Abstract
The HIV-1 gene nef is important for progression toward AIDS and cellular depletion of the infected thymus. Expression of the Nef protein alone impairs human thymopoiesis. Here, we performed a structure-function analysis of the Nef protein by comparing the effect on T-cell development of different nef alleles, either wild type or defective for selected functions, expressed by human thymocytes. We show that Nef-mediated impaired thymopoiesis is not due to altered surface marker trafficking, nor dependent on oligomerization of Nef. By contrast, mutations in the myristoylation site and in signaling sites of Nef, ie, sites important for interaction with phosphofurin acidic cluster sorting protein-1 (PACS-1), Src homology domain 3 (SH3) domains, and p21-activated kinase 2 (PAK2), were found to be critical for its effect on T-cell development. These results point to sites in Nef to target therapeutically for restoration of thymopoiesis in HIV infection.
Introduction
At the end of 2002, approximately 42 million people were infected with HIV worldwide.1 HIV infection is associated with a progressive loss of CD4+ T cells, leading to an acquired immunodeficiency syndrome (AIDS). The CD4+ T-cell depletion is due to the direct destruction of infected CD4+ T cells, as well as to an impaired production of T cells.2 De novo generation of naive human T cells occurs in the thymus, seeded with CD34+ progenitor cells, migrating from the bone marrow. These CD3–CD8–CD4– triple-negative thymocytes differentiate into immature single-positive CD3–CD8–CD4+ (ISP4+) cells and in a next step into CD4+CD8+ double-positive (DP) cells. During these early developmental stages, T-cell receptor (TCR) rearrangements occur, and the TCR is expressed on the cell surface. Generation and survival of these DP cells and ultimately differentiation into functionally mature CD4+ and CD8+ single-positive (SP) cells requires positive selection, which is mediated by major histocompatibility complex (MHC)–specific TCR interactions. Thymocytes expressing potentially self-reactive TCRs die by apoptosis (negative selection).3,4 Several groups provided evidence that HIV can infect the thymus and compromise its function, which results in a decreased thymic output, contributing to the general decline in peripheral CD4+ T-cell numbers.5-8 Thymocytes at various developmental stages are susceptible to HIV-1 infection.5,6,9,10
Several studies show Nef to be an important determinant of AIDS pathogenesis. Some long-term nonprogressors were found to carry HIV strains encoding a modified or deleted nef gene.11,12 Experiments using the human fetal thymus/liver transplants in severe combined immunodeficiency mice (SCID-hu) model demonstrate that nef-deleted HIV-1 causes lower levels of infection and pathogenicity, although thymocyte depletion is still induced.13-15 In the same model and the in vitro system fetal thymic organ culture (FTOC) Nef functions as an independent pathogenic factor, causing CD4+ T-cell depletion in the human thymus, in the absence of replication activity.16 In addition, in transgenic (Tg) mice, Nef expression in T cells leads to immunodeficiency, depletion of CD4+ thymocytes and peripheral T cells, and altered T-cell activation responses.17-20
The 27-kDa Nef protein is posttranslationally modified by phosphorylation and myristoylation, which targets Nef to cellular membranes. Structurally, Nef consists of a flexible N-terminal arm (residues 1-70), followed by a well-conserved and stably folded core domain (residues 71-203), containing a 30–amino acid disordered loop (residues 148-178) bulging out of the core.21 Oligomerization of Nef has been demonstrated and is thought to be crucial for function of Nef.22 Nef has multiple and distinct functions: it down-regulates cell surface receptors, eg, CD423 and MHC class I,24 interferes with signal transduction pathways, and enhances virion infectivity and viral production.25 Mutational analysis revealed that, at least in cell lines, the multiple actions of Nef have structural correlates. In general, down-regulation of CD4 requires conserved amino acid residues located in the N-terminal arm and the disordered loop of Nef, whereas down-regulation of MHC class I involves a cluster of acidic amino acid residues, as well as proline residues that are part of the Src homology domain 3 (SH3) binding surface in the core domain of the Nef protein. A large number of cellular partners have been identified, and for some of these proteins the binding sites in Nef have been mapped (reviewed by Geyer et al26 ).
Although over the past decade a wealth of information has been generated on the HIV protein Nef, no data exist on the mechanism that Nef uses to induce human T-cell depletion in the HIV-infected thymus. Here, we analyzed the capacity of a panel of mutants of different nef alleles and an inducible nef, expressed by retroviral gene transfer, to disturb human T-cell development. Our data show that mutations in the myristoylation site, the acidic cluster, the polyproline helix II, and in elements important for the association of Nef with p21-activated kinase 2 (PAK2) abolished the nef-induced impaired T-cell development. By contrast, Nef-mediated down-regulation of CD4, CD8β, or MHC class I does not have a gross effect on thymocyte generation in our system.
Materials and methods
Plasmid constructions
All plasmid constructs were made into the retroviral vector LZRS-IRES-EGFP as described before.16 The nef genes used in this study are listed in Table 1: NL4-3 wild type (WT), kindly provided by the AIDS Research and Reference Reagent Program (NIH, Bethesda, MD); NL4-3 mutants, kindly provided by W. Greene (Gladstone Institute of Virology and Immunology, San Francisco, CA); LAI WT and mutants, kindly provided by Y. Collette (Experimental Oncology Laboratory INSERM U119, Marseille, France), O. Schwartz (Institut Pasteur, Paris, France) and R. Benarous (Institut Cochin de Génétique Moléculaire INSERM U529, Paris, France). The double mutants NA-7DAEDAA and NA-7DALLAA were made by introducing a point mutation (Asp86 to Ala) by polymerase chain reaction (PCR) overlap extension mutagenesis using NA-7EDAA and NA-7LLAA, respectively, as DNA template for the primary PCR reactions. The SfcI-digested PCR fragments were ligated together. Enzymes used were from Roche Diagnostics (Penzberg, Germany), except SfcI was from New England Biolabs (Beverly, MA).
Mutated nef alleles and respective effects on CD4/MHC class I down-regulation, based on currently available data
Allele . | Mutation . | Amino acid position . | Affected domain (function) . | CD4* . | MHC-1* . | Reference . |
|---|---|---|---|---|---|---|
| NA-7 | WT | None | None | + | + | Schwartz et al,24 |
| NL4-3 | Mariani and Skowronski27 | |||||
| LAI | ||||||
| LAI | G2A | Gly2 | Myristoylation (membrane anchoring) | +/- | - | Aiken et al28 |
| NL4-3 | WLAA | Trp-Leu57-58 | CD4 binding site, HIV-1 protease cleavage site | - | + | Mangasarian et al29 |
| NL4-3 | E4A | Glu-Glu-Glu-Glu62-63-64-65 | PACS-1 binding site | + | - | Piguet et al30 |
| NA-7 | PPAA | Pro72, Pro75 | Polyproline helix type II (SH3 domain binding surface, interaction with Vav) | + | +/- | Saksela et al,31 |
| LAI | Greenberg et al32 | |||||
| NA-7 | D86A | Asp86 | SH3 binding site | + | + | Greenberg et al,33 |
| Iafrate et al34 | ||||||
| NL4-3 | R106A | Arg106 | PAK2 association | + | +/- | Iafrate et al,34 Sawai et al,35 present study |
| LAI | D123G | Asp123 | Oligomerization | - | - | Liu et al36 |
| NA-7 | LLAA | Leu-Leu164-165 | Clathrin adaptor interaction site | - | + | Greenberg et al,32 |
| LAI | LLGG | Bresnahan et al,37 | ||||
| Craig et al38 | ||||||
| NA-7 | DALLAA | Asp86, Leu-Leu164-165 | SH3 binding site, clathrin adaptor interaction site | - | + | Present study |
| NA-7 | EDAA | Glu-Asp174-175 | Vacuolar ATPase interaction site | - | + | Iafrate et al,34 |
| LAI | DDGA | Asp-Asp 174-175 | Aiken et al39 | |||
| NA-7 | DAEDAA | Asp86, Glu-Asp174, 175 | SH3 binding site, vacuolar ATPase interaction site | - | + | Present study |
Allele . | Mutation . | Amino acid position . | Affected domain (function) . | CD4* . | MHC-1* . | Reference . |
|---|---|---|---|---|---|---|
| NA-7 | WT | None | None | + | + | Schwartz et al,24 |
| NL4-3 | Mariani and Skowronski27 | |||||
| LAI | ||||||
| LAI | G2A | Gly2 | Myristoylation (membrane anchoring) | +/- | - | Aiken et al28 |
| NL4-3 | WLAA | Trp-Leu57-58 | CD4 binding site, HIV-1 protease cleavage site | - | + | Mangasarian et al29 |
| NL4-3 | E4A | Glu-Glu-Glu-Glu62-63-64-65 | PACS-1 binding site | + | - | Piguet et al30 |
| NA-7 | PPAA | Pro72, Pro75 | Polyproline helix type II (SH3 domain binding surface, interaction with Vav) | + | +/- | Saksela et al,31 |
| LAI | Greenberg et al32 | |||||
| NA-7 | D86A | Asp86 | SH3 binding site | + | + | Greenberg et al,33 |
| Iafrate et al34 | ||||||
| NL4-3 | R106A | Arg106 | PAK2 association | + | +/- | Iafrate et al,34 Sawai et al,35 present study |
| LAI | D123G | Asp123 | Oligomerization | - | - | Liu et al36 |
| NA-7 | LLAA | Leu-Leu164-165 | Clathrin adaptor interaction site | - | + | Greenberg et al,32 |
| LAI | LLGG | Bresnahan et al,37 | ||||
| Craig et al38 | ||||||
| NA-7 | DALLAA | Asp86, Leu-Leu164-165 | SH3 binding site, clathrin adaptor interaction site | - | + | Present study |
| NA-7 | EDAA | Glu-Asp174-175 | Vacuolar ATPase interaction site | - | + | Iafrate et al,34 |
| LAI | DDGA | Asp-Asp 174-175 | Aiken et al39 | |||
| NA-7 | DAEDAA | Asp86, Glu-Asp174, 175 | SH3 binding site, vacuolar ATPase interaction site | - | + | Present study |
PACS-1 indicates phosphofurin acidic cluster sorting protein-1; PAK2, p21-activated kinase 2; ATPase, adenosine triphosphatase.
+ indicates down-regulation; +/-, partial down-regulation; and -, no down-regulation.
Direct sequencing (ABI, Foster City, CA) confirmed the integrity of the constructs and nef genes.
Production of retroviral supernatant
The Phoenix-Amphotropic packaging cell line was transfected with the LZRS-IRES-EGFP and the LZRS-(insert)-IRES-EGFP plasmids to produce retroviral supernatant as described before.40
Monoclonal antibodies, flow cytometry, and cell sorting
Mouse antihuman monoclonal antibodies (mAbs) used were CD8α (OKT8 fluorescein isothiocyanate [FITC]–conjugated; American Type Culture Collection [Manassas, VA] or SK1 phycoerythrin [PE] or allophycocyanin [APC]; Becton Dickinson, Erembodegem, Belgium), CD8β (2ST8.5H7 PE; Coulter, Miami, FL), HLA-A, -B, -C (G46-2.6 PE; Becton Dickinson) and as described previously.40 Negative controls included isotype mAbs conjugated with the corresponding fluorochrome. The cells were analyzed by flow cytometry as described before.41 Cell sorting was done on a FACSVantage (Becton Dickinson).
Cell culture and purification of progenitor cells
SupT1 (AIDS Research and Reference Reagent Program, NIH, Bethesda, MD) and other cells were cultured in Iscove modified Dulbecco medium (IMDM), supplemented with penicillin (100 IU/mL), streptomycin (100 μg/mL), and 10% heat-inactivated fetal calf serum (complete IMDM, Invitrogen, Merelbeke, Belgium).
Child thymus tissue, removed during cardiac surgery, was obtained and used following the guidelines of the Medical Ethical Commission of Ghent University Hospital. Informed consent was provided according to the Declaration of Helsinki. CD34+ thymus cells were isolated by positive selection using super-paramagnetic MicroBeads (MACS; Miltenyi Biotec, Bergisch Gladbach, Germany). Isolated CD34+ thymocytes were prestimulated for 24 hours in complete IMDM supplemented with stem cell factor (10 ng/mL; R&D Systems, Abingdon, United Kingdom) and interleukin-7 (10 ng/mL; R&D Systems) prior to transduction.
Retroviral gene transfer
For transduction of cell lines, cells were mixed with retroviral supernatant, preincubated for 10 minutes with Dotap (Roche Diagnostics). To increase transduction efficiency, cells were spun (90 minutes, 950g, 32°C).42 Purified, precultured CD34+ cells were transduced as reported previously.16 Transduction efficiency was evaluated by flow cytometry 48 hours after transduction. Expression of Nef protein was verified by Western blotting (Figure S1 on the Blood website; see the Supplemental Figure link at the top of the online article).
FTOC and thymocyte generation ratio
Isolation of thymic lobes from fetal nonobese diabetic (NOD)–SCID mice, hanging drops, and FTOC were performed as described previously.40,41 During hanging drop each lobe was placed in culture with 5 to 10 × 103 cells, transduced 24 hours earlier. The excess of transduced progenitor cells, which were not used for hanging drop, were kept in culture for another 24 hours, to determine the transduction efficiency (varied between 10% and 20%). In the inducible system 1 μM 4-hydroxytamoxifen (4-HT) (Sigma, St Louis, MO) was added to the culture medium. After culturing for 22 days (for CD34+) or 15 days (for ISP4+), the lobes were analyzed. To assess thymic depletion because of Nef expression, we calculated the thymocyte generation ratio; ie, the ratio of the percentage of EGFP+ thymocytes harvested to the percentage of EGFP+ progenitors (varied between 10% and 20%) that were put in FTOC.
Statistical analysis
To test whether the difference between means was significant, the one-way analysis of variance (ANOVA) test with Bonferroni correction was used.
Results
Disturbance of human T-cell development is a conserved property of HIV-1 Nef
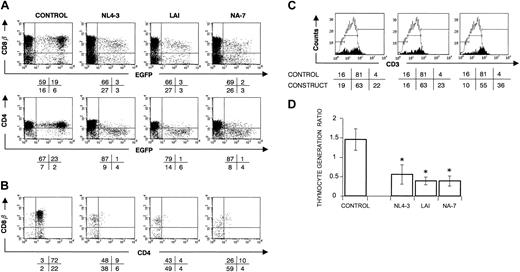
We previously reported that T-cell precursors, expressing HIV-1 NL4-3 nef after retroviral gene transfer, were impaired in generating T cells.16 To determine whether this Nef-mediated T-cell depletion was also found with other alleles, 2 additional HIV-1 nef alleles, LAI and NA-7, were introduced into sorted CD34+ thymocytes by retroviral gene transfer with bicistronic expression of Nef and EGFP (enhanced green fluorescent protein). T-cell development from the transduced progenitor cells was evaluated using FTOC. As a control, we included a control virus, only encoding for EGFP. As EGFP expression does not hamper T-cell development,41 CD34+ cells, retrovirally transduced with the control virus, developed after 3 weeks of FTOC into DP cells with comparable surface marker expression levels of CD4, CD8β (Figure 1A-B), and TCR-associated CD3 as nontransduced cells. In contrast, both nef alleles LAI and NA-7 resulted in a similar reduced CD4 and CD8β surface expression level as was observed with NL4-3 (Figure 1A-B). Thymocytes expressing higher Nef levels, as measured by the EGFP level, were skewed toward lower CD4 and CD8β levels (Figure 1A) and higher CD3 levels (Figure 1C) and showed MHC class I down-regulation (data not shown). Constructs expressing WT Nef protein generated a lower number of thymocytes. As shown in Figure 1D, all Nef WT constructs showed a mean thymocyte generation ratio, which differed significantly (P < .05) from the control.
nef WT-transduced CD34+ progenitors show a strongly impaired T-cell development in FTOC. Bivariate dot plots of flow cytometric measurement of Nef– (control) and Nef+ (NL4-3, LAI, NA-7) transduced cells. (A) CD8β-PE, CD4-APC versus EGFP expression of transduced CD34+ thymocytes at day 22 of FTOC. (B) CD8β-PE versus CD4-APC expression from thymocytes shown in panel A, gated on EGFP+ cells. Quadrants were set to include 99% of cells stained with isotypic controls and EGFP– cells in lower left quadrant. Values indicate percentage (rounded off to a whole number) of cells present in corresponding quadrants. (C) Filled histograms show the CD3-APC expression profile of EGFP+ thymocytes. For reference, the staining profile of the control is overlaid (open histogram). Gates are placed arbitrarily, and the numbers indicate the percentage of cells within the respective gates, although the amount of CD3high cells is often very low (< 100 events). (D) Thymic generation ratio was calculated as indicated in “Materials and methods.” The figure shows mean values of the thymic generation ratio and their standard deviations calculated from the data generated in at least 3 independent experiments. An asterisk (*) over a bar indicates a statistically significant difference between the experimental construct and control data sets (P < .05).
nef WT-transduced CD34+ progenitors show a strongly impaired T-cell development in FTOC. Bivariate dot plots of flow cytometric measurement of Nef– (control) and Nef+ (NL4-3, LAI, NA-7) transduced cells. (A) CD8β-PE, CD4-APC versus EGFP expression of transduced CD34+ thymocytes at day 22 of FTOC. (B) CD8β-PE versus CD4-APC expression from thymocytes shown in panel A, gated on EGFP+ cells. Quadrants were set to include 99% of cells stained with isotypic controls and EGFP– cells in lower left quadrant. Values indicate percentage (rounded off to a whole number) of cells present in corresponding quadrants. (C) Filled histograms show the CD3-APC expression profile of EGFP+ thymocytes. For reference, the staining profile of the control is overlaid (open histogram). Gates are placed arbitrarily, and the numbers indicate the percentage of cells within the respective gates, although the amount of CD3high cells is often very low (< 100 events). (D) Thymic generation ratio was calculated as indicated in “Materials and methods.” The figure shows mean values of the thymic generation ratio and their standard deviations calculated from the data generated in at least 3 independent experiments. An asterisk (*) over a bar indicates a statistically significant difference between the experimental construct and control data sets (P < .05).
As immature CD4 single-positive cells (CD4+CD3–CD8–; ISP4+) are one of the most immature thymocytes susceptible to HIV infection,6 we also analyzed development of nef-transduced ISP4+ cells in FTOC (see also Verhasselt et al16 ). As similar Nef-induced disturbances in phenotype and cellularity of the thymus were observed with CD34+ and ISP4+ cells (data not shown), we continued our experiments with the more easily transducible CD34+ population.
Nef-induced CD4 and CD8β down-modulation alone does not explain disturbed T-cell development
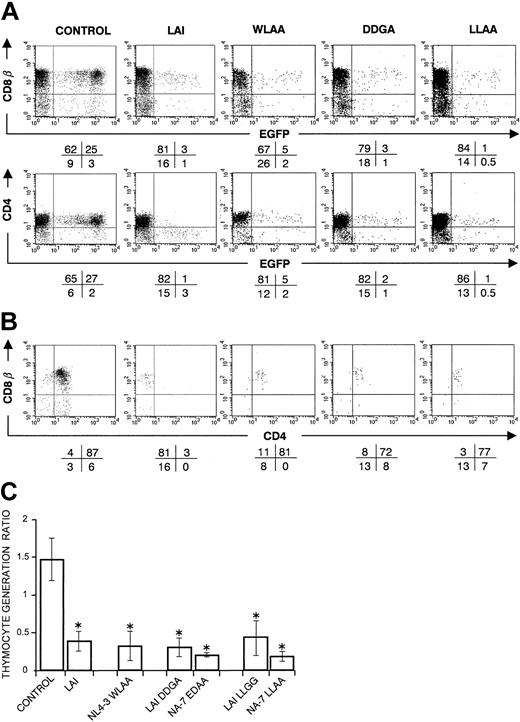
To investigate the importance of Nef-mediated CD4 and CD8β down-modulation in disturbance of T-cell development, we performed experiments with Nef proteins mutated in elements that are important for CD4 down-modulation. Thymocytes retrovirally transduced with the Nef mutants WLAA, DDGA/EDAA, or LLGG/LLAA (Table 1), lacked both CD4 and CD8β down-regulation (Figure 2A,) but still showed down-regulated MHC class I surface expression (data not shown). Accordingly, mutant Nef+ thymocytes showed the normal CD4+CD8+ phenotype (Figure 2B). However, after 3 weeks of FTOC, thymocyte generation was strongly impaired by these mutants with a ratio comparable to that seen with WT Nef (Figure 2C). This shows that down-regulation of CD4 and CD8β does not explain per se the impaired T-cell development, and other functions of Nef are needed for its interference in thymopoiesis.
CD34+ progenitors transduced with nef, mutated in regions important for CD4/CD8β down-regulation, still show an impaired T-cell development in FTOC. Bivariate dot plots of flow cytometric measurement of control, Nef WT (LAI), and mutant Nef (WLAA, DDGA, LLAA) transduced cells. LAI WT is representative for the other Nef WT constructs. (A) CD8β-PE, CD4-APC versus EGFP expression of transduced thymocytes at day 22 of FTOC. (B) CD8β-PE versus CD4-APC expression from thymocytes shown in panel A, gated on EGFP+ cells. Quadrants were set to include 99% of cells stained with isotypic controls and EGFP– cells in lower left quadrant. Values indicate percentage (rounded off to a whole number) of cells present in corresponding quadrants (< 100 events are shown in some plots). (C) The figure shows the mean thymocyte generation ratio and standard deviations calculated from the data generated in at least 3 independent experiments. An asterisk (*) over a bar indicates a statistically significant difference between the experimental construct and control data sets (P < .05).
CD34+ progenitors transduced with nef, mutated in regions important for CD4/CD8β down-regulation, still show an impaired T-cell development in FTOC. Bivariate dot plots of flow cytometric measurement of control, Nef WT (LAI), and mutant Nef (WLAA, DDGA, LLAA) transduced cells. LAI WT is representative for the other Nef WT constructs. (A) CD8β-PE, CD4-APC versus EGFP expression of transduced thymocytes at day 22 of FTOC. (B) CD8β-PE versus CD4-APC expression from thymocytes shown in panel A, gated on EGFP+ cells. Quadrants were set to include 99% of cells stained with isotypic controls and EGFP– cells in lower left quadrant. Values indicate percentage (rounded off to a whole number) of cells present in corresponding quadrants (< 100 events are shown in some plots). (C) The figure shows the mean thymocyte generation ratio and standard deviations calculated from the data generated in at least 3 independent experiments. An asterisk (*) over a bar indicates a statistically significant difference between the experimental construct and control data sets (P < .05).
Dimerization of Nef is not essential for Nef-mediated thymic depletion
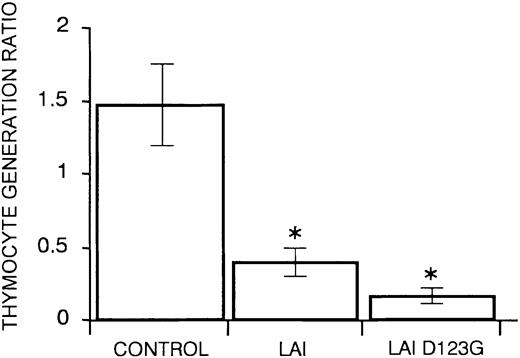
Nef oligomers have been reported by several groups,43,44 but their importance in Nef activities has not been elucidated yet. In the Nef oligomers, Asp123 is interacting with the conserved di-arginine motif Arg105Arg106 of another protomer.22 Mutant D123G, reported to be incapable to oligomerize,36 was tested in FTOC culture. Calculation of the mean thymocyte generation ratio of the D123G mutant showed an impaired T-cell development (Figure 3). This suggests that Nef dimerization is not essential to hamper T-cell development.
CD34+ progenitors transduced with nef, mutated in a region important for dimerization, still show a strongly impaired T-cell development in FTOC. The figure shows mean values of thymocyte generation ratio and their standard deviations calculated from the data generated in at least 3 independent experiments. An asterisk (*) over a bar indicates a statistically significant difference between the experimental construct and control data sets (P < .05).
CD34+ progenitors transduced with nef, mutated in a region important for dimerization, still show a strongly impaired T-cell development in FTOC. The figure shows mean values of thymocyte generation ratio and their standard deviations calculated from the data generated in at least 3 independent experiments. An asterisk (*) over a bar indicates a statistically significant difference between the experimental construct and control data sets (P < .05).
Mutations in the membrane anchor and the core domain of Nef abrogate Nef-mediated impairment of T-cell development
As Nef-mediated T-cell depletion could not solely be explained by CD4/CD8β down-modulation or dimerization, nef genes, mutated in other conserved regions (or amino acid residues), were tested in our model.
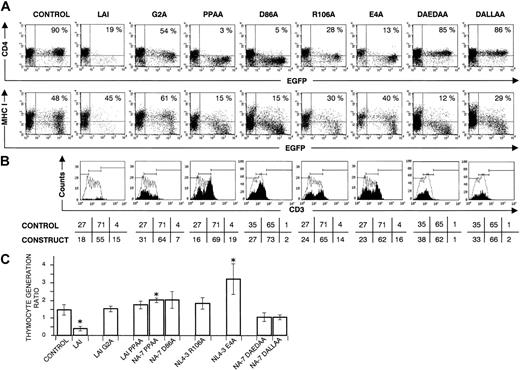
Membrane anchoring. Loss of the myristoylation signal in the mutant G2A resulted in a normal process of thymopoiesis (mean thymocyte generation ratio 1.52) (Figure 4). Similar to observations in cell lines,28 mutation of this residue resulted in only a minor Nef activity (eg, residual CD4 down-regulation in EGFP++ cells).
CD34+ progenitors transduced with nef, mutated in myristoylation site or elements in the core domain, that are important for Nef's interaction with signal transduction machinery, generate normal numbers of thymocytes in FTOC. Bivariate dot plots of flow cytometric measurement of Nef– (control), Nef WT (LAI), and mutant Nef (G2A, PPAA, D86A, R106A, E4A, DAEDAA, DALLAA) transduced cells. LAI WT is representative for the other Nef WT constructs; few transduced cells were recovered, limiting the accuracy of the percentages shown. (A) CD4-APC and MHC class I–PE versus EGFP expression of transduced thymocytes at day 22 of FTOC. Quadrants were set to include 99% of cells stained with isotypic controls and EGFP– cells in lower left quadrant. Values indicate percentage of EGFP+ cells positive for CD4 and MHC I. Filled histograms in (B) show the CD3-APC expression profile of EGFP+ thymocytes. For reference, the staining profile of the control is overlaid (open histogram). Gates are placed arbitrarily, and the numbers indicate the percentage (rounded to a whole number) of cells within the respective gates, although the amount of CD3high cells is often very low (< 100 events). (C) The figure shows mean thymocyte generation ratio values and their standard deviations calculated from the data generated in at least 3 independent experiments. An asterisk (*) over a bar indicates a statistically significant difference between the experimental construct and control data sets (P < .05).
CD34+ progenitors transduced with nef, mutated in myristoylation site or elements in the core domain, that are important for Nef's interaction with signal transduction machinery, generate normal numbers of thymocytes in FTOC. Bivariate dot plots of flow cytometric measurement of Nef– (control), Nef WT (LAI), and mutant Nef (G2A, PPAA, D86A, R106A, E4A, DAEDAA, DALLAA) transduced cells. LAI WT is representative for the other Nef WT constructs; few transduced cells were recovered, limiting the accuracy of the percentages shown. (A) CD4-APC and MHC class I–PE versus EGFP expression of transduced thymocytes at day 22 of FTOC. Quadrants were set to include 99% of cells stained with isotypic controls and EGFP– cells in lower left quadrant. Values indicate percentage of EGFP+ cells positive for CD4 and MHC I. Filled histograms in (B) show the CD3-APC expression profile of EGFP+ thymocytes. For reference, the staining profile of the control is overlaid (open histogram). Gates are placed arbitrarily, and the numbers indicate the percentage (rounded to a whole number) of cells within the respective gates, although the amount of CD3high cells is often very low (< 100 events). (C) The figure shows mean thymocyte generation ratio values and their standard deviations calculated from the data generated in at least 3 independent experiments. An asterisk (*) over a bar indicates a statistically significant difference between the experimental construct and control data sets (P < .05).
Core domain. The core domain is the only part of Nef that adopts a stable tertiary structure. A conserved motif in this core domain, important for interaction with SH3-containing Src-family tyrosine kinases, is the proline-rich sequence Pro72xxPro75.31 Another element in the core, the di-arginine Arg105Arg106 sequence, is required for the interaction with p21-activated kinase 2.45,46 As reported by Manninen et al,47 both domains contribute to Nef's ability to induce cellular activation in T-cell lines. In our FTOC experiments, mutation of the SH3 binding surface (PPAA and D86A) or the PAK2 interaction element (R106A) of Nef, resulted in an undisturbed T-cell development (Figure 4C), whereas the capacity to down-modulate CD4 (Figure 4A) and CD8β (data not shown) was not abrogated. Together with data mentioned earlier, this demonstrates that CD4 or CD8β down-regulation is not the cause of thymic depletion. Further, also MHC class I down-regulation was maintained, but to a lesser extent (Figure 4A), and CD3 expression levels were comparable to, or even higher than the control (Figure 4B). To determine whether the CD4dim/–CD8β+/dim surface marker phenotype of the harvested thymocytes, transduced with these mutants, was due to down-regulation of surface marker expression and not to a developmental arrest in the CD4dim/–CD8β+/dim stage, double mutants NA-7DAEDAA and NA-7DALLAA were constructed. Both constructs contain the D86A mutation, that disrupts the SH3 domain interaction surface, combined with an additional mutation (EDAA or LLAA) that disrupts the CD4 and CD8β down-regulation in T-cell lines32 and thymocytes (this report). Consequently, these double mutants were impaired in their capacity to bind SH3 domains as well as to down-regulate CD4 (Figure 4A) and CD8β. FTOC experiments revealed a normal mean thymocyte generation ratio compared with the control (Figure 4C). In summary, human T-cell development was not hampered by reduced CD4/CD8β expression, and disturbance of the Nef-mediated cellular activation complex resulted in a restored T-cell development.
MHC I down-regulation. To address this issue, we used a mutant of the acidic amino acid stretch EEEE62-65 of Nef, localized close to the core domain. This Nef motif is reported to play a key role in MHC class I internalization in cell lines, by connecting the cytoplasmic tail of MHC class I with the PACS-1–mediated trans-Golgi network retrieval pathway in the endosomes.30 Mutation of this motif (E4A) completely abolished down-modulation of MHC class I, as evaluated on SupT1 cells. In contrast, the capacity to down-modulate other surface markers was maintained (data not shown). When cultured in FTOC, E4A-mutated Nef+ thymocytes were CD4–CD8low but retained MHC class I expression (Figure 4A). Remarkably, the mean thymocyte generation ratio of this construct was significantly higher as compared with the control (Figure 4C). Importantly, restored thymopoiesis was also seen with the double mutants NA-7DAEDAA and NA-7DALLAA, still strongly down-regulating MHC class I (Figure 4). Together these data demonstrated that MHC class I down-modulation could not explain the observed disturbance in T-cell development induced by Nef.
Induction of Nef expression during T-cell development shows thymocyte depletion at all maturational stages
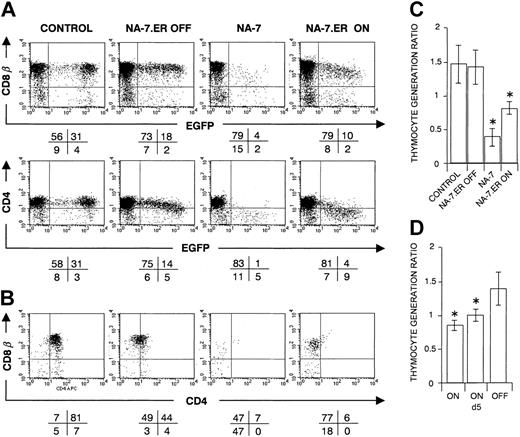
To elucidate the maturational stage at which Nef affects thymic development, we induced Nef expression at different stages of T-cell development. In LZRS-NA-7.ER-I-EGFP–transduced cells, a biologically inactive NA-7.ER (estrogen receptor) fusion protein is expressed together with the EGFP reporter protein. Binding of the membrane-permeable estrogen analog 4-hydroxytamoxifen (4-HT) to the ER relieves a sterical hindrance, which results in the normal activity of Nef NA-7.48 Anti-Nef immunoblotting confirmed the integrity of NA-7.ER (approximately 60 kDa) (Figure S1).
Neither 4-HT without NA-7.ER (data not shown) nor the expression of NA-7.ER in the absence of 4-HT (OFF in Figure 5A-C) affected T-cell development. In the latter condition, a minor down-regulation of CD4, but not of CD8β, was observed. The thymocyte generation ratios of the authentic protein NA-7 and the activated chimeric protein NA-7.ER, switched on at day 0 of FTOC, were significantly lower (P < .05) than the control or switched off NA-7.ER (Figure 5A-C). Together, these results show that both NA-7 and activated NA-7.ER impair thymopoiesis, although not with the same efficacy.
CD34+ progenitors transduced with NA-7.ER in FTOC. Bivariate dot plots of flow cytometric measurement of Nef– (control), NA-7– or NA-7.ER–transduced cells, analyzed at day 22 of FTOC. (A) CD8β-PE, CD4-APC versus EGFP expression of transduced thymocytes at day 22 of FTOC. (B) CD8β-PE versus CD4-APC expression from thymocytes shown in panel A, gated on EGFP+ cells. Quadrants were set to include 99% of cells stained with isotypic controls and EGFP– cells in lower left quadrant. Values indicate percentage (rounded to a whole number) of cells present in corresponding quadrants. (C) Thymocyte generation ratios for the different experimental conditions as indicated and (D) for NA-7.ER, switched on at day 0 or day 5 of organ culture. Panels C and D show mean values, and their standard deviations are calculated from the data generated in at least 3 independent experiments. An asterisk (*) over a bar indicates a statistically significant lower ratio of the experimental construct compared with the control (P < .05).
CD34+ progenitors transduced with NA-7.ER in FTOC. Bivariate dot plots of flow cytometric measurement of Nef– (control), NA-7– or NA-7.ER–transduced cells, analyzed at day 22 of FTOC. (A) CD8β-PE, CD4-APC versus EGFP expression of transduced thymocytes at day 22 of FTOC. (B) CD8β-PE versus CD4-APC expression from thymocytes shown in panel A, gated on EGFP+ cells. Quadrants were set to include 99% of cells stained with isotypic controls and EGFP– cells in lower left quadrant. Values indicate percentage (rounded to a whole number) of cells present in corresponding quadrants. (C) Thymocyte generation ratios for the different experimental conditions as indicated and (D) for NA-7.ER, switched on at day 0 or day 5 of organ culture. Panels C and D show mean values, and their standard deviations are calculated from the data generated in at least 3 independent experiments. An asterisk (*) over a bar indicates a statistically significant lower ratio of the experimental construct compared with the control (P < .05).
To determine at what maturational stage Nef affects human T-cell development, we switched on NA-7.ER after different durations of FTOC. Evaluation of thymocyte generation ratios showed that switching on at day 5, a moment when thymocytes are mainly ISP4+ (data not shown), still resulted in a significantly lower (P < .05) ratio than the off status (Figure 5D). Switching on Nef at later culture days showed a trend of reduced cellularity, but this trend did not reach statistical significance. The median EGFP expression of the harvested EGFP+ cells showed a similar decrease (data not shown), demonstrating that preferentially cells with high NA-7.ER protein levels were affected.
Discussion
Nef expression, in the absence of other HIV genes, hampers thymic T-cell development.16 As Nef has been extensively studied by mutational analysis, some structure-function relationships have been identified in T-cell lines (reviewed by Geyer et al26 ). Here, a structure-function analysis of Nef on human T-cell development was done to determine which functional elements in the Nef protein mediate the disruption of human thymopoiesis.
Besides NL4-3 and LAI nef alleles, both derived from the corresponding HIV-1 laboratory-adapted strains, also the nef allele of NA-7, an HIV-1 isolate derived from an individual infected with HIV-1 who was asymptomatic,27 was included in this study. In our T-cell development experiments, there was no significant difference between the laboratory strain–derived nef alleles (LAI, NL4-3) and the patient strain–derived nef (NA-7). This finding indicates that the capacity of Nef to disrupt T-cell development is a highly conserved property of nef.
The best-known and characterized Nef effect is internalization of the CD4 receptor. Although several theories have been published as to the role of Nef-mediated CD4 down-regulation in replication and pathogenicity of HIV,49-52 the importance of this Nef effect in vivo remains unclear. As reported previously and currently under investigation, Nef also causes down-regulation of the CD8β-chain on the cell surface of human thymocytes, which is in contrast to the rather unaffected expression of the CD8α-chain.16 In accordance with these results, a clearly decreased level of CD8β, but not CD8α, has been observed in peripheral blood lymphocytes of individuals infected with HIV.53 Moreover, several groups have found CD8+ cells to be infected by HIV-1.6,54,55 Whether Nef indeed has a role in HIV-infected CD8+ T cells is currently unknown. The CD4 and CD8 coreceptors, both targeted by Nef, are known to play a role in thymopoiesis. During T-cell development, TCRβ selection and positive and negative selection occur, causing a massive cell death and leaving only a small fraction of newly generated T cells for export to the periphery. In theory, Nef could affect all these processes, as it affects coreceptor surface expression, as well as T-cell signaling. In mice, the strength of signaling by the CD4 and CD8 coreceptor cytoplasmic tails determines the number of thymocytes undergoing positive selection, but not their ultimate CD4/CD8 phenotype.56 Because in our experiments, Nef+-transduced thymocytes showed a decreased expression of CD4, as well as of CD8β, a depletion of Nef+ human thymocytes might be caused by inefficient positive selection. However, inefficient positive selection cannot explain the sparse generation of CD3dim putative CD4+CD8+ cells. Moreover, thymocytes transduced with nef mutants, impaired in their capacity to down-regulate both CD4 and CD8β, but competent in other Nef functions, still hampered T-cell development. In addition, some mutations, retaining the CD4/CD8β down-modulating properties but severely impairing other reported Nef effects, resulted in a restored thymopoiesis. So, although extremely pronounced, the Nef-mediated CD4/CD8β down-regulation does not appear to have a biologic significance in the observed thymic depletion. Furthermore, because the few cells that are generated during FTOC are in part CD3high, we conclude that TCRβ selection is possible to occur in these nef+ thymocytes.
In this context, Stoddart et al57 recently demonstrated by using nef-mutated HIV that some, but not all, Nef mutants that are defective in CD4 down-regulation do not induce thymic depletion in the SCID-hu model, whereas mutants in the polyproline stretch still deplete the thymus. However, as an infectious virus was used in this study and as HIV-1 replication is enhanced by Nef-mediated CD4 down-regulation,52 this model did not allow us to consider the cytopathic effect of Nef as such on the thymus. This limitation can explain our opposite results as to the role of CD4 down-regulation in thymic depletion.
Because neither positive selection nor TCRβ-selection could explain the observed disruption of thymocyte development by Nef, we investigated whether increased apoptosis or early negative selection could be responsible for this Nef effect. Hanna et al20 and others demonstrated in Nef Tg thymus that Nef+ thymocytes were in a state of activation and hyperresponsiveness on TCR triggering.17 Because Nef has been reported to be part of, and to act through, a TCR-associated multiprotein complex,58 one putative cause of thymocyte depletion may be the interference of Nef with the signaling capacity of the TCR-CD3 complex in developing thymocytes. However, in agreement with our previous report, we did not find increased apoptosis in Nef-expressing thymocytes,16 most likely because of fast transition of these cells toward elimination. An immediate toxic effect as a result of protein overexpression in the CD34+ progenitors can be excluded, as switching on the inducible Nef.ER system at day 5 of organ culture also resulted in a significantly lower thymocyte generation ratio. The interaction of Nef with SH3 domains of Src-family tyrosine kinases results in an interference of Nef with signal transduction.31 The Pro72 and Pro75 residues are critical components of the polyproline PPII helix of Nef that make contact with the SH3 domain, whereas the Asp86 residue contacts the SH3 variable loop (RT-loop). This latter interaction determines the specificity of Nef for a subset of Src family SH3 domains and stabilizes the overall interaction.59,60 The SH3 domain-binding function of HIV-1 Nef is also required for association with PAK2.45 Mutating the polyproline helix (PPAA, D86A) resulted in a restored T-cell development in combination with a preserved CD4 and CD8β down-regulation. This finding was corroborated by a recent study using Nef Tg mice, demonstrating that mutation of the SH3 ligand-binding domain of Nef completely abolished the pathogenic potential, although still a partial down-regulation of the CD4 cell surface expression was observed.61
Because CD3 expression is a major marker for maturing thymocytes and T cells, CD3 expression profiles of Nef wild type and mutants were taken into consideration.Although the amount of CD3high cells was often very low, some increases were found. This was shown for the mutants PPAA, R106A, and E4A, which, although not resulting in a decreased cellularity, still did not give a completely normal thymopoiesis, as also illustrated by the generation of a CD4–CD8βlow population. In the progression toward AIDS, the decreased output by the thymus is of great concern. In HIV-infected thymus, as seen in specimens from patients and SCID-hu mice, the hallmark of HIV-induced pathology is the extreme reduction in thymic cellularity. Therefore, we argue that cellularity as read-out is the most relevant evaluation marker of thymopoiesis in the study of Nef effects on human thymocytes.
Besides a TCR-dependent mechanism, also a TCR-independent mechanism could underlie the Nef effect observed in thymocytes. More specifically, different Nef alleles could cooperate with the Ras/mitogen-activated protein kinase (MAPK) pathway to activate the transcription factor nuclear factor of activated T cells (NFAT).62 Membrane association and the SH3-binding function of Nef are required for this. In addition, Manninen et al47 demonstrated that several other residues, not involved in SH3 binding but important for PAK2 association, are required for activation of NFAT by Nef.47 Comparing our results with those of Manninen et al47 revealed that there is a correlation between the Nef domains involved in NFAT activation and those involved in hampered T-cell development. WT Nef, and mutants Leu164Leu165 and Asp174Asp175 induce activation of NFAT47 and disturb T-cell development, whereas mutations affecting membrane association (aa 2 converted), the SH3 binding (aa 72,75 converted), and the PAK2 interaction (aa 106 converted) inactivate both effects. Phorbol ester treatment or the addition of inhibitors (wortmannin, LY294002, 2-APB, cyclosporin A) was difficult to evaluate (data not shown), because the inherent effect of these agents on complex systems, like FTOC, has not been investigated yet or has only been examined in mice.63
In our experiments, the observed disturbance in thymopoiesis by Nef could not be explained by Nef dimerization or by the ability of Nef to down-regulate MHC class I. Down-regulation of the latter receptor requires an acidic amino acid cluster and membrane anchoring, as well as an intact core domain in Nef. In our experiments, mutation of these domains resulted in a restored thymocyte generation. Remarkably, mutating the acidic amino acid cluster resulted in a significantly higher thymic output. However, the intrinsic importance of MHC class I down-regulation was refuted by the FTOC results obtained with our double mutants DAEDAA and DALLAA.
When 4-hydroxytamoxifen was added to the inducible system NA-7.ER, we clearly saw a drop in EGFP expression level intensity in our FTOC, suggesting that preferentially the Nef++ thymocytes disappeared (data not shown). This inducible system also demonstrated that Nef, when switched on at day 5 of FTOC, is able to deplete ISP4+ thymocytes. This finding confirms our current and previous results on ISP4+ cells.
In this thymopoiesis study, we determined by structure-function analysis of Nef which functional elements in the Nef protein are responsible for disturbed thymocyte generation. We show that the property of Nef to down-regulate surface marker expression of CD4, CD8β, and MHC class I has a minor, if any, importance in Nef-mediated impaired T-cell development. We demonstrate that the interference with functional elements in Nef that link to signaling pathways in thymocytes, ie, sites important for interaction with PACS-1, SH3 domains, and PAK2, impairs thymocyte generation. Because the knowledge of these Nef sites may form the basis for therapeutic strategies to restore T-cell development in patients infected with HIV, further research on the exact molecular mechanism is warranted.
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2003-03-0833.
Supported by grants from the Fund for Scientific Research-Flanders (Belgium) and the US Public Health Service (IA-42561). V.S. and C.S. are research assistants and B.V. a Senior Clinical Investigator for the Fund for Scientific Research-Flanders.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank I. Vandewalle, N. De Cabooter, E. Demecheleer, C. De Boever, C. Collier, and M.-J. De Bosscher for technical assistance; the Department of Cardiac Surgery, Ghent University Hospital, for the supply of human thymus; H. Spits and collaborators (The Netherlands Cancer Institute, Amsterdam) for the gift of the IRES-EGFP construct; G. P. Nolan (Stanford University School of Medicine, Stanford, CA) for the gift of the Phoenix-NA packaging cell line and the LZRS vector; O. Schwartz, Y. Collette, W. Greene, R. Benarous, and the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID for the gift of the nef WT and mutant constructs. We also thank J. Hoxie for providing the hybridoma cell line EH-1, T. Loeys for help with statistics, M. De Smedt for expert advice, K. Smits and G. Dhondt for their help, and J. Skowronski for stimulating discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal