Abstract
Nelarabine, prodrug of arabinosylguanine (ara-G), has demonstrated T-lymphoblastic antileukemic activity in cell lines and in the clinic. To investigate the mechanism for lineage-specific toxicity, the effects of ara-G were compared in CEM (T-lymphoblast), Raji (B-lymphoblast), and ML-1 (myeloid) cell lines. CEM cells were the most sensitive to ara-G–induced apoptosis and accumulated the highest levels of ara-G triphosphate (ara-GTP). However, compared with myeloid and B-lineage cell lines, CEM cells incorporated fewer ara-G molecules—which were at internucleotide positions in all 3 cell lines— into DNA. Ara-G induced an S-phase arrest in both Raji and ML-1, while in CEM the S-phase cells decreased with a concomitant increase in the sub-G1 population. Within 3 hours of ara-G treatment, the levels of soluble Fas ligand (sFasL) in the medium increased significantly in CEM cultures. In parallel, an induction of FasL gene expression was observed by real-time reverse transcriptase–polymerase chain reaction (RT-PCR). Pretreatment of CEM cells with a Fas antagonistic antibody inhibited ara-G–mediated cell death. These results demonstrate that high ara-GTP accumulation in T cells results in an S phase–dependent apoptosis induced by ara-G incorporation into DNA, which may lead to a T cell–specific signal for the induction and liberation of sFasL. Subsequently, the sFasL induces an apoptotic response in neighboring non–S-phase cells. In contrast, myeloid and B cells accumulated lower levels of ara-GTP and arrested in S phase, blocking any apoptotic signaling.
Introduction
The discovery of purine nucleoside phosphorylase (PNP) deficiency, a naturally occurring disease, and the elucidation of its pathophysiology provided the rationale for the development of deoxyguanosine (dGuo) analogs as it was revealed that T cells and B cells responded differently to the derangement of purine metabolism. Specifically, the absence of PNP, the enzyme responsible for the catabolism of dGuo, manifests as a profound dGuo triphosphate (dGTP)–mediated T-cell lymphopenia without concomitant decrease in B cells.1-6 Mechanistically, elevated levels of dGTP in T cells resulted in a dGTP-mediated inhibition of DNA synthesis and cell death.3,7,8 Taken together, these data provided a rationale for the development of PNP inhibitors or PNP-resistant dGuo analogs, such as ara-G for the treatment of leukemias in general and T-cell acute lymphoblastic leukemia (T-ALL) in particular.9-11
The proximal cytotoxic metabolite of ara-G is its 5′-triphosphate, arabinosylguanine triphosphate (ara-GTP). The rate-limiting step in the formation of ara-GTP is the initial conversion to monophosphate by both deoxycytidine (dCyd) kinase and dGuo kinase.12 Several studies demonstrated that ara-G was specifically toxic to normal T cells13-15 and malignant T-lymphoid cells compared with non-T, non-B (null) cells, or B-lymphoid cells.16-20 Consistent with the cell line data, the toxicity of ara-G in freshly isolated primary leukemia cells showed T-lymphoblast selectivity.17 The differential accumulation and consequent selective cytotoxicity was postulated to be due to the higher phosphorylation and slower elimination of ara-GTP. Clinical investigations in humans using nelarabine (compound GW506U78, a prodrug of ara-G) also demonstrated a T-lymphoid lineage–specific accumulation of ara-GTP during therapy.21,22 This was associated with clinical responses in this patient population suggesting a role of ara-GTP in T-cell selective cytoreductiveness.
Similar to other nucleoside analogs,13-15,23-28 the cytotoxicity of ara-GTP is dependent on incorporation of the fraudulent nucleotide into DNA.29 For this incorporation, intracellular ara-GTP competes with its native endogenous nucleotide, dGTP.30 After incorporation, DNA primers containing ara-GTP are resistant to further elongation resulting in an effective chain termination in vitro.31 This replication block by incorporated ara-G results in inhibition of DNA synthesis9,16,32 and death of the target cell.29
Based on these data, we hypothesized that the T-cell selective cytotoxicity of ara-G should be related to quantitative and qualitative differences in incorporation of ara-G residues into DNA. For example, the number of arabinosylguanine monophosphate (ara-GMP) molecules in DNA may be higher in T cells, and the location of these moieties into DNA may vary between cell types. Furthermore, because the incorporation of ara-G into DNA was dependent on S-phase population, cell cycle–specific differences may be associated with the observed T-cell hypersensitivity. Finally, in addition to S phase–specific cell death, additional determinants may be responsible for lineage-specific cytotoxicity. The present work was conducted to elucidate the mechanisms that account for the observed differences in response to ara-G treatment in cell lines.
Materials and methods
Cell lines
CCRF-CEM, Raji, and Jurkat cell lines were obtained from American Type Culture Collection (Manassas, VA). ML-1 cell line was a gift from Dr M. Kastan (Memphis, TN). The cells were cultured in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum (GIBCO, Grand Island, NY) and maintained at 37°Cin5%CO2. Cells were routinely tested for Mycoplasma by using a commercially available kit and following the manufacturer's recommendations (Gen-Probe, San Diego, CA).
Cell proliferation assays
Cells were incubated with the desired concentration of ara-G (R.I. Chemical, Orange, CA) for 3 hours and washed 3 times in warmed drug-free medium before assaying. For the growth inhibition assay, cells were allowed to grow for 72 hours, counted on a Coulter counter (Coulter Electronics, Miami, FL), and expressed as a percent of control. For MTS assay, cells were allowed to grow for 72 hours, MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulphophenyl)-2H-tetrazolium, inner salt) reagent (Promega, Madison, WI) was added, and the absorbance was measured at 490 nm (Dynatech, Chantilly, VA). For clonogenicity, cells were embedded in 0.35% agar and after 10 days the colonies were stained with 0.02% p-iodo-nitrotetrazolium violet for 24 hours. Colonies composed of more than 50 cells were scored, and results were expressed as a percentage of colonies formed in untreated samples. Cloning efficiency ranged from 30% to 47%.
Accumulation and elimination of ara-GTP
Exponentially growing cells were incubated with ara-G for the indicated concentrations and times. To determine the kinetics of ara-GTP elimination, cells were incubated with 100 μM ara-G for 3 hours, washed 3 times in medium, and then resuspended to 1 × 105 cells per milliliter. At various times, aliquots were taken and the cells were pelleted. Nucleotides were extracted from cell pellet using perchloric acid method and analyzed by high-pressure liquid chromatography (HPLC).29 The intracellular concentrations of nucleotides and ara-GTP in the extracts were calculated from a given number of cells of a determined mean cell volume (Coulter channelizer, Coulter Electronics). This calculation assumes the nucleotides are uniformly distributed in the total cell volume. The lower limit of sensitivity of this assay is 10 pmol in an extract of 5 × 106 cells, corresponding to a cellular concentration of 1 μM.
Effect of ara-G on DNA synthesis
Cells were incubated for 2.5 hours with the indicated concentrations of ara-G followed by the addition of [3H]thymidine (2 μCi/mL [0.074 MBq/mL]) (Moravek Biochemicals, Brea, CA). DNA synthesis was measured using a MultiScreen assay system (Millipore, Bedford, MA) as described.29
Determination of ara-GMP incorporation into DNA
The cells were incubated with varying concentrations of [8-3H]ara-G (Moravek Biochemicals) for 3 hours, washed with phosphate-buffered saline (PBS), and the DNA was isolated.33 The amount of ara-GMP incorporated into DNA was determined by scintillation counting (Packard Instrument, Meridian, CT).
To determine the location of the ara-GMP in DNA, the cells were incubated either with 1, 10, 100 μM [8-3H]ara-G or with 0.025 μM[3H]dAdo for 3 hours. The isolated DNA was dissolved in H2O and denatured. The nucleic acids were then degraded to internal nucleotides and 3′-terminal nucleosides by the sequential action of micrococcal nuclease and spleen phosphodiesterase as described.34 The digests were separated by reverse-phase HPLC using an YMC-Pack ODS-AQ S-5 120A reverse-phase column (4.6 × 250 mm; YMC, Tokyo, Japan), and the radioactivity associated with the respective 3′-monophosphates and nucleosides was quantitated by online scintillation counting. The experiment was done twice in triplicate. Six data points depicting percent incorporation were used to calculate mean and SD.
Determination of Fas/FasL levels
Membrane-bound Fas and Fas ligand (FasL) were measured on cells (1 × 106) treated with ara-C, adriamycin, or ara-G; washed; and then stained with fluorescein isothiocyanate (FITC)–conjugated mouse antihuman Fas monoclonal antibody (Becton Dickinson, San Jose, CA) and biotin-conjugated mouse antihuman FasL monoclonal antibody (Becton Dickinson) in the dark for 15 minutes at 37°C. The double-labeled cells were stained with streptavidin-phycoerythrin conjugate (PharMingen, San Jose, CA) and analyzed using an Epics Profile II (Coulter Electronics), a FACScan, or a FACSCalibur (Becton Dickinson) flow cytometer.
The level of soluble FasL (sFasL) in the media was quantitated using Fas ligand enzyme-linked immunosorbent assay (ELISA) (Oncogene Research Products, Boston, MA). The quantity of FasL in the culture media from untreated control cells was compared with test samples.
Analyses of Fas and FasL gene expression
RNA was extracted from ara-G–treated CEM and Jurkat cells.35 The levels of Fas and FasL expression were measured using a TaqMan real-time reverse transcriptase–polymerase chain reaction (RT-PCR) assay on an ABI Prism 7700 and 7900 Sequence Detection system (Applied Biosystems, Foster City, CA). Gene expression levels were measured in 200 ng RNA using predeveloped assay reagents for Fas and FasL as described by the manufacturer (Applied Biosystems), and the relative quantities were calculated using standard curves generated from known dilutions of doxorubicin-treated CEM cell RNA36 and normalized to endogenous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels. The GAPDH-specific probe and primer were as follows: forward primer, 5′-GAAGGTGAAGGTCGGAGT-3′; reverse primer, 5′-GAAGATGGTGATGGGATTTC-3′; GAPDH probe, 5′-VIC-CAAGCTTCCCGTTCTCAGCC-TAMRA-3′.
Effect of Fas antagonist or Fas agonist on ara-G–induced cell death
Cells were incubated with 1 μg/mL ZB4 Fas antagonist antibody for 1 hour and then incubated with 100 μM ara-G for an additional 3 hours. ZB4 alone and untreated cells served as negative controls. Positive controls included 10 μg/mL CH-11 Fas agonist antibody for 24 hours or 10 μM ara-C for 4 hours. Cells were analyzed by flow cytometry for cell cycle and apoptosis using the terminal deoxyuridine triphosphate nick-end labeling (TUNEL) assay kit, APO-DIRECT (Phoenix Flow Systems, San Diego, CA). The percentages of TUNEL-positive and -negative cells and the cell cycle profile were obtained using Elite (Coulter Electronics) and Multicycle software (Phoenix Flow Systems), respectively.
Cell cycle analysis
Aliquots of untreated and ara-G–treated cells were pelleted, fixed in ice cold 70% ethanol, stained with propidium iodide (PBS containing 0.5% Tween 20, 15 μg/mL propidium iodide, and 5 μg/mL DNase-free RNase), and analyzed by flow cytometry (FACSCalibur, BD CellQuestPro v4.0, Becton Dickinson).
Statistical analysis
The accumulation and elimination kinetics of ara-GTP was determined using GraphPad Prism (GraphPad Software, San Diego, CA). The Kolmogorov-Smirnov (K-S) statistic was used for all flow analyses to compare mean intensity fluorescence to determine significance. P values more than .16 were considered significant. For sFas ligand levels, the values in untreated versus treated cells were compared using Student 1-tailed t test. The statistical significance of changes in gene expression levels was determined using Prism software to perform a 2-tailed, unpaired t test.
Results
T cell–specific cytotoxicity of ara-G
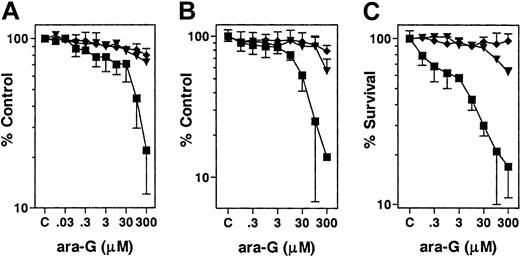
To gain insight into the mechanism of ara-G–mediated cytotoxicity and T-cell selectivity, the leukemic cell lines CEM, Raji, and ML-1 were used to represent T-, B-lymphoid, and myeloid lineages, respectively. Three separate tests demonstrated T-cell selectivity of ara-G in these cells. First, the cell proliferation assay suggested T-lymphoblastic CEM cells are the most sensitive to the effects of ara-G (Figure 1A). The inhibitory concentration of 25% (IC25) for CEM was 3 μM, whereas for Raji and ML-1 it was 300 μM. The second method, the MTS assay (Figure 1B), resulted in findings similar to the cell growth inhibition assay, because CEMs were the most sensitive with an IC25 and IC50 of 3 μM and 30 μM, respectively, while 300 μM appeared to be the IC50 and IC25 for Raji and ML-1, respectively. Finally, clonogenicity measurements demonstrated that in CEM cells there is a rapid loss of clonogenicity even at low concentrations of ara-G (Figure 1C). For instance, under these conditions, there was a 25% loss of clonogenicity in this cell line at 0.1 μM ara-G; IC50 was less than 10 μM. In contrast, an IC50 was not achieved in Raji or ML-1 cells despite incubations of up to 300 μM ara-G. T-cell selectivity was also observed when other cell lines, such as MOLT-4, were used (data not shown).
T cell–specific cytotoxicity of ara-G. Exponentially growing cultures of CEM (▪), Raji (▾), and ML-1 (♦) were incubated with the indicated concentration of ara-G for 3 hours. (A) Cells were washed free of drug, resuspended in warmed drug-free medium for 72 hours and, at the end of incubation, the cell number was counted and expressed as a percent of control. Data points represent mean of 2 independent experiments conducted in triplicate. (B) Cells were resuspended in drug-free medium for 72 hours. At the end of incubation, MTS reagent was added and the obtained absorbance at 490 nm was measured and expressed as a percent of control. Data points represent mean of 2 independent experiments conducted in quadruplicate. (C) Cells were washed free of drug, embedded in soft agar, and expressed as a percentage of colonies formed in untreated samples. Data points represent mean of 2 independent experiments conducted in triplicate.
T cell–specific cytotoxicity of ara-G. Exponentially growing cultures of CEM (▪), Raji (▾), and ML-1 (♦) were incubated with the indicated concentration of ara-G for 3 hours. (A) Cells were washed free of drug, resuspended in warmed drug-free medium for 72 hours and, at the end of incubation, the cell number was counted and expressed as a percent of control. Data points represent mean of 2 independent experiments conducted in triplicate. (B) Cells were resuspended in drug-free medium for 72 hours. At the end of incubation, MTS reagent was added and the obtained absorbance at 490 nm was measured and expressed as a percent of control. Data points represent mean of 2 independent experiments conducted in quadruplicate. (C) Cells were washed free of drug, embedded in soft agar, and expressed as a percentage of colonies formed in untreated samples. Data points represent mean of 2 independent experiments conducted in triplicate.
Accumulation and elimination kinetics of ara-GTP
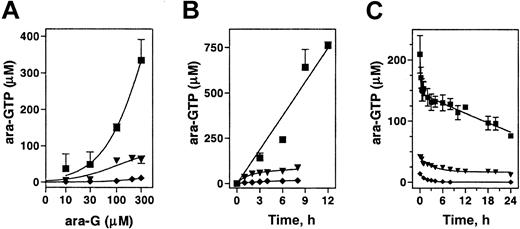
To determine if the observed differential cytotoxicity in model cell lines was related to the levels of ara-GTP, the concentration- and time-dependent accumulation of ara-GTP was compared. In CEM cells, incubation with up to 300 μM ara-G resulted in a proportional increase in ara-GTP accumulation. In B-lineage Raji or myeloid ML-1, the accumulation of ara-GTP was a log order lower (Figure 2A). The accumulation appeared to saturate by 100 μM in Raji and was never higher than 20 μM at any concentration tested in ML-1.
Accumulation and elimination of intracellular ara-GTP in 3 leukemia cell lines. Exponentially growing cultures of CEM (▪), Raji (▾), and ML-1 (♦) were incubated either with the indicated concentration of ara-G for 3 hours (A), or with 100 μM ara-G for up to 12 hours (B), or with 100 μM ara-G for 3 hours, washed, and incubated in drug-free medium (C). The extracted nucleotides were analyzed by HPLC. Data points represent mean of at least 3 independent experiments conducted in duplicate (A-B) or mean of 5 independent experiments (C).
Accumulation and elimination of intracellular ara-GTP in 3 leukemia cell lines. Exponentially growing cultures of CEM (▪), Raji (▾), and ML-1 (♦) were incubated either with the indicated concentration of ara-G for 3 hours (A), or with 100 μM ara-G for up to 12 hours (B), or with 100 μM ara-G for 3 hours, washed, and incubated in drug-free medium (C). The extracted nucleotides were analyzed by HPLC. Data points represent mean of at least 3 independent experiments conducted in duplicate (A-B) or mean of 5 independent experiments (C).
To determine if the rate of ara-GTP was linear with time, cell lines were incubated with 100 μM ara-G for up to 12 hours (Figure 2B). This concentration of ara-G was selected because at the maximally tolerated dose of nelarabine patients achieved a median 116 μM plasma ara-G concentration.21 The rate of ara-GTP accumulation in CEM cells was 60 μM/h and remained linear for up to 12 hours. The initial rate (0 to 2 hours) in Raji cells was approximately 30 μM/h but was saturated by 2 hours, and after 8 hours the intracellular ara-GTP concentration was 60 μM. In myeloid ML-1 cells, the initial rate of accumulation was low and remained low (less than 3 μM/h); at 8 hours, the intracellular ara-GTP concentration was only 18 μM.
Previous work investigating the differential cytotoxicity to ara-G has suggested that B cells eliminate ara-GTP more rapidly than T cells.20 Although the accumulation of ara-GTP was different among these cell lines, all 3 cell lines demonstrated prolonged retention of ara-GTP (Figure 2C). After a 3-hour incubation with 100 μM ara-G, the data in CEM demonstrated a biphasic elimination of ara-GTP. The initial elimination (t1/2 α) was rapid and lasted approximately 60 minutes. The terminal elimination (t½ β) was prolonged (more than 24 hours). The kinetics in B-lymphoid Raji and ML-1 myeloid cells was also biphasic and showed a prolonged retention of ara-GTP with terminal half-life of more than 24 hours.
Ara-G–induced DNA synthesis inhibition
To determine if differential accumulation of ara-GTP reflects on ara-G–mediated inhibition of DNA synthesis, the thymidine incorporation assay was employed. All 3 cell lines have a dose-dependent decrease in thymidine incorporation; however, the extent of inhibition was different (Figure 3). The IC50 for CEM is approximately 1 μM, and more than 90% inhibition of thymidine incorporation is evident at 30 μM. Raji and ML-1, on the other hand, are less sensitive, with an IC50 of approximately 300 μM. There was no effect on RNA and protein synthesis by ara-G (data not shown), indicating that DNA was the primary target.
Inhibition of DNA synthesis after ara-G treatment. Cell cultures of CEM (▪), Raji (▾), and ML-1 (♦) were exposed to various concentrations of ara-G for 3 hours and pulsed with [3H]thymidine. The [3H]DNA levels were measured and expressed as percent of control cells. Data points represent 3 independent experiments conducted in quadruplicate; bars, SE.
Inhibition of DNA synthesis after ara-G treatment. Cell cultures of CEM (▪), Raji (▾), and ML-1 (♦) were exposed to various concentrations of ara-G for 3 hours and pulsed with [3H]thymidine. The [3H]DNA levels were measured and expressed as percent of control cells. Data points represent 3 independent experiments conducted in quadruplicate; bars, SE.
Incorporation of ara-GMP into DNA
Previously we have demonstrated that the incorporation of ara-G nucleotide into DNA is critical for its cytotoxic effects.29 To test if the differential cytotoxicity was related to the number of ara-G moieties in the DNA, the levels of ara-GMP in the DNA was quantitated (Table 1). For all 3 cell lines, there was a dose-dependent increase in ara-GMP molecules in DNA. However, the number of molecules was different from one cell type to other at a given concentration. For example, in Raji, a 3-hour incubation with 100 μM ara-G resulted in 40 ± 2 pmol ara-GMP per milligram of DNA. Similar incubations in ML-1 cells resulted in statistically a similar number of ara-GMP residues in the DNA (30 ± 7 pmol ara-GMP per milligram of DNA; P > .05). Paradoxically, CEM incorporated the least amount of ara-GMP into DNA (10 ± 2 pmol ara-GMP per milligram of DNA), and this was different than either Raji or ML-1 at all but the lowest concentrations of ara-G (P < .05). Taken together, the number of ara-GMP molecules into DNA is not a determinant for T-cell selective cytotoxicity.
Incorporation of [3H]ara-G into DNA of leukemia cell lines after a 3-hour incubation with ara-G
. | Ara-GMP in DNA (pmol/mg DNA), mean ± SD . | . | . | ||
|---|---|---|---|---|---|
| Ara-G, μM . | CEM . | Raji . | ML-1 . | ||
| 0.01 | 0.009 ± 0.0003 | 0.005 ± 0.0008 | 0.003 ± 0.0009 | ||
| 0.1 | 0.01 ± 0.002 | 0.07 ± 0.01 | 0.03 ± 0.01 | ||
| 1 | 0.1 ± 0.01 | 0.7 ± 0.2 | 0.4 ± 0.2 | ||
| 10 | 1 ± 0.4 | 6 ± 1 | 4 ± 1 | ||
| 100 | 10 ± 2 | 40 ± 2 | 30 ± 7 | ||
. | Ara-GMP in DNA (pmol/mg DNA), mean ± SD . | . | . | ||
|---|---|---|---|---|---|
| Ara-G, μM . | CEM . | Raji . | ML-1 . | ||
| 0.01 | 0.009 ± 0.0003 | 0.005 ± 0.0008 | 0.003 ± 0.0009 | ||
| 0.1 | 0.01 ± 0.002 | 0.07 ± 0.01 | 0.03 ± 0.01 | ||
| 1 | 0.1 ± 0.01 | 0.7 ± 0.2 | 0.4 ± 0.2 | ||
| 10 | 1 ± 0.4 | 6 ± 1 | 4 ± 1 | ||
| 100 | 10 ± 2 | 40 ± 2 | 30 ± 7 | ||
While the absolute quantity of ara-GMP in DNA may not reflect its cytotoxic effect, the position of ara-GMP in DNA may be responsible for the observed differential sensitivity. Because there are 2 types of linkages in a DNA strand, internal and terminal, where ara-GMP may be incorporated, we identified the position of ara-GMP in DNA in each cell type. HPLC analysis of nuclease-digested DNA isolated from CEM, Raji, or ML-1 cells incubated with 100 μM[3H]ara-G 34 for 3 hours showed that 89.9% ± 11.2%, 89.9% ± 11.3%, and 89.6% ± 13.2% of the incorporated ara-GMP residues were detected as internal nucleotides, respectively. This was similar to the incorporation of [3H]dAdo (which served as a positive control), which was 84.9% ± 14.9%, 85.3% ± 15.6%, and 84.8% ± 15.2%, respectively. Between 89% and 93% internal incorporation was observed in each cell line at 1 and 10 μM ara-G. These findings strongly suggest that, in whole cells, the DNA with incorporated ara-GMP is extended by DNA polymerases.
Cell cycle distribution and apoptosis in response to ara-G
The number of ara-GMP molecules in DNAis dependent on the number of cells in S phase. To determine the cell cycle distribution in these cell lines, cells were incubated with ara-G and analyzed by flow cytometry (Table 2). The cell cycle profile was similar in untreated cells from all 3 lines, but the distribution differently changed after ara-G treatment. In CEM cells, there was an increase in the sub-G1 population from 3% to 32% between 0 and 24 hours with a corresponding decrease in the S-phase fraction from 28% to 7%. In contrast, Raji and ML-1 demonstrated approximately 2% sub-G1 DNA content over the same period, with an increase in the percentages of S-phase fraction from about 30% to 60%, indicative of S-phase arrest after drug treatment.
Effect of 100 μM ara-G on the cell cycle
. | G1 . | S . | G2M . | sub-G1 . |
|---|---|---|---|---|
| CEM, % total | ||||
| 0 h | 56 | 28 | 13 | 3 |
| 3 h | 39 | 19 | 17 | 25 |
| 6 h | 54 | 11 | 17 | 18 |
| 12 h | 60 | 4 | 9 | 27 |
| 24 h | 53 | 7 | 8 | 32 |
| Raji, % total | ||||
| 0 h | 45 | 28 | 25 | 2 |
| 3 h | 32 | 55 | 11 | 2 |
| 6 h | 34 | 49 | 15 | 2 |
| 12 h | 46 | 45 | 6 | 3 |
| 24 h | 20 | 64 | 13 | 3 |
| ML-1, % total | ||||
| 0 h | 45 | 29 | 24 | 2 |
| 3 h | 37 | 51 | 10 | 2 |
| 6 h | 37 | 52 | 9 | 2 |
| 12 h | 40 | 50 | 7 | 3 |
| 24 h | 28 | 56 | 14 | 2 |
. | G1 . | S . | G2M . | sub-G1 . |
|---|---|---|---|---|
| CEM, % total | ||||
| 0 h | 56 | 28 | 13 | 3 |
| 3 h | 39 | 19 | 17 | 25 |
| 6 h | 54 | 11 | 17 | 18 |
| 12 h | 60 | 4 | 9 | 27 |
| 24 h | 53 | 7 | 8 | 32 |
| Raji, % total | ||||
| 0 h | 45 | 28 | 25 | 2 |
| 3 h | 32 | 55 | 11 | 2 |
| 6 h | 34 | 49 | 15 | 2 |
| 12 h | 46 | 45 | 6 | 3 |
| 24 h | 20 | 64 | 13 | 3 |
| ML-1, % total | ||||
| 0 h | 45 | 29 | 24 | 2 |
| 3 h | 37 | 51 | 10 | 2 |
| 6 h | 37 | 52 | 9 | 2 |
| 12 h | 40 | 50 | 7 | 3 |
| 24 h | 28 | 56 | 14 | 2 |
Up-regulation of Fas after ara-G treatment
The Fas/FasL pathway is associated with the deletion of cells of lymphoblasts and myeloblasts. Furthermore, the expression of FasL is up-regulated in response to genotoxic stresses, which result in cell death.37 To test if ara-G affects the cell surface expression of either Fas or FasL, cells were stained with fluorescent antibodies and analyzed by flow cytometry (data not shown). After 3 hours of treatment, there was no increase in either Fas or FasL in any of the 3 cell lines tested, while a slight increase of both Fas and FasL was observed in CEM, Raji, and ML-1 cells after 24 hours. Treatment with 2 μM adriamycin for 24 hours served as the positive control (data not shown).
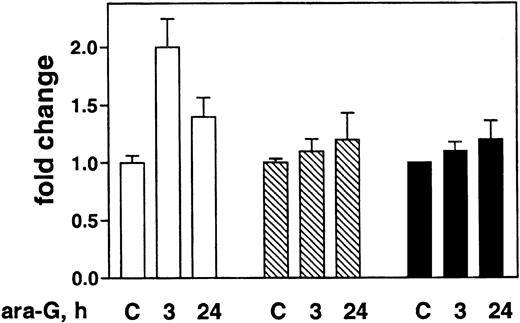
Although no increase in Fas or FasL was observed in response to ara-G, reports that T cells can die by apoptosis via a paracrine mechanism through Fas38-40 as a result of metalloproteinase-mediated release of human Fas ligand from the surface of the cell41 prompted an evaluation of soluble FasL in the medium (Figure 4). After 3 hours of incubation with 100 μM ara-G, a significant 2-fold increase in sFasL in the medium of CEM cells (P = .03) was detected by ELISA. After 24 hours, the level remained elevated over that seen in control, but this value was not significant. In contrast, neither Raji nor ML-1 showed an increase in sFasL in the medium at any time during the incubation.
Effect of ara-G incubation on sFasL in 3 leukemia cell lines. Exponentially growing cultures of CEM (open bar), Raji (striped bar), and ML-1 (filled bar) were treated with 100 μM ara-G for the indicated time. Medium was collected and analyzed for sFasL by ELISA. Data bars represent mean of 3 independent experiments; bars, SD.
Effect of ara-G incubation on sFasL in 3 leukemia cell lines. Exponentially growing cultures of CEM (open bar), Raji (striped bar), and ML-1 (filled bar) were treated with 100 μM ara-G for the indicated time. Medium was collected and analyzed for sFasL by ELISA. Data bars represent mean of 3 independent experiments; bars, SD.
Ara-G induces an increase in Fas and FasL gene expression
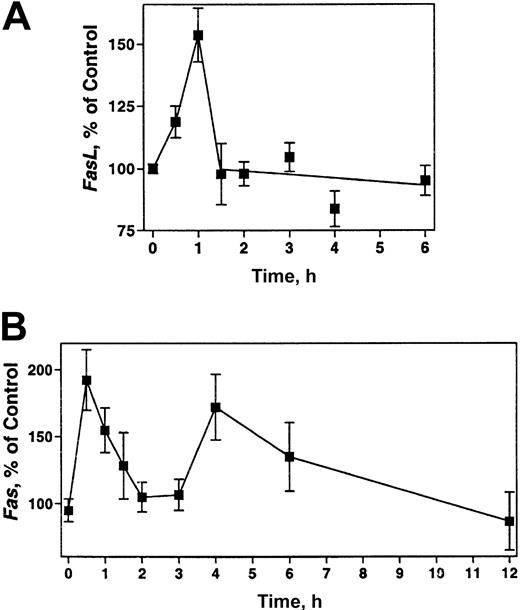
To determine if the ara-G–mediated increase in the FasL protein in CEM cells is related to changes in its gene expression, the levels of FasL and Fas mRNA were assessed by TaqMan real-time RT-PCR. The analysis of FasL mRNA levels consistently displayed a significant spiked increase in gene expression that peaked 1 hour after ara-G treatment (Figure 5A, P < .0001, n = 20). The amount of FasL mRNA returned to control levels by 1.5 hours of treatment (P = .78, n = 8) and remained at this level for at least 6 hours of treatment (2 hours: P = .70, n = 20; 3 hours: P = .44, n = 19; 6 hours: P = .38, n = 14). This transient increase in the transcript was consistent with the increase in soluble FasL protein levels and indicates that expression of FasL is controlled at the gene level.
Induction of FasL and Fas mRNA in CEM cells. Total RNA was isolated from 3 × 105 CEM cells after treatment with 100 μM ara-G at the indicated times. The relative mRNA levels of FasL (A) and Fas (B) were measured by real-time RT-PCR, normalized to the levels of GAPDH mRNA, and expressed as percentage of the levels in untreated cells.
Induction of FasL and Fas mRNA in CEM cells. Total RNA was isolated from 3 × 105 CEM cells after treatment with 100 μM ara-G at the indicated times. The relative mRNA levels of FasL (A) and Fas (B) were measured by real-time RT-PCR, normalized to the levels of GAPDH mRNA, and expressed as percentage of the levels in untreated cells.
Analysis of the Fas mRNA also revealed a reproducible induction of gene expression (Figure 5B). This induction peaked to 200% of control within 30 minutes of ara-G treatment (P = .0001, n = 16), decreased at a slower rate (150% at 1 hour: P = .0016, n = 23), and reached control levels by 2 hours of treatment (P = .5334, n = 22; 3 hours: P = .3857, n = 22). Unlike FasL, a second induction of Fas mRNA was found with the levels rising to 180% of control at 4 hours (P = .0003, n = 9) and returning to control levels by 6 hours (P = .086, n = 16). This Fas mRNA induction differed with the Fas protein levels, suggesting possible translational and/or posttranslational control mechanisms. A similar induction of FasL and Fas mRNA was observed in the Jurkat T cells, in which the induction of both genes peaked at 2 hours of ara-G treatment (data not shown).
Role of FasL in T-cell selective cytotoxicity
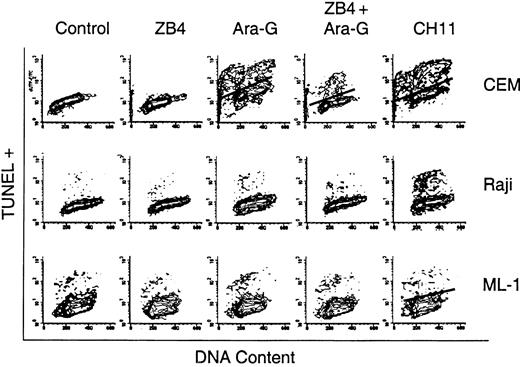
To test if the increase in the sFasL transcript and protein levels could play a role in ara-G–mediated T-cell selective cytotoxicity, cells were treated with 100 μM ara-G for 3 hours in the presence or absence of 1 μg/mL ZB4 Fas antagonist antibody (Figure 6). Untreated cells as well as cells treated for 4 hours with 1 μg/mL CH-11 Fas agonist antibody served as controls. The data from the lymphoid cell lines demonstrate that CH-11–mediated activation of Fas receptor resulted in about 41% apoptosis in CEM and about 23% in Raji while this same treatment did not result in an increase in apoptosis in myeloid ML-1. The cell cycle distribution shows that TUNEL-positive cells originate from all phases of the cell cycle in response to treatment with CH-11. The treatment of all 3 cell lines with ZB4 for 4 hours resulted in no change in the percentage of TUNEL-positive cells in comparison to control (less than 1%). Treatment with other nucleoside analogs, however, resulted in an S phase–specific increase in TUNEL positivity in all 3 cell lines, suggesting that these cells are sensitive to these agents (data not shown). Incubation of CEM with 100 μM ara-G for 3 hours resulted in 26% TUNEL-positive cells arising from all phases of the cell cycle, similar to the pattern observed with CH-11 treatment. In contrast, when CEM cells were preincubated with ZB4 and then exposed to 100 μM ara-G for 3 hours, there was an overall decrease in the percentage of TUNEL-positive cells to 13% where most of these TUNEL-positive cells appeared to arise from the S phase. Hence, in T cells, ara-G induces an S phase–dependent and S phase–independent (Fas death receptor mediated) cell death.
The effect of Fas antagonist ZB4 and Fas agonist CH11 antibodies on ara-G–mediated cell death. Exponentially growing cultures of CEM (upper panels), Raji (middle panels), and ML-1 (lower panels) were either untreated or treated with ZB4, the Fas antagonist antibody; ara-G; ZB4 for 1 hour followed by ara-G; or CH-11, the Fas agonist antibody. Cells were analyzed by flow cytometry for TUNEL positivity and their cell cycle profile.
The effect of Fas antagonist ZB4 and Fas agonist CH11 antibodies on ara-G–mediated cell death. Exponentially growing cultures of CEM (upper panels), Raji (middle panels), and ML-1 (lower panels) were either untreated or treated with ZB4, the Fas antagonist antibody; ara-G; ZB4 for 1 hour followed by ara-G; or CH-11, the Fas agonist antibody. Cells were analyzed by flow cytometry for TUNEL positivity and their cell cycle profile.
Discussion
Our model system cell lines demonstrated T-cell selective cytotoxicity with ara-G (Figure 1). As observed previously in primary leukemia cells in vitro16 and in phase 1 studies,21 the accumulation of ara-GTP is greatest in cells of T-lymphoid lineage (Figure 2).9,18 These data and the fact that ara-GTP levels are strongly related to response to nelarabine therapy suggest that triphosphate accumulation is one of the determinants for lineage-specific cytotoxicity. However, equimolar concentrations of ara-GTP were not equipotent for inhibition of colony formation in these cell lines. For example, 50 μM ara-GTP accumulation results in only 25% cell death in Raji but 75% in CEM cells, suggesting that ara-GTP accumulation is not the only determinant for cytotoxicity (not shown but based on Figures 1C and 2A).
Our previous work clearly demonstrated the requirement of ara-G incorporation into DNA for cell death.29 Therefore, we tested if the number of ara-G residues in DNA was greater in T cells compared with other cell lineages. Surprisingly, the CEM cell line, which was most sensitive to ara-G, incorporated the least number of ara-G molecules but had the greatest inhibition of DNA synthesis. In contrast, Raji and ML-1 accumulated a greater amount of ara-GMP in the DNA with lesser inhibition of DNA synthesis and cell death (Table 1). Although within a cell line ara-G incorporation into DNA appears to play a role in dose-dependent inhibition of DNA synthesis and cellular toxicity, between cell lines such correlation does not exist.
Because the incorporation of ara-GMP into DNA occurs only during the S phase, we sought to determine the dynamics of cell cycle distribution after ara-G treatment. Although the cell cycle distribution was similar in untreated cells, after treatment the proportion of S-phase cells dramatically decreased in CEM but increased in Raji and ML-1 cell lines (Table 2). This increase in the S-phase percentage will amplify the number of incorporated ara-GMP molecules of the whole population. Hence, we recalculated the number of ara-GMP incorporation into DNA by normalizing with the S-phase population. Even after this normalization, the number of ara-G residues was not greater in CEM cells; at 100 μM ara-G, the numbers were 53, 71, and 48 pmol/mg of S-phase DNA in CEM, Raji, ML-1 cells, respectively. Taken together, these observations suggest that there are additional parameters responsible for differential cytotoxicity.
In vitro DNA primer extension assays using purified DNA polymerases have established termination of DNA synthesis after ara-GMP incorporation. It is feasible that the greater number of ara-GMP in the terminal position may reflect on inhibition of DNA synthesis leading to cell death. Hence, we quantitated the ara-G residues at terminal versus internal position in these cell lines. Similar to dAMP incorporation, most ara-GMP molecules were found in the internal linkage in the 3 model cell lines. Clearly, this internucleotide incorporation of ara-G does not play a role for T cell–specific sensitivity.
As described above, a similar number of ara-GMP molecules per S-phase cells in internal linkage results in separate consequences in T-versus non–T-cell types. In T cells, this event was lethal while in non-T cells this incorporation was associated with S-phase arrest. This arrest may be transient and represent a cell cycle checkpoint initiated to repair the damage caused by ara-GMP molecules in DNA. In such a scenario, if the drug is removed, cells may repair DNA and proceed with a transit through S phase and normal proliferation rate. Our Raji and ML-1 cloning data (Figure 1C) suggest that this postulate is correct because there was only a minimal loss of colony formation capability of these cells after a 3-hour ara-G treatment.
An additional possibility exists for the resistance of non-T cells to ara-G. This could be due to up-regulation of survival pathways, increased levels of antiapoptotic proteins, or inherent attenuation of cell death pathways. In such a case, these cells would be resistant to other nucleoside analogs. However, these cells were hypersensitive to many purine and pyrimidine analogs such as gemcitabine, ara-C, and fludarabine.24,42,43 Furthermore, molecules such as p53, which has been shown to be involved in various forms of apoptosis induced by cellular “stress,”44 were not a factor in our experimental systems. The p53 is heterozygous mutated in CEM45 and Raji46 while ML-1 is p53 wild type,47 suggesting that p53 is not a component of the observed ara-G selectivity.
The mitochondria have been implicated in driving the apoptotic cascade by releasing cytochrome c from the intermembrane space to the cytoplasm.After release, cytochrome c binds to Apaf1 in the presence of deoxyadenosine triphosphate (dATP), and this complex (the apoptosome) then activates a caspase cascade with caspase 9 at its apex.48,49 Recently, it has been shown that ara-GTP can substitute for dATP in an in vitro assay system using cytochrome c and Apaf1 proteins.50 The Michaelis constant (Km) value of ara-GTP in this reaction is about 200 μM. In T cells, the intracellular level of ara-GTP is more than 250 μM, suggesting that in whole cells ara-GTP molecules may substitute for dATP in apoptosome. Such mechanism may amplify the death signal; however, the initial trigger for apoptosis in response to ara-G occurs only after incorporation of ara-GMP into DNA.
There is evidence for an active role of the Fas/FasL system in the process of drug-induced apoptosis including nucleoside analogs.36,51-58 Because T, B, and myeloid cells are hematopoietic cells derived from a single pluripotent stem cell59-61 and because of the demonstrated importance of Fas/FasL in the normal development of T and B cells,62 we evaluated the role of Fas and FasL in ara-G–mediated apoptosis in CEM, Raji, and ML-1. A 3-hour incubation with ara-G did not result in an increase in the expression of surface Fas or FasL in the cell types tested but resulted in the liberation of soluble FasL into the medium of only CEM cells (Figure 4). This was preceded by a transient but consistent increase in the transcript level of FasL (Figure 5A). This finding suggested a role for soluble FasL in the observed T-cell hypersensitivity to ara-G. Prompted by our previous data,63 Mansson et al64 further confirmed involvement of the Fas pathway in ara-G–mediated T-cell cytotoxicity by demonstrating a 3-fold to 6-fold reduction in Fas protein levels and no detectable levels of Fas mRNA in ara-G–resistant T-cell lines. An important feature of Fas-mediated apoptosis is the mechanism of autocrine suicide or paracrine death initially described for activated T cells.40 The data from the TUNEL assay demonstrate that by 3 hours ara-G is able to induce apoptosis in CEM from all phases of the cell cycle. When ZB4 is added to CEM cells prior to the addition of ara-G, only S-phase cells remain susceptible to ara-G–induced apoptosis. This suggests that the T-cell selective cytotoxicity observed in CEM is due to ara-G incorporation into the DNA of S-phase cells, stimulating the release of sFasL into the medium (Figure 4) and triggering death receptor–mediated cell death in the bystander cells (Figure 6). This mechanism of action might also explain the selectivity observed in immature T cells both in vitro and in vivo.
Several transcription factors are known to regulate FasL gene expression65 ; in T cells it is mediated by nuclear factor–κB (NF-κB) and activator protein-1 (AP-1),66 while hepatocellular carcinoma cells induce it via the Jun kinase/stress-activated protein kinase pathway and AP-1 activation.67 Interestingly, CEM and Jurkat cells autoamplify sFasL, tumor necrosis factor (TNF)–related apoptosis-inducing ligand, TRAIL, and TNF-α levels in response to Fas stimulation with a low level of the Fas agonistic antibody, αAPO-1.68 Under these conditions, cell killing was blocked only about 30% by FasL neutralizing antibodies and required anti–TNF-α antibodies to achieve a high degree of apoptosis inhibition. Apoptosis was blocked by more than 85% with FasL neutralizing antibodies in our system and primarily only an S-phase cytotoxicity remains, suggesting the activation of other death receptor pathways may not play a major role in ara-G–mediated cell killing.
In conclusion, the present study illustrates that several factors are responsible for ara-G–induced cytotoxicity in the T-cell lineage. First, the pharmacokinetics of ara-GTP accumulation is favorable, resulting in higher levels of ara-GTP. Second, incorporation of this fraudulent nucleotide into replicating DNA initiates S phase–specific cell death. Third, the signal for apoptosis may be enhanced by cytosolic ara-GTP by binding to apoptosome complexes. Finally, the transcriptional and translational up-regulation of sFasL may account for S phase–independent cell death. Taken together, these parameters may be responsible for T-cell selective ara-G toxicity.
Prepublished online as Blood First Edition Paper, May 15, 2003; DOI 10.1182/blood-2003-01-0317.
Supported in part by grant CA57629 from the National Cancer Institute, Department of Health and Human Services.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors are thankful to Dr Elizabeth Hileman and Dr Deepa Sampath for assistance with flow cytometry and Mary Ayres for deoxynucleotide analyses.

![Figure 3. Inhibition of DNA synthesis after ara-G treatment. Cell cultures of CEM (▪), Raji (▾), and ML-1 (♦) were exposed to various concentrations of ara-G for 3 hours and pulsed with [3H]thymidine. The [3H]DNA levels were measured and expressed as percent of control cells. Data points represent 3 independent experiments conducted in quadruplicate; bars, SE.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/5/10.1182_blood-2003-01-0317/6/m_h81734861003.jpeg?Expires=1767734726&Signature=MTBhfMgJzh-snxyhXdeepad0T3hTkPcQbTg8BQUYno5lwfj9xQluIfYQevS4pHzlYC0ilQRBBI3q0L~OKI6aQ~tG5dZ7qavG6VPh8V6Bhv5oSTnjj7fsozrQ49gCIZxQnqPqRBVaGXYVnrQf5NDt5pN91CXLkQPQ1Txr1ZW4QbDHrjzQa6kq2Hu9MCtTYewUxELKAm1TUN1NjlrSxfRgw~jO7eHj0WiLSKR2TCAUAhQ4A9BiSkuuvKr~Zhdf7NrByJ~IOuZkATRiRX52zKoBFT~xWipvjpL0KdU4qVm8q0DalYUcJf7k3qBvUz5R~gmWOLyRAAwpVLWGVjBneaaWXw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal