2 × 104 | 4.5 × 10-5 | 1.6 × 109 | 6.4 × 10-10 |
| HybE | 232 ± 52 | 0.9 × 104 | 9.8 × 10-5 | 0.08 × 109 | 0.01 × 10-10 |
| HybF | 2199 ± 150 | 8.0 × 104 | 3.0 × 10-5 | 2.7 × 109 | 3.8 × 10-10 |
| 9B6 | 2440 ± 112 | 1.2 × 104 | 9.1 × 10-5 | 1.3 × 109 | 7.9 × 10-10 |
| 2C7 | 2290 ± 87 | 5.9 × 104 | 3.8 × 10-5 | 1.6 × 109 | 6.5 × 10-10 |
The difference between the relative units measured after and before injection of mAbs directed against p97 are presented (ΔRU) as well as the apparent association (Ka) and dissociation (Kd) constants. The affinity (KA) and dissociation (KD) constants were calculated from the Ka and Kd.
When pro-uPA and tPA (0.05 μg/μL) were injected over immobilized p97, protein interaction occurred between pro-uPA and p97 but not between tPA and p97 (Figure 3A). Around 8 to 10 ng/mm2 of PAI-1 was also immobilized onto another well of a sensor chip surface using NHS/EDC coupling conditions. No interaction between p97 and immobilized PAI-1 could be detected (Figure 3A). However, a strong interaction could be observed when tPA was injected over PAI-1, indicating that PAI-1 can still interact with tPA following immobilization (data not shown). In addition, plasminogen, plasmin, and angiostatin (0.05 μg/μL) were injected over immobilized p97 (Figure 3B). According to the SPR, plasminogen also interacts with immobilized p97, whereas plasmin and angiostatin, 2 plasminogen fragments, do not. The kinetic data obtained from binding of pro-uPA or plasminogen to immobilized p97 biosensor surface were evaluated using both the 1:1 Langmuir binding model and the 2-state conformational change model. Interestingly, the 2-state conformational change model was a better fit than the 1:1 Langmuir binding model when comparing a single concentration of either pro-uPA and plasminogen over p97 biosensor surface. Kinetic data obtained with the 2-state conformational model are presented in Table 2. Kinetic data for the interaction between pro-uPA and p97 show an association constant (ka1) of 0.6 × 104 M–1s–1 and a dissociation rate constant (kd1) of 1.7 × 10–3s–1. Furthermore, the forward rate constant (ka2 = 3.2 × 10–3s–1) and backward rate constant (kd2 = 7.1 × 10–4s–1) for the conformational change provide an apparent equilibrium dissociation constant [(KD = kd1/ka1)/(kd2/ka2)] of 65 nM. The kinetic analysis of plasminogen interaction with p97 shows an association constant (ka1) of 2.1 × 104 M–1s–1. The dissociation rate constant (kd1 = 43.0 × 10–3s–1), as well as the forward rate constant (ka2) of 6.0 × 10–3s–1 and backward rate constant (kd2) of 1.1 × 10–3s–1, are different from those seen for the pro-uPA interaction with p97. However, the apparent equilibrium dissociation constant (KD) between p97 and plasminogen is 350 nM, which is different from that observed for the interaction of pro-uPA with immobilized p97.
Molecular interactions of p97 and various components of the PA:plasmin system. (A) Pro-uPA and tPA (0.05 μg/μL), diluted in Ringer-HEPES, were injected onto immobilized p97 on a sensor chip at a flow rate of 5 μL/min. The SPR response for these proteins was plotted in RU as a function of time. p97 (0.05 μg/μL) was also injected over immobilized PAI-1 (p97/PAI-1). (B) Plasminogen, plasmin, or angiostatin (0.05 μg/μL) was injected onto immobilized p97. The SPR response for these proteins was plotted in RU as a function of time. The results indicate that pro-uPA and plasminogen interact with p97. After each injection the sensor chip surface with immobilized p97 was regenerated by injecting 10 mM glycine, pH 2.2, for 2 minutes.
Molecular interactions of p97 and various components of the PA:plasmin system. (A) Pro-uPA and tPA (0.05 μg/μL), diluted in Ringer-HEPES, were injected onto immobilized p97 on a sensor chip at a flow rate of 5 μL/min. The SPR response for these proteins was plotted in RU as a function of time. p97 (0.05 μg/μL) was also injected over immobilized PAI-1 (p97/PAI-1). (B) Plasminogen, plasmin, or angiostatin (0.05 μg/μL) was injected onto immobilized p97. The SPR response for these proteins was plotted in RU as a function of time. The results indicate that pro-uPA and plasminogen interact with p97. After each injection the sensor chip surface with immobilized p97 was regenerated by injecting 10 mM glycine, pH 2.2, for 2 minutes.
Kinetics of interaction between immobilized p97 and pro-uPA or plasminogen using the 2-state conformational model
Immobilized protein . | Ligand . | ka1, × 104 M-1 s-1 . | ka2, × 10-3 s-1 . | kd1, × 10-3 s-1 . | kd2, × 10-4 s-1 . | KD, × 10-9 M . |
|---|---|---|---|---|---|---|
| p97 | Pro-uPA | 0.6 | 3.2 | 1.7 | 7.1 | 65 |
| p97 | Plasminogen | 2.1 | 6.0 | 43.0 | 11.2 | 350 |
Immobilized protein . | Ligand . | ka1, × 104 M-1 s-1 . | ka2, × 10-3 s-1 . | kd1, × 10-3 s-1 . | kd2, × 10-4 s-1 . | KD, × 10-9 M . |
|---|---|---|---|---|---|---|
| p97 | Pro-uPA | 0.6 | 3.2 | 1.7 | 7.1 | 65 |
| p97 | Plasminogen | 2.1 | 6.0 | 43.0 | 11.2 | 350 |
Kinetic parameters were based on a 2-state conformational change binding model using the biosensorgram shown in Figures 1 and 2. This model describes a 1:1 binding of analyte to immobilized ligand followed by a conformational change in the complex. It is assumed that the conformationally changed complex can dissociate only through the reverse of the conformational change: A + B = AB = ABx. The dissociation constants (KD) were derived using both association (ka) and dissociation (kd) rates [KD = (kd1/ka1) (kd2/ka2)]. The parameters are ka1, association rate constant for A + B1 = AB1 (M-1 s-1); kd1, dissociation rate constant for AB1 = A + B1 (s-1); ka2, forward rate constant for AB = ABx (s-1); kd2, backward rate constant for AB = ABx (s-1). Mean χ2 values for the sensorgram fits were less than 0.4.
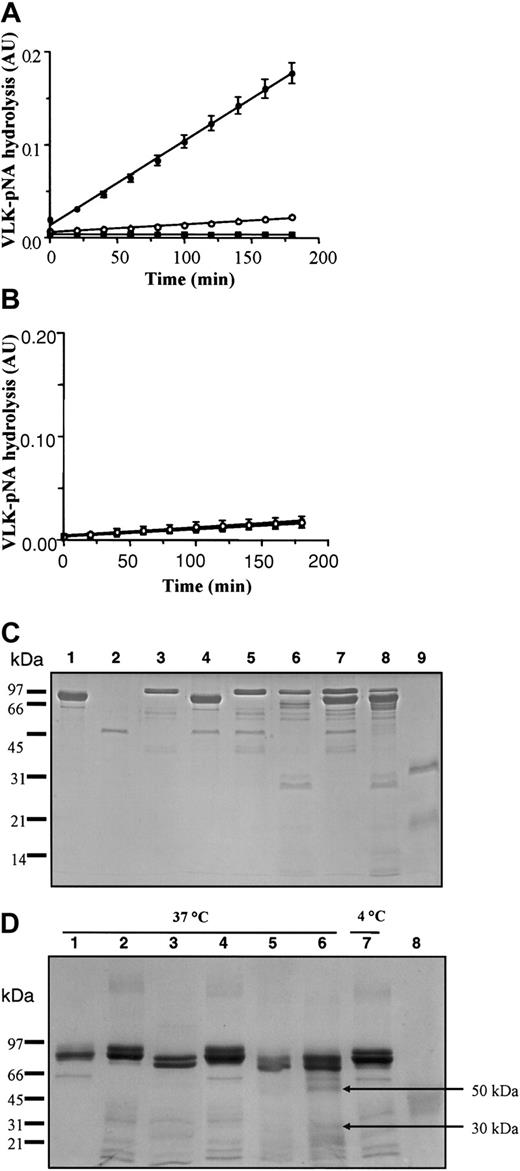
To evaluate the effect of p97 interaction on pro-uPA, we measured the serine activity (VLK-pNA hydrolysis) of pro-uPA and tPA using a colorimetric assay both with and without p97 (Figure 4). In the absence of p97, only a slight activity was measured for both pro-uPA and tPA. However, after 180 minutes, the VLK-pNA hydrolysis by pro-uPA goes from less than 0.02 absorbance unit (AU) in the absence of p97 to more than 0.18 AU when p97 is added into the incubation (Figure 4A). Addition of p97 to tPA elicits no observable effect and p97 alone had no proteolytic activity (Figure 4B). The results from both SPR and enzymatic activity indicate that the change in pro-uPA conformation induced by p97 increased its ability to degrade the plasmin substrate. To determine whether interaction with p97 leads to a cleavage of pro-uPA, the proteins were coincubated for 5 minutes at 37°C (Figure 4C). They were then separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions, using a 12.5% acrylamide gel, and stained with standard Coomassie blue. Under these conditions, p97 and uPA migrated as 97-kDa and 33-kDa bands, respectively, whereas pro-uPA migrated as a single band at 55 kDa. No major degradation of either protein could be detected, indicating that the incubation of pro-uPA with p97 under the conditions used to perform the VLK-pNA hydrolysis did not cleave either protein. Even after 6 hours' incubation at 37°C, both proteins were stable (data not shown). In the presence of plasminogen, pro-uPA was cleaved after an incubation of 5 minutes at 37°C and 2 major fragments of 33 kDa and 29 kDa could be observed. When p97 was added to the incubation medium, the generation of these fragments did not change. We further estimated the impact of p97 on plasminogen fragmentation by pro-uPA using 6 hours' incubation at 37°C (Figure 4D). As controls, the proteins used for this experiment (p97, Glu-plasminogen, or Lys-plasminogen), in the same amount (3 μg), were incubated for the same length of time at 37°C. Proteins were separated on a 7.5% acrylamide gel under nonreducing conditions and stained with Coomassie blue. When p97 was added to Glu-plasminogen no apparent fragment was generated. In contrast, the addition of a low amount (10 ng) of pro-uPA, which could not be detected using standard Coomassie blue staining, induced degradation of Glu-plasminogen with the appearance of fragments that migrated at the same molecular weight as Lys-plasminogen. Moreover, when p97 is added to Glu-plasminogen and pro-uPA, the degradation profile of Glu-plasminogen is changed. In the presence of p97 with Glu-plasminogen and pro-uPA, higher levels of bands migrating at the same molecular weight as Lys-plasminogen were observed and 2 other fragments appeared at 50 and 30 kDa. These fragments do not seem to be related to angiostatin, since they migrated at a different molecular weight than did the control angiostatin at 42 kDa. These results suggest that p97 alters the cleavage of Glu-plasminogen by pro-uPA.
Effect of p97 on pro-uPA, tPA, and plasminogen. The serine activity of 90 nM pro-uPA (A) and 75 nM tPA (B) were measured in the absence (○) or presence (•) of 70 nM p97 without plasminogen. The reaction was performed in a final volume of 200 μL as described in “Materials and methods.” In both panels A and B, controls were also performed with p97 (▪) but without pro-uPA or tPA (n = 9 for pro-uPA; n = 6 for tPA). (C) The effect of p97 on pro-uPA was evaluated after incubation in the presence or absence of plasminogen. 2 μg p97 (lane 1), 1 μg pro-uPA (lane 2), and 2 μg plasminogen (lane 3) were incubated for 5 minutes at 37°C alone as controls. Pro-uPA (2 μg) was incubated at 37°C for 5 minutes with 2 μg p97 (lane 4). Plasminogen and pro-uPA were added without incubation (lane 5) and with 5 minutes' incubation at 37°C (lane 6). Pro-uPA with 2 μg of both p97 and plasminogen was added without incubation (lane 7) or with 5 minutes' incubation at 37°C (lane 8). Tc-uPA (2 μg) was also loaded as a control (lane 9). Proteins were then separated by SDS-PAGE, using a 12.5% acrylamide gel under reducing conditions. After electrophoresis, proteins were visualized by staining the gel with Coomassie blue. (D) Effect of p97 on plasminogen degradation by pro-uPA. Quantities (3 μg) of p97 (lane 1), Glu-plasminogen (lane 2), and Lys-plasminogen (lane 3) were incubated alone for 6 hours at 37°C. In lane 4, 3 μg of both glu-plasminogen and p97 was also incubated for 6 hours at 37°C. Pro-uPA (20 ng) was added to plasminogen for the same period of incubation at 37°C (lane 5). p97 was added to pro-uPA and plasminogen for 6 hours at 37°C (lane 6) or 4°C (lane 7). In lane 8, 3 μg angiostatin was also added as a control. Proteins were then separated by SDS-PAGE, using a 7.5% acrylamide gel under nonreducing conditions. After electrophoresis, proteins were visualized by staining the gel with Coomassie blue.
Effect of p97 on pro-uPA, tPA, and plasminogen. The serine activity of 90 nM pro-uPA (A) and 75 nM tPA (B) were measured in the absence (○) or presence (•) of 70 nM p97 without plasminogen. The reaction was performed in a final volume of 200 μL as described in “Materials and methods.” In both panels A and B, controls were also performed with p97 (▪) but without pro-uPA or tPA (n = 9 for pro-uPA; n = 6 for tPA). (C) The effect of p97 on pro-uPA was evaluated after incubation in the presence or absence of plasminogen. 2 μg p97 (lane 1), 1 μg pro-uPA (lane 2), and 2 μg plasminogen (lane 3) were incubated for 5 minutes at 37°C alone as controls. Pro-uPA (2 μg) was incubated at 37°C for 5 minutes with 2 μg p97 (lane 4). Plasminogen and pro-uPA were added without incubation (lane 5) and with 5 minutes' incubation at 37°C (lane 6). Pro-uPA with 2 μg of both p97 and plasminogen was added without incubation (lane 7) or with 5 minutes' incubation at 37°C (lane 8). Tc-uPA (2 μg) was also loaded as a control (lane 9). Proteins were then separated by SDS-PAGE, using a 12.5% acrylamide gel under reducing conditions. After electrophoresis, proteins were visualized by staining the gel with Coomassie blue. (D) Effect of p97 on plasminogen degradation by pro-uPA. Quantities (3 μg) of p97 (lane 1), Glu-plasminogen (lane 2), and Lys-plasminogen (lane 3) were incubated alone for 6 hours at 37°C. In lane 4, 3 μg of both glu-plasminogen and p97 was also incubated for 6 hours at 37°C. Pro-uPA (20 ng) was added to plasminogen for the same period of incubation at 37°C (lane 5). p97 was added to pro-uPA and plasminogen for 6 hours at 37°C (lane 6) or 4°C (lane 7). In lane 8, 3 μg angiostatin was also added as a control. Proteins were then separated by SDS-PAGE, using a 7.5% acrylamide gel under nonreducing conditions. After electrophoresis, proteins were visualized by staining the gel with Coomassie blue.
Plasminogen activation by p97
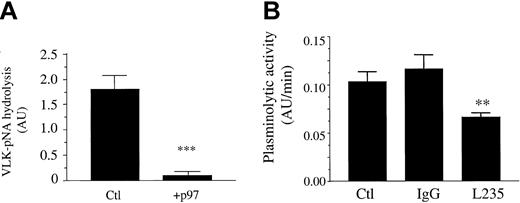
We futher characterized the interaction of p97 with pro-uPA by measuring the activation of plasminogen by pro-uPA in the presence of p97 (Figure 5). When p97 is added to pro-uPA and plasminogen, the VLK-pNA hydrolysis is 4-fold higher after 180 minutes (Figure 5A). Control experiments performed with p97 indicated that this protein alone does not generate plasmin when it is added to plasminogen. We also measured the plasmin activity in the presence of various concentrations of p97 (Figure 5B). Since the generation of plasmin proceeds at a constant rate under the assay conditions used, plotting the experimental data as a function of time (t)2 allowed us to determine the initial rate of plasmin formation (data not shown). From these linear curves, the initial plasmin activity measured in the absence of p97 was subtracted from the activities obtained in the presence of various p97 concentrations. Thus, the data represent the initial rates of plasmin activity (corresponding to the slopes) in the presence of various p97 concentrations. p97 stimulates the plasminogen cleavage by pro-uPA in a dose-dependent manner, with half-maximal stimulation occurring at 25 ± 6 nM. We further measured the effect of p97 on plasmin activity in the presence of various concentrations of plasminogen (Figure 5C). Initial rates of plasmin activity calculated at several plasminogen concentrations were plotted as a function of plasminogen concentrations. The resulting experimental data were fitted using nonlinear regression analysis. p97 decreased the apparent Michaelis constant (Km) of pro-uPA for plasminogen from 188 ± 22 to 102 ± 17 nM and increased the maximal velocity (Vmax) from 6.9 ± 0.4 to 8.9 ± 0.6 mAU/min. These results indicate that p97 positively affects the activation of plasminogen by pro-uPA by increasing the catalytic efficiency by a factor of 2.4.
Effect of p97 on plasminolytic activity induced by pro-uPA. (A) The plasminolytic activity of 1 nM pro-uPA was measured without (○) or with (•) 70 nM p97 in the presence of 30 nM plasminogen. The reaction was performed in a final volume of 200 μL as described in “Materials and methods.” As a control, the enzymatic activity in the presence of p97 alone was also measured (▪). (B) Plasmin activity induced by pro-uPA was measured in the presence of various p97 concentrations. (C) Plasmin activity induced by pro-uPA was measured without (○) or with (•) 250 nM p97 and various concentrations of plasminogen. (D) Inhibition by the mAb L235 of the increase in plasminolytic activity induced by p97. The plasminolytic activity of pro-uPA was measured in the presence of 70 nM p97 and 65 nM of either mAb L235 (○) or nonspecific mouse IgG (•). One representative experiment is shown and data represent the means ± SDs (n = 3)
Effect of p97 on plasminolytic activity induced by pro-uPA. (A) The plasminolytic activity of 1 nM pro-uPA was measured without (○) or with (•) 70 nM p97 in the presence of 30 nM plasminogen. The reaction was performed in a final volume of 200 μL as described in “Materials and methods.” As a control, the enzymatic activity in the presence of p97 alone was also measured (▪). (B) Plasmin activity induced by pro-uPA was measured in the presence of various p97 concentrations. (C) Plasmin activity induced by pro-uPA was measured without (○) or with (•) 250 nM p97 and various concentrations of plasminogen. (D) Inhibition by the mAb L235 of the increase in plasminolytic activity induced by p97. The plasminolytic activity of pro-uPA was measured in the presence of 70 nM p97 and 65 nM of either mAb L235 (○) or nonspecific mouse IgG (•). One representative experiment is shown and data represent the means ± SDs (n = 3)
To determine whether the induction of plasmin formation by p97 was specific, we measured the formation of plasmin by pro-uPA in the presence of either the mAb L235 (directed against p97) or a nonspecific IgG (Figure 5D). MAb L235 (50 nM) inhibited the pro-uPA activation induced by p97 by 50%. These results suggest that the effect of p97 upon pro-uPA's activation of plasminogen is specific and may involve the epitope recognized by the mAb L235.
Inhibition of cell migration by mAb L235
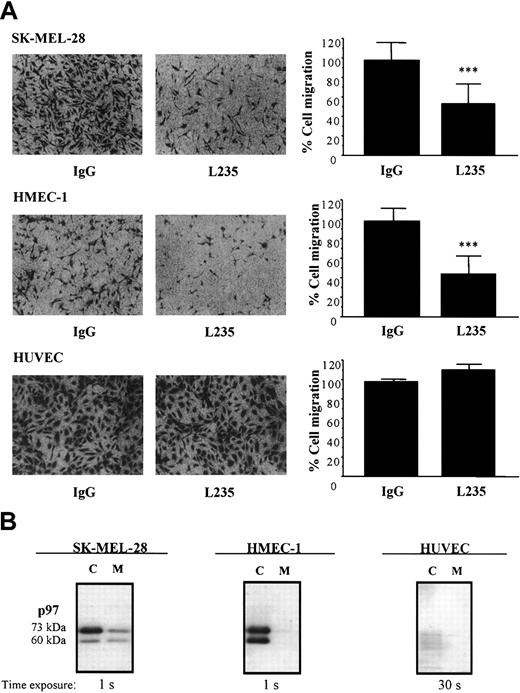
Since p97 affects the activation of plasminogen in vitro and since the uPA/uPAR system is important in cell migration, we further investigated whether endogenous p97 might be associated with this process. Migration of HMECs-1 and HUVECs was evaluated in the presence of either mAb L235 or nonspecific mouse IgG by using modified Boyden chambers (Figure 6A). In addition, because p97 was first identified in melanoma cells,3 we also measured the impact of the mAb L235 on the migration of human melanoma (SK-MEL-28) cells (Figure 6A). In the presence of mAb L235 (50 nM), the migration of HMEC-1 and SK-MEL-28 cells was inhibited by 54% and 48%, respectively. However, cell migration of HUVECs was unaffected by this concentration of mAb L235. Endogenous p97 was immunodetected in lysates from HMEC-1, SK-MEL-28, and HUVEC cells as well as in their conditioned culture media (Figure 6B). p97 migrated under unreduced conditions at 73 and 60 kDa, as previously observed.28 It was highly expressed in lysates from HMEC-1 and SK-MEL-28 cells and at lower levels in their respective conditioned culture media. In HUVECs, p97 was, however, almost undetectable. In fact, the exposure time was at least 30 times greater to detect a much lower level of p97 in HUVECs compared with HMEC-1 and SK-MEL-28 cells. These results indicate that mAb L235, by interacting with endogenous p97, inhibits the migration of HMEC-1 and SK-MEL-28 cells. This also suggests that the endogenous p97 in these cells is involved in cell migration.
Effect of mAb L235 on cell migration. (A) Cell migration of HMEC-1, SK-MEL-28, or HUVEC cells was measured using modified Boyden chambers as described in “Materials and methods.” Cells that had migrated to the lower surface of the filters were fixed and stained with crystal violet. Images obtained from a representative experiment are shown. Cells that had migrated in the presence of 50 nM mAb L235 or a nonspecific mouse IgG were also counted. The results were expressed as the percentage of the control measured in the presence of a nonspecific mouse IgG and represent means ± SDs (n = 5 for HMEC-1; n = 4 for SK-MEL28; n = 3 for HUVEC). ***Statistically significant differences (P < .001; Student t test). (B) Detection of endogenous p97 by Western blot analysis. p97 was immunodetected in lysates or serum-deprived culture media (18 hours) from HMEC-1, SK-MEL28, and HUVEC-1 cells. Proteins were separated by SDS-PAGE and were electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes. p97 was detected by Western blotting, using mAb L235 and a secondary antimouse IgG linked to peroxidase.
Effect of mAb L235 on cell migration. (A) Cell migration of HMEC-1, SK-MEL-28, or HUVEC cells was measured using modified Boyden chambers as described in “Materials and methods.” Cells that had migrated to the lower surface of the filters were fixed and stained with crystal violet. Images obtained from a representative experiment are shown. Cells that had migrated in the presence of 50 nM mAb L235 or a nonspecific mouse IgG were also counted. The results were expressed as the percentage of the control measured in the presence of a nonspecific mouse IgG and represent means ± SDs (n = 5 for HMEC-1; n = 4 for SK-MEL28; n = 3 for HUVEC). ***Statistically significant differences (P < .001; Student t test). (B) Detection of endogenous p97 by Western blot analysis. p97 was immunodetected in lysates or serum-deprived culture media (18 hours) from HMEC-1, SK-MEL28, and HUVEC-1 cells. Proteins were separated by SDS-PAGE and were electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes. p97 was detected by Western blotting, using mAb L235 and a secondary antimouse IgG linked to peroxidase.
Effect of exogenous p97 on cell migration
In addition, we estimated whether exogenous p97 could affect the migration of HMEC-1 and SK-MEL-28 cells (Figure 7A-B). Exogenous p97, at 10 nM and 100 nM, inhibited the migration of HMEC-1 cells by 34% and 50% (Figure 7C). The migration of SK-MEL-28 cells was inhibited by 44% and 70% in the presence of 10 and 100 nM p97. Migration of HUVECs was unaffected by these concentrations of p97 (data not shown). Moreover, this inhibition of cell migration is not related to a reduction of endothelial or melanoma cell adhesion, since the same concentrations of p97 did not affect adhesion on gelatin of either HMEC-1 or SK-MEL-28 cells (data not shown).
Effect of exogenous p97 on cell migration and plasminolytic activity. HMEC-1 (A) and SK-MEL-28 (B) cell migration was performed using modified Boyden chambers as described in “Materials and methods.” Cells that had migrated in the presence or absence of p97 (100 nM) to the lower surface of the filters were fixed and stained with crystal violet. Photos (orginial magnification, × 100) obtained from a representative experiment are shown. (C) Cells that had migrated were also counted and expressed as a percentage of the control cells, measured in the absence of p97 (n = 4 for HMEC-1; n = 3 for SK-MEL28). Data represent means ± SDs. ***P < .01 (student t test).
Effect of exogenous p97 on cell migration and plasminolytic activity. HMEC-1 (A) and SK-MEL-28 (B) cell migration was performed using modified Boyden chambers as described in “Materials and methods.” Cells that had migrated in the presence or absence of p97 (100 nM) to the lower surface of the filters were fixed and stained with crystal violet. Photos (orginial magnification, × 100) obtained from a representative experiment are shown. (C) Cells that had migrated were also counted and expressed as a percentage of the control cells, measured in the absence of p97 (n = 4 for HMEC-1; n = 3 for SK-MEL28). Data represent means ± SDs. ***P < .01 (student t test).
Inhibition of plasminolytic activity at the cell surface by soluble p97 and mAb L235
HMECs-1 were incubated for 18 hours with or without p97. Following this treatment, the plasmin activity at the surface of control and treated cells was measured. When cells were treated with p97 (100 nM), plasminogen activation was inhibited by 95% (Figure 8A). This marked reduction in the plasminolytic capacity of these cells by soluble p97 could explain the inhibition of HMEC-1 migration. Moreover, when HMECs-1 were treated with the mAb L235, the plasminolytic activity was inhibited by more than 50% compared with nonspecific mouse IgG (Figure 8B). This inhibition by the mAb L235 suggests that endogenous, membrane-bound p97 could participate in plasminogen activation in HMECs-1.
Inhibition of plasminolytic activity at the cell surface of HMECs-1 by soluble p97 and mAb L235. (A) Effect of p97 on plasminolytic activity. HMECs-1 were treated for 18 hours with 100 nM p97 (+p97) or Ringer solution (Ctl). Following this treatment the plasminolytic activity was measured using standard conditions, as described in “Materials and methods.” (B) Effect of mAb L235 on plasminolytic activity of HMECs-1. HMECs-1 (1 × 105 cells) were preincubated for 1 hour at 37°C with Ringer solution (Ctl) or with 250 nM mAb L235 or nonspecific mouse IgG. Following this preincubation, the plasminolytic activity was measured for 6 hours by adding pro-uPA (1 nM) and plasminogen (50 nM) using standard conditions, as described in “Materials and methods.” The plasminolytic activity of HUVECs was also measured, using 1 × 105 cells under the same conditions. Data represent the means ± SDs of 3 independent experiments performed in triplicate. **P < .01, ***P < .001 (Student t test).
Inhibition of plasminolytic activity at the cell surface of HMECs-1 by soluble p97 and mAb L235. (A) Effect of p97 on plasminolytic activity. HMECs-1 were treated for 18 hours with 100 nM p97 (+p97) or Ringer solution (Ctl). Following this treatment the plasminolytic activity was measured using standard conditions, as described in “Materials and methods.” (B) Effect of mAb L235 on plasminolytic activity of HMECs-1. HMECs-1 (1 × 105 cells) were preincubated for 1 hour at 37°C with Ringer solution (Ctl) or with 250 nM mAb L235 or nonspecific mouse IgG. Following this preincubation, the plasminolytic activity was measured for 6 hours by adding pro-uPA (1 nM) and plasminogen (50 nM) using standard conditions, as described in “Materials and methods.” The plasminolytic activity of HUVECs was also measured, using 1 × 105 cells under the same conditions. Data represent the means ± SDs of 3 independent experiments performed in triplicate. **P < .01, ***P < .001 (Student t test).
Discussion
Using an in vitro model of the BBB, we have previously shown that RAP, which is a folding chaperone for LRP,29 inhibits transcytosis of p97.13 In this previous study we also observed that p97 inhibited the transcytosis of lactoferrin, which, in an earlier study, had been reported to be LRP-dependent.30 From these results, we suggested that LRP is involved in the transport of p97.13 However, because the members of the LDL-receptor family (which includes LRP) transport similar substrates,29 we cannot exclude the possibility that other receptors of this family could also bind and transport p97. In the present study, the BIAcore technology shows no interaction between p97 and RAP, indicating that the transport inhibition caused by this LRP ligand is not caused through its interaction with p97. Because LRP recognizes a large variety of ligands and is associated with the homeostasis of proteinases and proteinase inhibitors,31 we further investigated the potential interactions between other LRP ligands and p97. Our data clearly show that both pro-uPA and plasminogen interact with p97 and that these interactions are specific, since no interaction between p97 and other proteins, including tPA, PAI-1, plasmin, angiostatin, BSA, and ovalbumin, could be measured. These results are the first to describe potential interactions between p97 and proteins of the uPA system.
In addition to its interaction with pro-uPA and plasminogen, p97 stimulates plasminogen activation by decreasing the Km of pro-uPA for plasminogen and by increasing the Vmax of the reaction. The conversion of pro-uPA to Tc-uPA occurs by proteolytic cleavage of a single peptide bond (Lys158-Ile159 in human uPA).32 This conversion can be catalyzed by plasmin or several other proteases, such as plasma kallikrein, blood coagulation factor XIIa, cathepsin B, cathepsin L, and prostate-specific antigen.33 In the present work the SPR assay, the enzymatic assay, and electrophoresis experiments all suggest that p97 induces a conformational change that increases pro-uPA activity without any apparent cleavage of pro-uPA. The 2-state conformational model gave the best fits for the interactions of both pro-uPA and plasminogen with immobilized p97 on the BIAcore. Such good fits of experimental data to a multistate model of interaction are an indication that a conformational change may be taking place. Other approaches, such as limited proteolysis, circular dichroism spectrometry, or magnetic nuclear resonance (MNR) technology, would be helpful in confirming and providing additional information on the interaction between p97 and either pro-uPA or plasminogen. Interestingly, the fragments of plasminogen generated by adding p97 were different from the plasminogen degradation by pro-uPA alone. These biochemical analyses further suggest that p97 could also be seen as a cofactor in uPA-dependent plasminogen activation.
The uPA/uPAR system has been involved in several pathologic and physiologic processes thsat require cell migration, such as tumor cell invasion and metastasis. Several reports have shown that the uPA/uPAR system plays a key role in signal transduction as well as in regulation of melanoma cell migration and angiogenesis.34-37 A recent study showed that p97 promotes in vivo angiogenesis and HMEC-1 migration when used as a chemoattractant.38 In our conditions, when p97 is added to both compartments of the Boyden chamber, migration of HMECs-1 is inhibited by more than 50%. Thus, given the important role of plasmin,39 a protein like p97 that targets the formation of plasmin and acts on the migration of endothelial cells as well as of SK-MEL-28 cells might be expected to affect angiogenesis and cancer progression. We also observed that the basal capacity for plasminogen activation by HMECs-1 decreased following p97 treatment. A recent study demonstrated that the expression of LDL receptor–related protein 1B (LRP1B), a new member of the LDL receptor family, leads to an accumulation of uPAR on the cell surface, which event inhibits the migration of Chinese hamster ovary (CHO) cells.40,41 From these results, it was proposed that LRP1B negatively regulates uPAR regeneration and function, whereas the net results of uPAR regeneration seems to depend on the relative expression of the 2 receptors. Since p97 transcytosis may involve LRP and p97 interacts with pro-uPA, further experiments are required to determine the effects of p97 treatment on both LRPs.
Recently, it was shown that when Glu-plasminogen is bound to cell surfaces, plasmin generation by plasminogen activators is markedly stimulated compared with the reaction in solution.42 This is a key element for cell migration, where the process of “grip and go” would play an important role.39,42 The process of plasminogen activation system is regulated by 2 different mechanisms: (1) cell surface–binding sites that facilitate the productive catalytic interactions with plasminogen and thereby increase plasmin generation, and (2) protein inhibitors such as serpin inhibitors, which restrict the activities of the proteases.43 In light of this, soluble p97 could participate in the activation of plasminogen without being in the pericellular environment (Figure 9A). Our data also indicate that the migration and the plasminolytic activity of cells expressing p97 are inhibited by mAb L235, suggesting that endogenous, membrane-bound p97 may be involved in these processes, which are associated with cancer and angiogenesis (Figure 9B). Moreover, both the migration of HMECs-1 and the plasminolytic activity are diminished when exogenous p97 is added, suggesting that soluble p97 affects the regulation of plasminogen activation at the cell surface (Figure 9C). Thus, by breaking the equilibrium between soluble p97 and membrane-bound p97, it is possible to affect cell migration of HMEC-1 and SK-MEL-28 cells. Further studies are required to establish the impact of p97 treatment on components of the uPA system at the cellular membrane in order to elucidate the molecular events by which both L235 and exogenous p97 affect cell migration.
Schematic representation of p97 regulation of plasminogen and cell migration. This schematic representation summarizes the results obtained for p97 in the present study. (A) The interaction of pro-uPA and plasminogen with soluble activity increases the activation of plasminogen. This induction could be inhibited by the mAb L235, which recognizes a conformational epitope on p97. (B) The addition of mAb L235 reduces the plasminolytic activity on HMEC-1 cell surface and results in an inhibition of cell migration. (C) The interaction of plasminogen and pro-uPA with membrane-bound p97 (mb p97) is diminished when exogenous, competing human recombinant p97 is added. This also caused a decrease in the activation of plasminogen (plg) and leads to an inhibition of cell migration. This representation of the interaction between p97 and pro-uPA indicates that the balance between membrane bound and soluble p97 may be crucial for cell migration.
Schematic representation of p97 regulation of plasminogen and cell migration. This schematic representation summarizes the results obtained for p97 in the present study. (A) The interaction of pro-uPA and plasminogen with soluble activity increases the activation of plasminogen. This induction could be inhibited by the mAb L235, which recognizes a conformational epitope on p97. (B) The addition of mAb L235 reduces the plasminolytic activity on HMEC-1 cell surface and results in an inhibition of cell migration. (C) The interaction of plasminogen and pro-uPA with membrane-bound p97 (mb p97) is diminished when exogenous, competing human recombinant p97 is added. This also caused a decrease in the activation of plasminogen (plg) and leads to an inhibition of cell migration. This representation of the interaction between p97 and pro-uPA indicates that the balance between membrane bound and soluble p97 may be crucial for cell migration.
In conclusion, these are the first results indicating that p97 can potentially interact with pro-uPA as well as with plasminogen and regulate the activation of plasminogen by pro-uPA. We are also reporting that migration of HMEC-1 and SK-MEL-28 cells is inhibited by mAb L235 and soluble p97, indicating that active and functional p97 participates in this process. Collectively, our results thus suggest that the balance between membrane-bound and soluble p97 could affect cell migration. Further studies are now under way to establish the molecular events characterizing both the soluble and membrane-bound p97 inhibitory actions and to determine the potential involvement of p97 in angiogenesis.
Prepublished online as Blood First Edition Paper, May 15, 2003; DOI 10.1182/blood-2003-01-0166.
Supported by grants from the Canadian Institutes of Health Research (R.B.). J.J. is a recipient of a scholarship from the Canadian Institutes of Health Research.
M.D. and Y.B. contributed equally to this study.
One of the authors (R.G.) has declared a financial interest in a company (Biomarin Pharmaceutical) whose potential product (p97) was studied in the present work. One of the authors (R.G.) is employed by and has financial interests in Biomarin Pharmaceutical.
The study was supported in part by research funding from Biomarin Pharmaceutical.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are very thankful to Dr Kennard for providing us with the human recombinant p97 and the different mAbs directed against p97 and to Dr Bu for providing us with RAP. We also thank Julie Poirier for her technical support. Many thanks to Ben Sulsky for his enlightened vision of science.

![Figure 1. Transcytosis of p97 across BBCEC monolayers. (A) Transcytosis experiments were performed at 37°C for 2 hours. [125I]-p97 (25 nM) was added to the upper side of the cell-covered filter in the absence or presence of RAP (650 nM) or BSA (5 μM). At the end of the experiment, radiolabeled proteins were measured in the lower chamber of each well by TCA precipitation. Results represent means ± SE (n = 6). (B) p97 was immobilized on a sensor chip surface (CM5) as described in “Materials and methods.” p97, RAP, and BSA (5 μg/100 μL) were injected over immobilized p97. One representative experiment is shown (n = 3).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/5/10.1182_blood-2003-01-0166/6/m_h81734874001.jpeg?Expires=1769120944&Signature=DvHP-aapXVa7dDs8XJGWWAPOH4di1nMkHY3f5J7FpJRmVaE3AY0Bu4zztFtqxIy9CDBm40yHQg0yUon0FrAqt8X8WTbHVIEfpxCWyyenuyecF3f49UMwOwYuJ6E0oQEZpWJRo4GRZ9u~v1qeqbHQBO0aaQpme4vbxB77KrU7RH957cFYWLVIviL7t6mclSPYmSybksIFCkL8RQj6i7HsQQA71wqBBQjzmjfpj4bdGjPF5PGy1bNFqLYks172Sz1e2P4T173~G5bCXJUq~99s1tXLo4sMF8w8kEgDSQ3PmTRW4hvBzWmD3ZOi6PIlk1Xu3wyG0ItFAqj9TjvVNWRnOg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Biospecific interaction analysis in real time between p97 and various anti-p97 mAbs. p97 was immobilized on a sensor chip (CM5) using standard coupling procedures incorporating NHS, EDC, and ethanolamine. Different mAbs directed against p97 (HybC, HybE, HybF, L235, 2C7, 9B6), diluted to 0.05 μg/μL in Ringer-HEPES, were injected into the BIAcore at a flow rate of 5 μL/min. The surface plasmon resonance response obtained for these mAbs was plotted (in relative units [RU]) as a function of time. After each injection, immobilized p97 was regenerated with 0.2M glycine at pH 2 for 2 minutes followed by a 2-minute injection of ferric ammonium citrate (1 mM) (n = 4).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/5/10.1182_blood-2003-01-0166/6/m_h81734874002.jpeg?Expires=1769120944&Signature=dWlsQguG1B7d1i3CiZv1QJF3ksw3AUIkZuSzg~l37qReXCEyV5f6pEgyGFjcErMau4VgIbGHb7CnPL1muZ1Ian2eq3smrZ8QHMB3sqskI8w~VlhCxncD85ZYNup3pEQzVqIy11LdKtYT2gjyb0ltRHA6PH~0H5wTv2P5dAWxva5Ycqcz-GiDxONlW0JC7OmbS3502q1ZHPWAgNHa3lXRmeeJtyAVXuPuJboQDfYmTHQAkXG3xvwcxNPHTAwZyn4o583yBX9JW67fo6SMC2o1F3RI5-sF8ItvzTvSvrBY4gFpOeY~hrSgPmXMkVh8AqIgrVdlsut-jFEns0c9yh~h8w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal