Abstract
Glutathione S-transferase P1 (GSTP1) is a phase 2 drug metabolism enzyme involved in the metabolism and detoxification of a range of chemotherapeutic agents. A single nucleotide polymorphism (Ile105Val) results in a variant enzyme with lower thermal stability and altered catalytic activity. We hypothesized that patients with the less stable variant have a decreased ability to detoxify chemotherapeutic substrates, including melphalan, and have an altered outcome following treatment for multiple myeloma. We have assessed the impact of GSTP1 codon 105 polymorphisms in 222 patients entered into the Medical Research Council (MRC) myeloma VII trial (comparing standard-dose chemotherapy with high-dose therapy). In the standard-dose arm, patients with the variant allele (105Val) had an improved progression-free survival (PFS) (adjusted hazard ratios for PFS were 0.55 for heterozygotes and 0.52 for 105Val homozygotes, compared with 105Ile homozygotes; P for trend = .04); this was supported by a trend to improved overall survival, greater likelihood of entering plateau and shorter time to reach plateau in patients with the 105Val allele. No difference in outcome by genotype was found for patients treated with high-dose therapy. However, the progression-free survival advantage of the high-dose arm was seen only in patients homozygous for 105Ile (P = .008). (Blood. 2003;102:2345-2350)
Introduction
Patients with multiple myeloma have a median survival of 3 to 4 years when treated with standard-dose chemotherapy.1 Recently it has been shown that high-dose therapy with stem cell rescue is associated with an improved survival of approximately 12 months, compared with standard treatment.2,3 One of the most powerful markers for risk stratification is the serum β2-microglobulin; this along with other standard prognostic markers has been incorporated into staging formulas to define more refined prognostic groups.4 These prognostic groups are not based on the biology of the tumor and do not take into account interindividual variation in response. Consequently, they lack power to accurately define prognosis or treatment-specific responses in individual patients. The development of host or tumor markers that take such biologic variability into account would be a major step forward.
The human genome mapping project has focused attention on interindividual genetic variation and its capacity to define variability in disease predisposition and outcome after treatment. Genetic variation governing the metabolism of chemotherapeutic agents used to treat tumors has been the focus of much research.5
Glutathione S-transferases (GSTs) are a polygenic group of enzymes involved in the phase 2 detoxification of a wide range of xenobiotics and chemotherapeutic agents by catalyzing the conjugation of reactive electrophiles to glutathione.6 GSTP1 is the most abundant GST found in many normal and malignant tissues and is known to be expressed in human plasma cells.7-9 By means of gene transfection and pharmacokinetic studies, many chemotherapeutic agents, including melphalan, cyclophosphamide, vincristine, adriamycin, cisplatin, etoposide, thiotepa, chlorambucil, and busulphan, have been shown to be substrates for GSTP1.10-17 GSTP1 is polymorphic with 2 single-nucleotide substitutions in the coding sequence (1578A>G, and 2293C>T), giving rise to Ile105Val and Ala114Val amino acid substitutions. Both amino acid residues lie within the substrate-binding site of GSTP1.7,18 The GSTP1 114Val variant occurs in approximately 10% of whites, whereas 33% of whites have the GSTP1 105Val variant, with 14% being homozygous.19 The GSTP1 105Val variant is associated with a lower thermal stability and altered catalytic activity to a variety of substrates compared with GSTP1 105Ile.20,21 The catalytic activity of GSTP1 105Val to Thiotepa is 2 times lower than that of GSTP1 105Ile while the difference is up to 15 times lower for chlorambucil.14,22 However, differences in metabolic activity may be substrate specific as it has been shown in vitro that, when cisplatin is the substrate, the GSTP1 105Val variant has a 3- to 4-fold greater cytoprotective effect than GSTP1 105Ile.23 These differences in catalytic activity have led investigators to study the association of inherited allelic variation in GSTP1 and outcome from therapy. Individuals with the GSTP1 105Val variant have been found to have a decreased risk of relapse from childhood acute lymphoblastic leukemia and an improved survival following treatment for breast cancer and colon cancer.24-27 Furthermore, following chemotherapy using drugs known to be GSTP1 substrates, an increased incidence of therapy-related acute leukemia has been found in patients with the GSTP1 105Val variant, further implicating GSTP1 polymorphisms as a modulator of drug response.28 In addition to its role in drug metabolism, GSTP1 is also involved in the detoxification of environmental toxins, and alteration in its function has been proposed as a risk factor for carcinogenesis.29 The GSTP1 105Val variant has been associated with an increased incidence of breast, prostate, bladder, and testicular cancer.30,31 The MRC myeloma VII trial compares standard-dose chemotherapy (adriamycin, BCNU [1,3-bis(2-chloroethyl)-1-nitrosourea], cyclophosphamide, and melphalan [ABCM]) with high-dose therapy (cyclophosphamide, vincristine, adriamycin, and methylprednisolone followed by high-dose melphalan [200 mg/m2] and stem cell rescue [CVAMP/HDM]).2 Most of these agents have been shown to be substrates for GSTP1.
Using samples and data from patients entered into this trial, we have examined the role of GSTP1 polymorphisms on the outcome of therapy and also evaluated its role in the incidence of myeloma.
Patients, materials, and methods
Samples were available on 233 of 401 patients enrolled in the Medical Research Council myeloma VII trial, which recruited patients with newly diagnosed myeloma younger than age 65 years, from 1993 to 2000. Patients were randomized to treatment with either ABCM or CVAMP/HDM.2 Samples, which consisted of fresh whole blood or bone marrow smears taken at presentation or at follow-up, were stored and archived following informed consent.
Follow-up data were collected prospectively using a form detailing course number and date given, response, remission status, and vital status measured at 3-month intervals.
For the case-control study, 440 age- and sex-matched controls were selected from a previously reported cohort.19 These patients had previously been genotyped for the GSTP1 105 polymorphism. Selection was blinded as to GSTP1 genotype status. In addition, 158 cases and 192 controls were also genotyped for the GSTP1 114 polymorphism.
Population description
Of the 233 patients whose samples were available, 222 had a genotype allocated (the DNA from 11 patients failed to amplify). Patient characteristics are shown in Table 1.
Comparison of presentation features between treatment arm
. | Treatment arm . | . | . | . | |
|---|---|---|---|---|---|
. | ABCM, no. (%) . | CVAMP/HDM, no. (%) . | Total, no. . | P . | |
| Total | 101 | 121 | 222 | ||
| Sex | |||||
| M | 52 (52) | 62 (51) | 114 | ||
| F | 49 (48) | 59 (49) | 108 | .51 | |
| Age, y | |||||
| 31-40 | 7 (7) | 1 (1) | 8 | ||
| 41-50 | 25 (24) | 27 (22) | 52 | ||
| 51-60 | 50 (50) | 68 (56) | 118 | ||
| 61-66 | 19 (19) | 25 (21) | 44 | .09 | |
| B2M level, mg/L | |||||
| Below 4 | 45 (45) | 52 (43) | 97 | ||
| 4-8 | 30 (29) | 37 (31) | 67 | ||
| More than 8 | 22 (22) | 28 (23) | 50 | ||
| Unknown | 4 | 4 | 8 | .93 | |
| Hemoglobin level, g/L | |||||
| Below 800 | 1300 (13) | 1300 (11) | 2600 | ||
| 800-1000 | 2700 (26) | 3600 (30) | 6300 | ||
| More than 1000 | 5900 (59) | 65 (54) | 12 400 | ||
| Unknown | 200 | 700 | 900 | .75 | |
| Creatinine level, μM | |||||
| Below 100 | 43 (43) | 57 (47) | 100 | ||
| 100-200 | 44 (43) | 49 (40) | 93 | ||
| More than 200 | 14 (14) | 15 (13) | 29 | .83 | |
. | Treatment arm . | . | . | . | |
|---|---|---|---|---|---|
. | ABCM, no. (%) . | CVAMP/HDM, no. (%) . | Total, no. . | P . | |
| Total | 101 | 121 | 222 | ||
| Sex | |||||
| M | 52 (52) | 62 (51) | 114 | ||
| F | 49 (48) | 59 (49) | 108 | .51 | |
| Age, y | |||||
| 31-40 | 7 (7) | 1 (1) | 8 | ||
| 41-50 | 25 (24) | 27 (22) | 52 | ||
| 51-60 | 50 (50) | 68 (56) | 118 | ||
| 61-66 | 19 (19) | 25 (21) | 44 | .09 | |
| B2M level, mg/L | |||||
| Below 4 | 45 (45) | 52 (43) | 97 | ||
| 4-8 | 30 (29) | 37 (31) | 67 | ||
| More than 8 | 22 (22) | 28 (23) | 50 | ||
| Unknown | 4 | 4 | 8 | .93 | |
| Hemoglobin level, g/L | |||||
| Below 800 | 1300 (13) | 1300 (11) | 2600 | ||
| 800-1000 | 2700 (26) | 3600 (30) | 6300 | ||
| More than 1000 | 5900 (59) | 65 (54) | 12 400 | ||
| Unknown | 200 | 700 | 900 | .75 | |
| Creatinine level, μM | |||||
| Below 100 | 43 (43) | 57 (47) | 100 | ||
| 100-200 | 44 (43) | 49 (40) | 93 | ||
| More than 200 | 14 (14) | 15 (13) | 29 | .83 | |
P values are associated with Pearson chi-square statistic. B2M indicates β2-microglobulin.
The groups were evenly matched between treatment arm, with no significant difference in age, sex, hemoglobin level, creatinine level, β2-microglobulin level, or GSTP1 105 genotype. There was also no significant difference in these characteristics among the 3 genotypic groups (data not shown). The subgroup of patients investigated for GSTP1 polymorphisms did not differ significantly from the trial group as a whole for presentation prognostic markers or protocol deviations (data not shown). There was no difference in overall or progression-free survival between the GSTP1 subgroup and the trial as a whole. For all patients in the myeloma VII trial, median overall survival (OS) was 48.5 months (95% confidence interval [CI], 42.2-56.3 months), and median progression-free survival (PFS) was 25.1 months (95% CI, 21.4-27.8 months), compared with 49.1 months (95% CI, 44.6-61.4 months) and 27.2 months (95% CI, 23.7-29.8 months), respectively, for the GSTP1 subgroup.
Genotyping
A polymerase chain reaction followed by restriction enzyme digestion (PCR-RFLP) for the GSTP1 105 polymorphism was performed by means of a previously published technique.32 Briefly, amplification leads to a 177-base pair (bp) product. The A to G polymorphism introduces a restriction site recognized by BsmA1 digestion enzyme (New England Biolabs, Beverly, MA). Digestion of the 177-bp amplicon results in either retention of the 177-base pair (bp) product or complete digestion to 93-bp and 84-bp fragments corresponding to individuals homozygous for the Ile or Val alleles respectively. The presence of all 3 fragments corresponded to individuals heterozygous at codon 105.
A modified allele-specific PCR (AS-PCR) technique was also used to determine GSTP1 genotypes for the codon 105 polymorphism.33 In 2 separate reactions, primers specific for the wild-type allele and the variant allele were combined with a common reverse primer. A common reverse primer sequence and specific forward primers for the wild-type and the variant allele were 5′TCAGTTGCCCGGGCACTG, 5′TGGTGTCTGGCAGGAGGC, and 5′TGGTGTCTGGCAGGAGGT, respectively. For each genotype determination, 10 ng DNA was amplified in a total reaction volume of 20 μL, consisting of 0.2 μmol each primer; 6 U stoffel gold DNA polymerase (Roche Molecular Systems, Pleasanton, CA); 1 × stoffel buffer (10 mM Tris-HCl [tris(hydroxymethyl)aminomethane)-HCl], 10 mM KCl at pH 8.0); 30 mM KCl; 2 mM MgCl2; 0.2 × Sybr green (Molecular Probes, Leiden, the Netherlands); 2 μM Rox (Molecular Probes); 5% dimethyl sulfoxide (DMSO); and 2.5% glycerol. All primers were amplified on an Applied Biosystems 7700 (Weiterstadt, Germany) under the following amplification conditions: 2 minutes at 50°C; 12 minutes at 95°C; followed by 40 cycles of 20 seconds at 58°C, 20 seconds at 95°C, with a final extension step of 20 minutes at 72°C. Presence or absence of amplicon was determined with the use of Sybr green as the reporter dye.
For the GSTP1 114 polymorphism, samples were genotyped with the use of RFLP as described elsewhere,34 in addition to AS-PCR as described above. Primer sequences for AS-PCR were 5′TCAGTTGCCCGGGCAGTG (common reverse), 5′TGGTGTCTGGCAGGC (forward wild type), and 5′TGGTGTCTGGCAGGAGGT (forward variant).
For the GSTP1 105 polymorphism, all samples were initially genotyped by means of PCR-RFLP, which was successful on 140 (60%) cases. Of those initially successful on PCR-RFLP, 56 (40%) had the genotype confirmed by repeat PCR-RFLP and 84 (60%) by AS-PCR. Of those that failed PCR-RFLP, 82 (88%) were successfully allocated a genotype with AS-PCR. All patients failing PCR-RFLP had AS-PCR performed twice. There were 5 discrepancies between the 2 techniques, all of which were consistent with failure of digestion on PCR-RFLP. All 5 were repeated with both techniques confirming the heterozygous state.
Statistical methods
The associations between GSTP1 genotype and patient presenting features were assessed by means of the chi-square test. Plateau and progression were defined according to published guidelines.35 The median time to plateau for the whole group was 260 days; differences in time to plateau between the genotypes were assessed as the number of patients reaching plateau prior to, or after, the median. Trend was evaluated by means of likelihood-ratio tests comparing models with and without a variable representing the number of variant alleles (0, 1, and 2); P values for trend tests are 2-sided. PFS was defined as time from randomization to death or progression, and OS was defined as time from randomization to death. Both PFS and OS were analyzed by means of Kaplan-Meier analysis and Cox proportional hazard models. Kaplan-Meier plots presented are unadjusted survival curves. Hazard ratios were calculated for relative risk of death or progression for patients either heterozygous for the GSTP1 105 polymorphism (Ile/Val) or homozygous for the variant allele (Val/Val) compared with patients homozygous for the wild-type allele (Ile/Ile). Hazard ratios were adjusted for prognostic factors including age and levels of B2M, hemoglobin, and creatinine. To evaluate the interaction of genotype and treatment arm, a likelihood-ratio test was performed comparing Cox proportional hazard models with and without terms for the interaction between genotype and treatment arm, interacting an indicator of the variant with treatment group. All analyses were performed on an intention-to-treat basis unless otherwise stated. For the case-control study odds, ratios (ORs) were calculated by means of logistic regression analysis. Estimation of Hardy-Weinberg equilibrium was carried out with chi-square analysis from the HWE software program (Linkage Utility Programs, Rockefeller University, New York, NY).
Results
Case-control study
For the GSTP1 105 polymorphism, 222 patients were assigned a genotype. There were 98 (44%) patients genotyped as Ile/Ile, 86 (39%) as heterozygous (Ile/Val), and 38 (17%) as Val/Val. There was no significant difference in genotype distribution between the cases and controls. For GSTP1 105, compared with Ile/Ile, ORs for Ile/Val and Val/Val were 1.02 (95% CI, 0.72-1.45) and 1.37 (95% CI, 0.83-2.22), respectively. For the GSTP1 114 polymorphism, ORs were 1.17 (95% CI, 0.63-1.97) and 1.23 (95% CI, 0.17-8.91) for heterozygotes and variant homozygotes, respectively, compared with wild-type homozygotes. Interestingly, the GSTP1 105 genotype was out of Hardy-Weinberg equilibrium for both cases and controls, with a deficit of heterozygotes in both populations; this was particularly marked in females.
Outcome data
In order to look at the impact of variation in GSTP1 105 genotype on therapy, we first looked at response, progression-free and overall survival, by arm. For the ABCM arm, individuals with 1 or 2 Val alleles had a significant trend to a greater likelihood of entering plateau (P = .04), a shorter time to reach plateau (P = .07), and fewer courses of ABCM received prior to entering plateau (P = .03) compared with patients homozygous for the Ile allele (Table 2).
Response to treatment by genotype for 222 patients with multiple myeloma treated with ABCM or CVAMP/HDM
. | GSTP1 105 genotype . | . | . | . | . | ||
|---|---|---|---|---|---|---|---|
. | Ile/Ile, no.(%) . | Ile/Val, no.(%) . | Val/Val, no.(%) . | Total, no.(%) . | P* . | ||
| ABCM arm | |||||||
| Plateau reached | |||||||
| Yes | 32 (66) | 25 (78) | 17 (89) | 74 (74) | |||
| No | 16 (34) | 7 (22) | 2 (11) | 25 (25) | .04 | ||
| No. of courses ABCM prior to plateau | |||||||
| 6 or fewer | 21 (65) | 19 (76) | 16 (94) | 56 (76) | |||
| More than 6 | 11 (35) | 6 (24) | 1 (6) | 18 (24) | .03 | ||
| Time to plateau | |||||||
| 260 d or fewer | 12 (38) | 12 (48) | 11 (65) | 35 (47) | |||
| More than 260 d | 20 (62) | 13 (52) | 6 (35) | 39 (53) | .07 | ||
| CVAMP/HDM arm | |||||||
| Plateau reached | |||||||
| Yes | 40 (80) | 42 (82) | 15 (79) | 97 (81) | |||
| No | 9 (20) | 9 (18) | 4 (21) | 22 (19) | .86 | ||
| Time to plateau | |||||||
| Fewer than 260 d | 24 (60) | 19 (45) | 8 (53) | 51 (53) | |||
| More than 260 d | 16 (40) | 23 (55) | 7 (47) | 46 (47) | .41 | ||
| Transplant received | |||||||
| Yes | 45 (90) | 41 (80) | 15 (79) | 101 (84) | |||
| No | 5 (10) | 10 (20) | 4 (21) | 19 (16) | .17 | ||
. | GSTP1 105 genotype . | . | . | . | . | ||
|---|---|---|---|---|---|---|---|
. | Ile/Ile, no.(%) . | Ile/Val, no.(%) . | Val/Val, no.(%) . | Total, no.(%) . | P* . | ||
| ABCM arm | |||||||
| Plateau reached | |||||||
| Yes | 32 (66) | 25 (78) | 17 (89) | 74 (74) | |||
| No | 16 (34) | 7 (22) | 2 (11) | 25 (25) | .04 | ||
| No. of courses ABCM prior to plateau | |||||||
| 6 or fewer | 21 (65) | 19 (76) | 16 (94) | 56 (76) | |||
| More than 6 | 11 (35) | 6 (24) | 1 (6) | 18 (24) | .03 | ||
| Time to plateau | |||||||
| 260 d or fewer | 12 (38) | 12 (48) | 11 (65) | 35 (47) | |||
| More than 260 d | 20 (62) | 13 (52) | 6 (35) | 39 (53) | .07 | ||
| CVAMP/HDM arm | |||||||
| Plateau reached | |||||||
| Yes | 40 (80) | 42 (82) | 15 (79) | 97 (81) | |||
| No | 9 (20) | 9 (18) | 4 (21) | 22 (19) | .86 | ||
| Time to plateau | |||||||
| Fewer than 260 d | 24 (60) | 19 (45) | 8 (53) | 51 (53) | |||
| More than 260 d | 16 (40) | 23 (55) | 7 (47) | 46 (47) | .41 | ||
| Transplant received | |||||||
| Yes | 45 (90) | 41 (80) | 15 (79) | 101 (84) | |||
| No | 5 (10) | 10 (20) | 4 (21) | 19 (16) | .17 | ||
Chi-square test for linear trend.
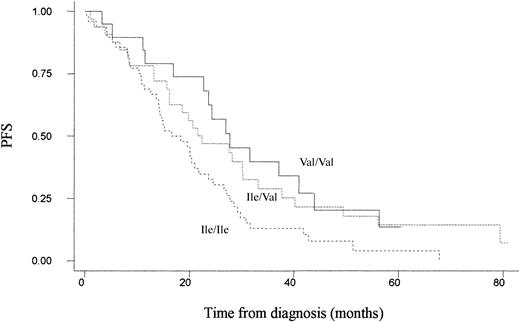
On an intention-to-treat basis, there was a significant improvement in progression-free survival for patients randomized to the ABCM arm who possessed at least one Val allele (compared with Ile/Ile, the adjusted hazard ratio for progression for Ile/Val was 0.55 [95% CI, 0.32-0.96] and for Val/Val was 0.52 [95% CI, 0.28-0.97], P = .04 for trend) (Figure 1; Table 3).
Unadjusted Kaplan-Meier plot for progression-free survival among patients treated with ABCM by GSTP1 105 genotype. Ile/Ile (n = 48), Ile/Val (n = 34), and Val/Val (n = 19).
Unadjusted Kaplan-Meier plot for progression-free survival among patients treated with ABCM by GSTP1 105 genotype. Ile/Ile (n = 48), Ile/Val (n = 34), and Val/Val (n = 19).
Effect of GSTP1 genotype on progression-free survival and overall survival for patients treated with ABCM and CVAMP/HDM
. | Progression-free survival . | . | . | Overall survival . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | Median survival, mo . | Adjusted hazard ratio* . | 95% CI . | Median survival, mo . | Adjusted hazard ratio* . | 95% CI . | ||||
| ABCM† | ||||||||||
| Overall | 21.0 | NA | NA | 44.7 | NA | NA | ||||
| Ile/Ile | 16.6 | 1 | NA | 39.3 | 1 | NA | ||||
| Ile/Val | 21.6 | 0.55 | 0.32-0.96 | 51.8 | 0.65 | 0.34-1.25 | ||||
| Val/Val | 27.8 | 0.52 | 0.28-0.96 | 48.5 | 0.78 | 0.39-1.58 | ||||
| CVAMP/HDM‡ | ||||||||||
| Overall | 34.3 | NA | NA | 58.5 | NA | NA | ||||
| Ile/Ile | 43.9 | 1 | NA | 65.2 | 1 | NA | ||||
| Ile/Val | 27.3 | 1.60 | 0.95-2.69 | 52.3 | 1.71 | 0.93-3.14 | ||||
| Val/Val | 29.4 | 1.60 | 0.81-3.17 | 49.1 | 1.76 | 0.78-3.98 | ||||
. | Progression-free survival . | . | . | Overall survival . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | Median survival, mo . | Adjusted hazard ratio* . | 95% CI . | Median survival, mo . | Adjusted hazard ratio* . | 95% CI . | ||||
| ABCM† | ||||||||||
| Overall | 21.0 | NA | NA | 44.7 | NA | NA | ||||
| Ile/Ile | 16.6 | 1 | NA | 39.3 | 1 | NA | ||||
| Ile/Val | 21.6 | 0.55 | 0.32-0.96 | 51.8 | 0.65 | 0.34-1.25 | ||||
| Val/Val | 27.8 | 0.52 | 0.28-0.96 | 48.5 | 0.78 | 0.39-1.58 | ||||
| CVAMP/HDM‡ | ||||||||||
| Overall | 34.3 | NA | NA | 58.5 | NA | NA | ||||
| Ile/Ile | 43.9 | 1 | NA | 65.2 | 1 | NA | ||||
| Ile/Val | 27.3 | 1.60 | 0.95-2.69 | 52.3 | 1.71 | 0.93-3.14 | ||||
| Val/Val | 29.4 | 1.60 | 0.81-3.17 | 49.1 | 1.76 | 0.78-3.98 | ||||
NA indicates not applicable.
Hazard ratios are adjusted for age, sex, and levels of hemoglobin, creatinine, and B2M.
P = .04 for adjusted hazard ratio for ABCM PFS and OS.
P = .16 for adjusted hazard ratio for CVAMP/HDM PFS and OS.
Furthermore, in multivariate analysis, taking into account the levels of B2M, hemoglobin, and creatinine did not affect the significance of the result. In support of this finding, we found an improved OS in this arm for patients with at least one valine allele, although this finding did not reach statistical significance (compared with Ile/Ile, adjusted hazard ratios for death were 0.65 [95% CI, 0.34-1.25] for Ile/Val and 0.78 [95% CI, 0.39-1.58] for Val/Val; P = .40 for trend) (Table 3). We found no effect of genotype on toxicity, as defined by number of course delays or length of delay in patients in this arm (number of courses delayed as a proportion of total courses given were 58 [25%] of 228 for Ile/Ile, 34 [20%] of 168 for Ile/Val, and 27 [31%] of 87 for Val/Val; P = .33).
Of patients randomized to the high-dose arm, 84% went on to receive a transplant. There was no significant difference in the likelihood of receiving a transplant between the 3 genotypic groups, nor was any effect of genotype on response seen (Table 2). No effect of genotype on PFS or OS was seen on either an intention-to-treat basis or when treatment actually received was analyzed for patients in this arm (for PFS on an intention-to-treat basis, compared with Ile/Ile, adjusted hazard ratios were 1.60 [95% CI, 0.95-2.69] for Ile/Val and 1.60 [95% CI, 0.81-3.17] for Val/Val; for OS, adjusted hazard ratios were 1.71 [95% CI, 0.94-3.14] and 1.76 [95% CI, 0.78-3.98], respectively) (Table 3).
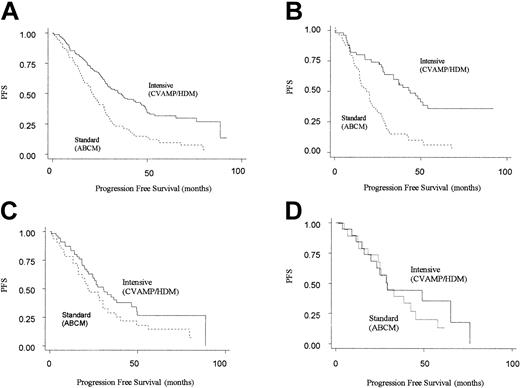
The next step in the analysis was to consider how the 2 different treatment regimens interacted with genotype. Patients randomized to CVAMP/HDM had an improved progression-free and overall survival compared with those in the ABCM arm, in keeping with the results of the MRC myeloma 7 trial as a whole (Table 3).2 However, an improved PFS for CVAMP/HDM compared with ABCM was seen only in patients homozygous for the GSTP1 105Ile allele (adjusted hazard ratio for PFS was 0.29 [95% CI, 0.18-0.49]). No difference in outcome was seen for the other 2 genotype groups. (For CVAMP/HDM compared with ABCM, adjusted hazard ratios for PFS are 0.76 [95% CI, 0.43-1.40] and 0.64 [0.25-1.63] for Ile/Val and Val/Val, respectively) (Table 4; Figure 2).
Hazard ratios for progression-free survival and overall survival within genotypic groups for patients treated with CVAMP/HDM compared with ABCM
. | Progression-free survival . | . | Overall survival . | . | ||
|---|---|---|---|---|---|---|
| Genotype . | Adjusted hazard ratio*† . | 95% CI . | Adjusted hazard ratio*† . | 95% CI . | ||
| Ile/Ile | 0.29 | 0.18-0.49 | 0.40 | 0.23-0.72 | ||
| Ile/Val | 0.76 | 0.43-1.40 | 1.00 | 0.52-1.95 | ||
| Val/Val | 0.64 | 0.25-1.63 | 0.67 | 0.20-2.16 | ||
. | Progression-free survival . | . | Overall survival . | . | ||
|---|---|---|---|---|---|---|
| Genotype . | Adjusted hazard ratio*† . | 95% CI . | Adjusted hazard ratio*† . | 95% CI . | ||
| Ile/Ile | 0.29 | 0.18-0.49 | 0.40 | 0.23-0.72 | ||
| Ile/Val | 0.76 | 0.43-1.40 | 1.00 | 0.52-1.95 | ||
| Val/Val | 0.64 | 0.25-1.63 | 0.67 | 0.20-2.16 | ||
Hazard ratios are for PFS and OS comparing outcome following CVAMP/HDM with ABCM and are adjusted for age, sex, and levels of hemoglobin, creatinine, and B2M.
P = .008.
P = .064.
Kaplan-Meier plots for progression-free survival on an intention-to-treat basis by arm. Solid line represents patients randomized to intensive (CVAMP/HDM) arm, and the dotted line represents patients randomized to standard-dose arm (ABCM). (A) All patients (n = 222). (B) GSTP1 105Ile homozygotes (n = 98). (C) GSTP1 105Ile/105Val heterozygotes (n = 86). (D) GSTP1 105Val homozygotes (n = 38).
Kaplan-Meier plots for progression-free survival on an intention-to-treat basis by arm. Solid line represents patients randomized to intensive (CVAMP/HDM) arm, and the dotted line represents patients randomized to standard-dose arm (ABCM). (A) All patients (n = 222). (B) GSTP1 105Ile homozygotes (n = 98). (C) GSTP1 105Ile/105Val heterozygotes (n = 86). (D) GSTP1 105Val homozygotes (n = 38).
Through a test of interaction between the effects of treatment arm and genotype on PFS, this difference in response between the 3 genotypes was found to be significant (P = .008). A similar though nonsignificant pattern was seen for overall survival (adjusted hazard ratios for OS for Ile/Ile, Ile/Val and Val/Val genotypes are 0.40 [95% CI, 0.23-0.72], 1.00 [95% CI, 0.52-1.93], and 0.67 [95% CI, 0.20-2.16]; P = .064 for interaction).
There was no association of the GSTP1 114 polymorphism with outcome in this patient sample (data not shown).
Discussion
Polymorphisms regulating the expression of tumor necrosis factor-α (TNF-α) have previously been shown to modulate survival following therapy for myeloma; however, like other standard prognostic markers, they are unable to discriminate between the effects of different treatment regimens.36,37 We have shown that GSTP1 105 genotype is an independent prognostic marker for PFS in patients with myeloma younger than 65 years treated with ABCM. This is further supported by a trend to improved overall survival for patients in this arm with 1 or 2 GSTP1 105Val-encoding alleles. This finding is consistent with the hypothesis that patients with the GSTP1 105Val variant enzyme have a reduced ability to detoxify chemotherapeutic agents, thereby increasing the effective dose of the drug within the cell. The trend to an increased chance of entering plateau and a greater likelihood that plateau is reached in a shorter time in patients with GSTP1 105Val further supports this hypothesis.
Differences in outcome for either progression-free or overall survival are not seen by genotype in patients treated with CVAMP/HDM. Of patients randomized to this arm, 14% did not receive a transplant, and this may affect the results; however, there was no effect of genotype on the likelihood of receiving a transplant. Furthermore, analysis on a treatment-received basis did not alter the overall findings in this arm (data not shown). The reasons for the differences in effect of GSTP1 polymorphisms on outcome in the 2 arms are unknown but may be due to the capacity for high-dose therapy to overcome functional differences between the genotypes or to differences in GSTP1 specificity for the chemotherapeutic agents used in each arm.
When differences in outcome between arms are analyzed by genotype, there is a marked improvement in progression-free and overall survival for patients homozygous for the GSTP1 105Ile wild-type enzyme when treated with CVAMP/HDM compared with ABCM. This treatment effect is greater than that seen within the other genotype groups, thus further supporting the hypothesis that patients with GSTP1 105Ile are effectively being underdosed by ABCM, and potentially identifying a group of patients (those homozygous for GSTP1 105Ile) who benefit most from high-dose therapy. While most agents used in the Myeloma VII trial are known to be GSTP1 substrates, the specific effect of GSTP1 polymorphisms on the metabolism of these agents is unknown, and further investigation is required.
These results are compatible with studies in breast and colon cancer showing improvements in survival for patients with GSTP 105Val following chemotherapy with drugs known to be GSTP1 substrates.26,27 Interestingly, one of these studies involved patients treated with oxaliplatin. Recent evidence has suggested that it is the GSTP1 105Val variant enzyme that has the greater cytoprotective effect against platinum-containing agents.23 Thus, there is the possibility that the survival differences observed between the variants may not be due solely to polymorphic functional differences in drug metabolism. The effect of having 1 or 2 underactive alleles on outcome is unknown. Our study and that of Stoehlmacher et al27 into colon cancer showed a survival advantage for patients with 1 or 2 alleles; however, the study by Sweeney et al26 into breast cancer showed an advantage only in patients homozygous for the variant allele. These differences may be related to the different diagnoses or to the different chemotherapy regimens used.
The detoxification of chemotherapeutic agents by the GSTP1 pathway is determined not only by functional GSTP1 polymorphisms but also by expression levels of the enzyme and a requirement for adequate levels of reduced glutathione. Furthermore, adequate cellular efflux of glutathione conjugates, which themselves may be toxic to cells, is required.38,39 Expression of GSTP1 shows wide interindividual variation, and thus high expression of an underactive variant may overcome functional differences owing to polymorphic change.40 Coordinated expression of GSTP1 in conjunction with multidrug resistance protein 1 (MRP1) (the main efflux pump involved in the removal of glutathione conjugates) and gamma-glutamate cysteine ligase (the rate-limiting enzyme in the production of reduced glutathione) has been shown to increase the detoxification of xenobiotic agents.17 Whether polymorphic variation in GSTP1 alters expression of MRP1 is unknown. However, transfection studies comparing allelic variants of the human GSTP1 gene have shown no difference in GSTP1 expression or levels of reduced glutathione.23 GSTP1 is also involved in the detoxification of reactive oxygen species,7 which may act as intermediaries in the cytotoxicity of many chemotherapeutic agents, and thus may have an effect on response to a specific drug even when the chemotherapeutic agent itself is not a substrate. A further function of GSTP1 is the inhibition of Jun N-terminal kinase (JNK) signaling, which may affect tumor cell biology.41 However, the effect of polymorphic variation on this function is unknown.
It is possible that the effects on outcome we have seen with the GSTP1 polymorphism are due to associated changes at other loci. The GSTP1 105Val variant has recently been shown to associate with hypermethylation of the promoter regions of a cyclin-dependent kinase inhibitor, P16ink4a, a tumor suppressor gene, and O6-methylguanine methyltransferase (O6MGMT), a DNA repair protein.42 However, DNA repair by O6MGMT is unlikely to play a major role in drug resistance in this group of patients as most drugs used in this study cause lesions that are not substrates for this DNA repair pathway. Loss of function of P16ink4a has been shown to be associated with an increased risk of developing malignancy, but studies on outcome in myeloma have shown conflicting results, with one study suggesting an improved progression-free and overall survival for patients with the unmethylated gene whereas another study reported no such difference.43,44
An increased risk of developing multiple myeloma has been suggested following exposure to a number of environmental agents such as benzene, dioxins, pesticides, tobacco smoke, and hair dyes.45 Many of these are known to be, or to contain, carcinogens that are substrates for GSTP1, including polyaromatic hydrocarbons. We found no association between GSTP1 105 or 114 genotype and an increased risk of myeloma.
Thus, the effect of GSTP1 genotype on outcome that we have seen may be due to the direct functional effect of the polymorphism on drug metabolism or due to associated changes in related enzymes. Further work is required to understand the exact mechanisms behind these differences. Furthermore, we have found no association with GSTP1 genotype and risk of toxicity in patients receiving ABCM, although only data on course delays were available to evaluate this. However, in support of the above hypothesis, an increased risk of secondary leukemia following chemotherapy in patients with the underactive allele has been shown.28 Further clarification of the effect of GSTP1 genotype and risk of toxicity following chemotherapy is required. However, our findings, along with those in breast and colon cancer, show polymorphic variation in GSTP1 to be a significant predictor of outcome following treatment with a wide range of chemotherapeutic agents and may be a step in the development of more individualized treatment regimens for myeloma based on host genetic factors.
Prepublished online as Blood First Edition Paper, June 5, 2003; DOI 10.1182/blood-2003-02-0444.
Supported by the Leukemia Research Fund and Yorkshire Cancer Research. F.E.D. is supported by the Department of Health, United Kingdom.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to acknowledge the help and support of the Medical Research Council Adult Leukaemia Working Party and the Northern and Yorkshire Clinical Trials and Research Unit.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal