Abstract
High hyperdiploidy, common in childhood acute lymphoblastic leukemia (ALL) with a favorable prognosis, is characterized by specific trisomies. Virtually nothing is known about its formation or pathogenetic impact. We evaluated 10 patients with ALL using 38 microsatellite markers mapped to 18 of the 24 human chromosomes to investigate the mechanisms underlying hyperdiploidy and to ascertain the parental origin of the trisomies. Based on the results, doubling of a near-haploid clone and polyploidization with subsequent losses of chromosomes could be excluded. The finding of equal allele dosage for tetrasomy 21 suggests that hyperdiploidy originates in a single aberrant mitosis, though a sequential gain of chromosomes other than 21 in consecutive cell divisions remains a possibility. Our study, the first to address experimentally the parental origin of trisomies in ALL, revealed no preferential duplication of maternally or paternally inherited copies of X, 4, 6, 9, 10, 17, 18, and 21. Trisomy 8 was of paternal origin in 4 of 4 patients (P = .125), and +14 was of maternal origin in 7 of 8 patients (P = .0703). Thus, the present results indicate that imprinting is not pathogenetically important in hyperdiploid childhood ALL, with the possible exception of the observed parental skewness of +8 and +14.
Introduction
Cytogenetic and molecular genetic analyses of childhood acute lymphoblastic leukemia (ALL) are today used routinely for diagnostic and prognostic purposes,1 and acquired chromosomal aberrations are detected in approximately 70% of patients. High hyperdiploidy—that is, more than 51 chromosomes—is the most common cytogenetic abnormality pattern.2-5 The hyperdiploid subgroup is characterized by a nonrandom gain of chromosomes, usually X, 4, 6, 10, 14, 17, 18, and 21—the latter often being tetrasomic3,6 —and has a superior prognosis,1,4 which in part may be explained by a propensity for the leukemic blasts to undergo apoptosis.7-9 Compared with other cytogenetic subtypes of childhood ALL, hyperdiploidy is often associated with good-risk features, such as a CD10+ early pre-B immunophenotype, low leukocyte count, and age between 2 and 10 years.2,10
Whereas the transforming effects of fusion genes, generated by neoplasia-associated balanced structural abnormalities such as inversions and reciprocal translocations, have been well elucidated,11,12 the molecular genetic and pathogenetic consequences of unbalanced changes are still poorly understood.13,14 In fact, in spite of the clinical importance and the frequent occurrence of hyperdiploidy in childhood ALL, next to nothing is known about how this chromosomal pattern arises or about its pathogenetic impact. A subset seems to be formed in a 2-step process by the doubling of a near-haploid set of chromosomes. However, these patients have 2 or 4 copies of each chromosome and lack the characteristic trisomies usually seen in hyperdiploid childhood ALL, and they should probably be classified within the near-haploid cytogenetic subgroup, which has an inferior prognosis.15,16 Alternative mechanisms for the formation of hyperdiploidy include polyploidization with subsequent losses of chromosomes, successive gains of individual chromosomes in consecutive cell divisions, and a simultaneous occurrence of trisomies in a single abnormal mitosis.17 The first of these possibilities would result in loss of heterozygosity (LOH) for one third of the disomic chromosomes, whereas the second would give rise to unequal allele dosages in most tetrasomic chromosomes. The only previous study addressing this issue showed no evidence of LOH in the disomic chromosomes, and parental chromosome 21 alleles were present in equal dosages in 10 of 11 patients with hyperdiploidy with tetrasomy 21, making a simultaneous gain the most likely mechanism.17 Recently, Panzer-Grümayer et al18 investigated the somatic recombination of the immunoglobulin heavy-chain gene at chromosome 14 and showed that the formation of trisomy 14 and, hence, presumably the hyperdiploid pattern was an early event during leukemogenesis, already occurring in utero. Regarding the pathogenetic impact of hyperdiploidy, Haas19 discussed the possibility of imprinting effects related to the parental origin of the duplicated chromosomes, suggesting that the extra copy of a maternally or paternally inherited allele is the transforming agent. This attractive hypothesis has, however, never been tested experimentally. In the present study, we have performed quantitative fluorescent polymerase chain reaction (QF-PCR)20 with a large set of microsatellite markers to investigate the allelic status and to ascertain the parental origin of the trisomic chromosomes in hyperdiploid childhood ALL.
Patients, materials, and methods
Patients
Study participants were 10 patients with hyperdiploid pre-B (HLA-DR+, CD10+, CD19+) childhood ALL, investigated using routine cytogenetics at diagnosis (Table 1). Some of the G-banding findings were confirmed, and a few additional trisomies were detected, using comparative genomic hybridization (CGH), performed as described previously.21 DNA was extracted according to standard methods22 from cultured bone marrow cells, obtained at the time of diagnosis, that had been stored in fixative (methanol/acetic acid, 3:1) for 1 to 9 years at –20°C. The DNA extracted from cells in fixative was, in some cases, fragmented. Although this had no impact on the QF-PCR analyses, the CGH studies were, to some extent, hampered. DNA was also extracted from peripheral blood samples from patients in remission and from their parents. The investigation was approved by the Research Ethics Committee of Lund University; informed consent was provided according to the Declaration of Helsinki.
Clinical and cytogenetic features of 10 patients with hyperdiploid childhood pre-B ALL
Patient . | Sex/age, y . | FAB . | WBC, ×109/L . | Karyotype* . | Gains detected by QF-PCR† . |
|---|---|---|---|---|---|
| 1 | F/1 | L1 | 10 | 56,XX, +X, +4, +6, +8, +9, +14, +16, +17, +19, +21[10]/46,XX[15] | X, 4, 6, 10, 14, 17, 18, 21 |
| 2 | M/2 | L1 | 2 | 52-54,XY, +8, +9, +10, +11, +14, +17, +19, +21[cp13]/46,XY[8] | 6, 10, 14, 17, 21 |
| 3 | F/4 | L1 | 1 | 53-57,XX,del(6)(q23), +17, +19, +20, +21, +3-7mar[cp6]/46,XX[13] 48,XX, +19, +20[cp3]/46,XX[13] (6 weeks after dx; clinical/morphologic remission) | 4, 6, 14, 17, 18, 21 |
| 4 | F/2 | L2 | 17 | 56,XX, +X, +4, +6, +8, +10, +14, +17, +18, +20, +21[33]/46,XX[9] | X, 4, 6, 8, 10, 14, 17, 18, 21 |
| 5 | M/1 | L1/L2 | 3 | 55,XY, +X, +4, +6, +10, +14, +15, +17, +18, +21[4]/46,XY[16] | 4, 6, 10, 14, 17, 18, 21 |
| 6 | M/7 | L2 | 14 | 50-54,XY, +4, +6, +10, +14, +17, +18, +20, +20, +mar[cp 18] | 4, 6, 9, 10, 17, 21 |
| 7 | F/13 | L1 | 4 | 50-53,XX, +4, +6, del(6)(q23)×2, +add(17)(q25), +18, +21, +21, +2-3mar[cp22]/46,XX[5] 46,XX,add(1)(q32)[5]/46,XX[18] (2 years after dx; clinical/morphologic remission) | 4, 14, 17, 18, 21 |
| 8 | M/2 | L1 | 41 | 51-54,XY, +?inv(3), +4, +5, +6, +8, +11, +13, +14, +17, +21[cp 13]/46,XY[9] | 4, 6, 8, 14, 17, 18, 21 |
| 9 | M/2 | L1 | 2 | 55-57,XY, +4, +5, +6, +7, +8, +10, +12, +15, +17 +mar[cp24] | 4, 6, 8, 9, 10, 14, 17, 18, 21 |
| 10 | M/5 | L2 | 2 | 58,XY, +X, +4, +5, +6, +8, +10, +13, +14, +15, +17, +18, +21[12]/46,XY[11] | 4, 6, 8, 10, 17, 21 |
Patient . | Sex/age, y . | FAB . | WBC, ×109/L . | Karyotype* . | Gains detected by QF-PCR† . |
|---|---|---|---|---|---|
| 1 | F/1 | L1 | 10 | 56,XX, +X, +4, +6, +8, +9, +14, +16, +17, +19, +21[10]/46,XX[15] | X, 4, 6, 10, 14, 17, 18, 21 |
| 2 | M/2 | L1 | 2 | 52-54,XY, +8, +9, +10, +11, +14, +17, +19, +21[cp13]/46,XY[8] | 6, 10, 14, 17, 21 |
| 3 | F/4 | L1 | 1 | 53-57,XX,del(6)(q23), +17, +19, +20, +21, +3-7mar[cp6]/46,XX[13] 48,XX, +19, +20[cp3]/46,XX[13] (6 weeks after dx; clinical/morphologic remission) | 4, 6, 14, 17, 18, 21 |
| 4 | F/2 | L2 | 17 | 56,XX, +X, +4, +6, +8, +10, +14, +17, +18, +20, +21[33]/46,XX[9] | X, 4, 6, 8, 10, 14, 17, 18, 21 |
| 5 | M/1 | L1/L2 | 3 | 55,XY, +X, +4, +6, +10, +14, +15, +17, +18, +21[4]/46,XY[16] | 4, 6, 10, 14, 17, 18, 21 |
| 6 | M/7 | L2 | 14 | 50-54,XY, +4, +6, +10, +14, +17, +18, +20, +20, +mar[cp 18] | 4, 6, 9, 10, 17, 21 |
| 7 | F/13 | L1 | 4 | 50-53,XX, +4, +6, del(6)(q23)×2, +add(17)(q25), +18, +21, +21, +2-3mar[cp22]/46,XX[5] 46,XX,add(1)(q32)[5]/46,XX[18] (2 years after dx; clinical/morphologic remission) | 4, 14, 17, 18, 21 |
| 8 | M/2 | L1 | 41 | 51-54,XY, +?inv(3), +4, +5, +6, +8, +11, +13, +14, +17, +21[cp 13]/46,XY[9] | 4, 6, 8, 14, 17, 18, 21 |
| 9 | M/2 | L1 | 2 | 55-57,XY, +4, +5, +6, +7, +8, +10, +12, +15, +17 +mar[cp24] | 4, 6, 8, 9, 10, 14, 17, 18, 21 |
| 10 | M/5 | L2 | 2 | 58,XY, +X, +4, +5, +6, +8, +10, +13, +14, +15, +17, +18, +21[12]/46,XY[11] | 4, 6, 8, 10, 17, 21 |
FAB indicates French-American-British classification; WBC, white blood cell count; F, female; M, male; and dx, diagnosis.
Original karyotypes have previously been published in Andreasson et al.2 Data for patients 2, 3, and 5 to 10 have been revised. Trisomies identified with CGH only are indicated by underlining. Note that, from a formal point of view, the karyotype of patient 10 is hypotriploid and should be written as 58,XXY, -1, -2, -3, -7, -9, -11, -12, -16, -19, -20, -22[12]/46,XY[11]. Trisomies detected by G-banding and investigated (see next table footnote) but not identified by QF-PCR are shown in bold type.
See Table 3 for details. Because of equal allele dosage, tetrasomy 21 could not be distinguished from disomy 21 using QF-PCR. Note that chromosomes 1, 3, 16, 19, 20, and Y were not included in the QF-PCR analyses and that gains of the X chromosome were investigated in girls only. Gains identified by QF-PCR, but not cytogenetically, are shown in bold type.
Microsatellite markers
The PowerPlex 16 System (Promega, Madison, WI), which includes 16 polymorphic loci, and 22 additional microsatellite markers—tetranucleotide repeats of variable lengths—localized to selected chromosomes were used (Table 2). The latter were chosen on the basis of high maximum heterozygosity, and the primer sequences were obtained from The Genome Database (http://www.gdb.org). In addition, analyses with microsatellite markers for 9p (Table 2), close to CDKN2A (p16),23 were performed to test the previously described preference for deletions in the maternal 9p in childhood ALL.23,24 For each patient except patient 8, for whom no remission sample was available, samples from diagnosis, remission, and both parents were used. All patients were investigated with at least one informative marker for chromosomes 4-6, 8-11, 14, 17, 18, 21, and 22.
Designation and chromosomal locations of the 38 microsatellite markers
Marker . | Location . |
|---|---|
| TPOX* | 2p23-pter |
| D2S1238 | 2q31 |
| D4S1508 | 4p14 |
| D4S2303 | 4q26 |
| FGA* | 4q28 |
| D5S818* | 5q23.3-32 |
| CSF1PO* | 5q33.3-34 |
| D6S967 | 6p25 |
| D6S1279 | 6p23-24 |
| D6S1053 | 6q12 |
| D7S820* | 7q11.21-22 |
| D8S639 | 8p22 |
| D8S347 | 8q24 |
| D8S1179* | 8q |
| D9S1679 | 9p21 |
| D9S746 | 9p13 |
| D10S1423 | 10p12 |
| D10S1214 | 10p12 |
| TH01* | 11p15.5 |
| vWA* | 12p12-pter |
| D12S811 | 12q24 |
| D13S317* | 13q22-31 |
| D14S122 | 14p11 |
| D14S139 | 14q13 |
| Penta E* | 15q |
| D17S1159 | 17p12 |
| D17S1293 | 17q12-21 |
| D17S846 | 17q21 |
| D18S499 | 18q21.32-21.33 |
| D18S51* | 18q21.3 |
| D18S386 | 18q22 |
| D21S11* | 21q11-21 |
| D21S1412 | 21q21 |
| D21S1411 | 21q22 |
| Penta D* | 21q |
| D22S417 | 22q13 |
| D22S526 | 22q13 |
| HUMARA | Xq11-12 |
Marker . | Location . |
|---|---|
| TPOX* | 2p23-pter |
| D2S1238 | 2q31 |
| D4S1508 | 4p14 |
| D4S2303 | 4q26 |
| FGA* | 4q28 |
| D5S818* | 5q23.3-32 |
| CSF1PO* | 5q33.3-34 |
| D6S967 | 6p25 |
| D6S1279 | 6p23-24 |
| D6S1053 | 6q12 |
| D7S820* | 7q11.21-22 |
| D8S639 | 8p22 |
| D8S347 | 8q24 |
| D8S1179* | 8q |
| D9S1679 | 9p21 |
| D9S746 | 9p13 |
| D10S1423 | 10p12 |
| D10S1214 | 10p12 |
| TH01* | 11p15.5 |
| vWA* | 12p12-pter |
| D12S811 | 12q24 |
| D13S317* | 13q22-31 |
| D14S122 | 14p11 |
| D14S139 | 14q13 |
| Penta E* | 15q |
| D17S1159 | 17p12 |
| D17S1293 | 17q12-21 |
| D17S846 | 17q21 |
| D18S499 | 18q21.32-21.33 |
| D18S51* | 18q21.3 |
| D18S386 | 18q22 |
| D21S11* | 21q11-21 |
| D21S1412 | 21q21 |
| D21S1411 | 21q22 |
| Penta D* | 21q |
| D22S417 | 22q13 |
| D22S526 | 22q13 |
| HUMARA | Xq11-12 |
Locations as given by http://www.gdb.org and http://www.ncbi.nlm.nih.gov/genome/guide/human/ except for those indicated with asterisks.
Included in the PowerPlex 16 System (Promega).
HUMARA assays
To investigate the methylation status of the X chromosome in the 4 girls (patients 1, 3, 4, and 7; Table 1) included in the study and to identify the parental origin of the gained X chromosome in patients 1 and 4, as detected by G-banding, human androgen-receptor gene AR (HUMARA) assays (Table 2) were performed.25 To ascertain methylation status, samples were digested with HpaII before PCR amplification of the first exon of the AR gene, using the forward primer 5′-HEX-CCGAGGAGCTTTCCAGAATC-3′ and the reverse primer 5′-TACGATGGGCTTGGGGAGAA-3′. Undigested DNA was amplified using the same primers to detect gains of the X chromosome. All PCR products were size-separated using fragment analysis as described in “QF-PCR.” Skewing was defined as an allelic ratio more extreme than 75:25 or 25:75.26
QF-PCR
For the PowerPlex 16 System (Promega), QF-PCR was performed in accordance with the manufacturer's instructions. For the other 24 microsatellite markers, PCR amplification was performed in a total volume of 50 μL using 20 ng genomic DNA in 1 × PCR buffer (20 mM Tris-HCl, pH 8.4, 50 mM KCl; Invitrogen, Carlsbad, CA), 1.25 to 1.75 mM MgCl2, 0.8 mM dNTP, 0.5 μM each primer, and 1 U Platinum Taq DNA polymerase (Invitrogen). One primer pair was used in each amplification, except for the 9p markers, which were coamplified. Forward primers were labeled with HEX or 6-FAM. After an initial denaturation step at 94°C for 5 minutes, 25 to 30 cycles of denaturation at 94°C for 1 minute, annealing at 56°Cto60°C for 1 minute, and extension at 72°C for 1 minute were carried out, followed by a final extension step at 72°C for 10 minutes. All samples were size-separated on an ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA) and were analyzed using GeneScan Analysis Software (Applied Biosystems) for fragment analysis.
Statistical analysis
Exact 2-tailed binomial probabilities were calculated for the distribution of additional parental alleles for each trisomy.27 A P value of less than .05 was considered significant.
Results
Chromosomes 2, 4 to 15, 17, 18, 21, and 22 could be successfully analyzed in all 10 patients with ALL (Table 3; Figure 1A-D). If any findings were noninformative or uncertain, additional markers were used until conclusive results could be obtained in all patients for chromosomes 4 to 6, 8 to 11, 14, 17, 18, 21, and 22. Ratios for the trisomic chromosomes ranged from 1.4 to 2.9, depending on the marker used and, most likely, on the fraction of diploid cells in the diagnostic samples. Overlapping ratio values for disomies and trisomies were not seen. Ratios varied more with the PowerPlex 16 System (Promega), probably because of multiplex PCR amplification. Three markers were therefore excluded, stricter cutoff criteria for trisomies (greater than 1.7) were applied, and all analyses were performed in duplicate. Because of the stricter criteria, some trisomies might have been misclassified as disomies for chromosomes investigated solely with this primer set. In total, 38 markers mapping to 18 of the 24 human chromosomes were used in this study (Table 2).
Gains and parental origin of duplicated chromosomes in 10 hyperdiploid childhood pre-B ALL
. | Chromosome . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | 2 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 17 . | 18 . | 21 . | 22 . | X . | |||||||||||||||||
| 1 | NI | M | D | P | D | D | D | M | D | NI | D | M | D | M | P | T | D | P/pat | |||||||||||||||||
| 2 | D | D | D | M | D | D | D | P | D | NI | D | M | NI | M | D/mut | T | D | — | |||||||||||||||||
| 3 | D | P/mut | D | P | D | D | D | D | D | D | D | P | D | M | M | T | D | D/NI | |||||||||||||||||
| 4 | D | M | D | P | D | P | D | P | D | D | D | M | D | P | M | T | D | P/mat | |||||||||||||||||
| 5 | LOH | M | D | M | D | D/mut | D | P | D | LOH | NI | M | D | M | P | T | D | — | |||||||||||||||||
| 6 | NI | M | D | P | NI | D | P | P | D | NI | D | D | D | M | D | T | D | — | |||||||||||||||||
| 7 | NI | P | D | D | NI | D | D | D | D | D | D | M | NI | P | P | M | D | D/mat | |||||||||||||||||
| 8 | D | P | D | M | D | P | LOH/d | D | D | D | D | M | D | P | P | P | D | — | |||||||||||||||||
| 9 | NI | M | D | M | D | P | M | M | LOH | D | D | M | D | P | P | T | D | — | |||||||||||||||||
| 10 | NI | P | D | M | D | P | LOH | P | D | D | D | D | MSI | M | D | T | D | — | |||||||||||||||||
| P | — | 1 | — | 1 | — | .125 | — | .453 | — | — | — | .0703 | — | .754 | .453 | — | — | .500 | |||||||||||||||||
. | Chromosome . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient . | 2 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | 13 . | 14 . | 15 . | 17 . | 18 . | 21 . | 22 . | X . | |||||||||||||||||
| 1 | NI | M | D | P | D | D | D | M | D | NI | D | M | D | M | P | T | D | P/pat | |||||||||||||||||
| 2 | D | D | D | M | D | D | D | P | D | NI | D | M | NI | M | D/mut | T | D | — | |||||||||||||||||
| 3 | D | P/mut | D | P | D | D | D | D | D | D | D | P | D | M | M | T | D | D/NI | |||||||||||||||||
| 4 | D | M | D | P | D | P | D | P | D | D | D | M | D | P | M | T | D | P/mat | |||||||||||||||||
| 5 | LOH | M | D | M | D | D/mut | D | P | D | LOH | NI | M | D | M | P | T | D | — | |||||||||||||||||
| 6 | NI | M | D | P | NI | D | P | P | D | NI | D | D | D | M | D | T | D | — | |||||||||||||||||
| 7 | NI | P | D | D | NI | D | D | D | D | D | D | M | NI | P | P | M | D | D/mat | |||||||||||||||||
| 8 | D | P | D | M | D | P | LOH/d | D | D | D | D | M | D | P | P | P | D | — | |||||||||||||||||
| 9 | NI | M | D | M | D | P | M | M | LOH | D | D | M | D | P | P | T | D | — | |||||||||||||||||
| 10 | NI | P | D | M | D | P | LOH | P | D | D | D | D | MSI | M | D | T | D | — | |||||||||||||||||
| P | — | 1 | — | 1 | — | .125 | — | .453 | — | — | — | .0703 | — | .754 | .453 | — | — | .500 | |||||||||||||||||
NI indicates not informative; M, maternal chromosome duplicated; D, disomic; P, paternal chromosome duplicated; T, tetrasomic; mut, mutation (difference in the length of the microsatellite repeat during transmission from parent to child); pat, paternal X active; mat, maternal X active; d, homozygous deletion; MSI, microsatellite instability; and —, not done.
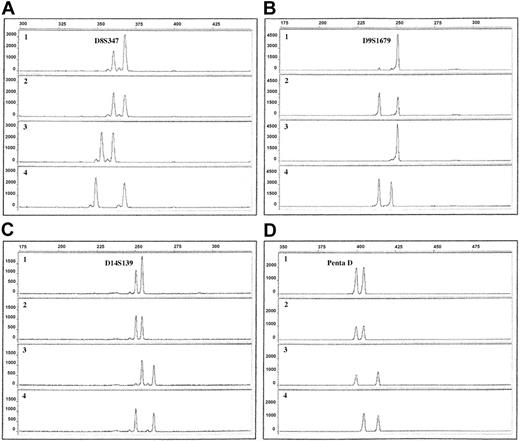
Electropherograms of QF-PCR products using 4 different microsatellite markers. The x-axis displays the lengths of the PCR products in base pairs, determined by the use of an internal size standard. The y-axis displays the fluorescence activity in arbitrary units. Row 1, ALL sample; row 2, remission sample; row 3, maternal sample; row 4, paternal sample. (A) Patient 9, marker D8S347, showing duplication of the paternal allele (ratio 1.8). (B) Patient 10, marker D9S1679, showing LOH with retention of the maternal allele. (C) Patient 5, marker D14S139, showing duplication of the maternal allele (ratio 1.6). (D) Patient 10, marker Penta D, showing tetrasomy 21, visible as equal allele dosage (ratio 1.0).
Electropherograms of QF-PCR products using 4 different microsatellite markers. The x-axis displays the lengths of the PCR products in base pairs, determined by the use of an internal size standard. The y-axis displays the fluorescence activity in arbitrary units. Row 1, ALL sample; row 2, remission sample; row 3, maternal sample; row 4, paternal sample. (A) Patient 9, marker D8S347, showing duplication of the paternal allele (ratio 1.8). (B) Patient 10, marker D9S1679, showing LOH with retention of the maternal allele. (C) Patient 5, marker D14S139, showing duplication of the maternal allele (ratio 1.6). (D) Patient 10, marker Penta D, showing tetrasomy 21, visible as equal allele dosage (ratio 1.0).
With the use of QF-PCR, trisomies for chromosomes 4, 6, 10, 14, 17, and 18 were found in 70% or more of the ALL patients, and gain of chromosome 8 was present in 4 of 10 patients, a pattern of extra chromosomes agreeing well with previous studies (Table 3).6 Some gains detected through QF-PCR were not identified cytogenetically and vice versa (Table 1). There are 2 main reasons for these discrepancies. First, as mentioned, the stricter cutoff criteria using the PowerPlex 16 System (Promega) might have caused misclassification of some trisomies as disomies. In fact, 13 of the 21 trisomies detected by G-banding/CGH but not with QF-PCR involved chromosomes investigated with the PowerPlex 16 System (Promega) only (Tables 1, 2). Second, the poor G-banding quality, which is common in hyperdiploid ALL, often results in unidentified chromosome markers and in erroneous karyotypic descriptions, and the results obtained by the QF-PCR are most likely more accurate. For chromosome 21, unequal allele dosages were found in patients 7 and 8 (Table 3), suggesting trisomy 21 only in these 2 ALL patients. Considering that cytogenetic analysis revealed trisomy 21 in most of the patients (Table 1) and that gains of this chromosome, often in the form of 2 extra copies, are detected in almost all hyperdiploid childhood ALL,6,12 it seems safe to conclude that the present findings should be interpreted as tetrasomy 21, with duplications of both homologues (Figure 1D), yielding equal allele dosages, in 8 of 10 of the investigated ALL patients.
For 3 of the 38 markers, differences in the lengths of the microsatellite repeats occurred during transmission from parent to child (Table 3), without any evidence of nonpaternity. Microsatellite instability was found in one patient for a single marker (Table 3). LOH was identified in 4 patients (Table 3). Patient 8 had LOH for D9S1679, with retention of the paternal allele, whereas D9S746 was homozygously deleted. Patient 10 displayed LOH for both these 9p markers, with retention of the maternal alleles (Figure 1B). Patient 9 showed LOH at 11p15, with retention of the paternally inherited allele. Patient 5 had LOH for markers on both the p and the q arms of chromosomes 2 and 12 (retention of the paternal and maternal alleles, respectively), suggesting that the LOH involved each whole chromosome. This finding could be attributed to monosomies or to uniparental disomies (UPDs)—that is, both copies of a chromosome were derived from only one parent. Because neither G-banding nor CGH analysis revealed any losses, the LOH in patient 5 was most likely caused by UPD for chromosomes 2 and 12.
Statistically significant differences in the parental origin of the duplicated chromosomes were not found for any of the trisomic chromosomes (Table 3). However, there was a trend toward significance regarding chromosomes 8 and 14; trisomy 8 (Figure 1A) was derived from the paternal homologue in all 4 patients with +8(P = .125), and +14 (Figure 1C) was of maternal origin in 7 of 8 ALL patients with gain of this chromosome (P = .0703). HUMARA assay was used for analysis in 4 patients. The X chromosome was of paternal origin in the 2 patients (patients 1 and 4; Table 1) with trisomy X (P = .500). The active X was paternal in the leukemic cells of patient 1 and maternal in patients 4 and 7. Patient 3 was not informative regarding inactivation status, probably because of a large fraction of diploid cells in the sample.
Discussion
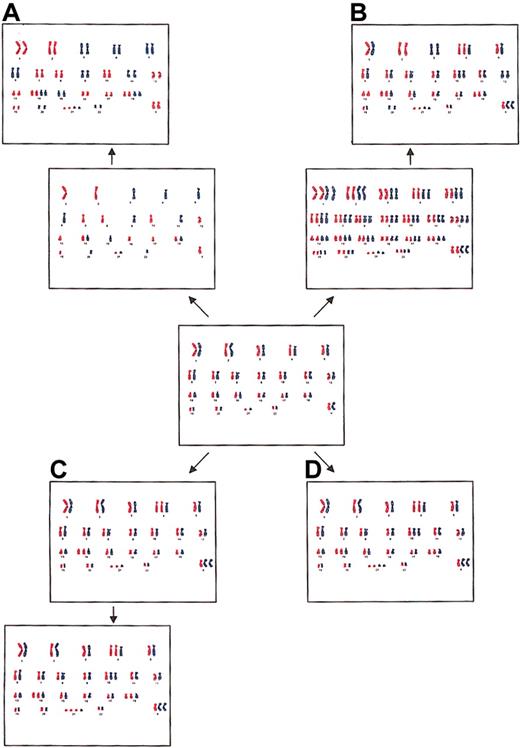
The hyperdiploid pattern in childhood ALL may originate by several different mechanisms: (1) by the doubling of a near-haploid set of chromosomes (Figure 2A), (2) by prior polyploidization with subsequent losses of chromosomes (Figure 2B), (3) by successive gains of chromosomes in consecutive cell divisions (Figure 2C), and (4) by a simultaneous occurrence of trisomies and tetrasomies in a single aberrant mitosis (Figure 2D).17 These 4 possibilities are not easily addressed experimentally, but analyses of allele dosages, as in this study, may provide some valuable clues. Hyperdiploidy formed by the first mechanism is characterized by a pattern of disomies and tetrasomies, in particular +14, +14, +18, +18, and +21, +21,12 with a lack of trisomies usually seen in the hyperdiploid subgroup, and it has UPDs for most of the disomic chromosomes, detectable as widespread LOH (Figure 2A).15,16 However, all the patients with ALL in the present study displayed the normal hyperdiploid abnormality pattern with trisomies (Table 1), and LOH was found only in 8 loci—2 each in chromosomes 2 and 12 in patient 5, one in chromosome 9 in patient 8, 1 in chromosome 11 in patient 9, and 2 in chromosome 9 in patient 10 (Table 3). Furthermore, in none of the patients were near-haploid clones observed cytogenetically. Thus, we deem it highly unlikely that the hyperdiploidy in the investigated ALL patients originated through a near-haploid pathway. The second alternative, the hyperdiploid pattern formed by prior polyploidization with subsequent losses of chromosomes, would have given rise to LOH as a consequence of UPD for approximately one third of the disomies (Figure 2B).17 Patient 5 probably had UPD for chromosomes 2 and 12—that is, for 2 of the 9 informative disomies (Table 3). It is hence possible that the hyperdiploidy in this patient originated through such a mechanism, whereas this scenario is implausible for the other patients included in the study.
Four possible mechanisms for the formation of hyperdiploidy. (A) Doubling of a near-haploid set of chromosomes, resulting in widespread LOH caused by UPD. (B) Initial polyploidization with subsequent losses of chromosomes, resulting in LOH for approximately one third of the disomic chromosomes and equal allele dosages for tetrasomic chromosomes. (C) Sequential gains of individual chromosomes in consecutive cell divisions, resulting in unequal allele dosages for two thirds of the tetrasomic chromosomes. (D) Simultaneous gain of chromosomes in a single abnormal mitosis, resulting in equal allele dosages for tetrasomic chromosomes.
Four possible mechanisms for the formation of hyperdiploidy. (A) Doubling of a near-haploid set of chromosomes, resulting in widespread LOH caused by UPD. (B) Initial polyploidization with subsequent losses of chromosomes, resulting in LOH for approximately one third of the disomic chromosomes and equal allele dosages for tetrasomic chromosomes. (C) Sequential gains of individual chromosomes in consecutive cell divisions, resulting in unequal allele dosages for two thirds of the tetrasomic chromosomes. (D) Simultaneous gain of chromosomes in a single abnormal mitosis, resulting in equal allele dosages for tetrasomic chromosomes.
Although cytogenetic investigations of childhood ALL have revealed that the hyperdiploid pattern is usually stable,6,12 arguing against sequential gains of chromosomes in consecutive cell divisions, it is still possible that intermediate forms—moderately hyperdiploid cells—exist but are not detectable because of a strong proliferative advantage for the major clone. Hyperdiploidy formed by sequential gains would display unequal allele dosages in most of the tetrasomic chromosomes (Figure 2C). However, the alleles on chromosomes 21 were found in equal proportions in 8 of 10 patients in the present investigation (Table 3) and in 10 of 11 patients in the study reported by Onodera et al.17 This pattern of allele dosages is to be expected if the hyperdiploidy originated through a simultaneous gain of all extra chromosomes in a single abnormal cell division (Figure 2D). It is possible that both copies of chromosome 21 were duplicated at the same time even if the hyperdiploid pattern as a whole arose sequentially. It is noteworthy that patient 7 displayed unequal allele dosages of loci situated at chromosome 21, despite a confirmed tetrasomy 21 (Tables 1, 3). This finding argues against a simultaneous gain. Taken together, however, the present results suggest that the hyperdiploidy in most of the investigated childhood ALL patients arose in a single step, though the possibility of sequential gains cannot be excluded with certainty.
Not only the underlying mechanism of hyperdiploidy but also its pathogenetic effects seem elusive. In fact, the biologic significance of genomically unbalanced changes, such as trisomies, in neoplastic disorders remains enigmatic.13,14 It has been suggested that such aberrations are always secondary to primary, cytogenetically balanced, fusion gene-forming rearrangements and that they contribute to tumor evolution rather than to tumorigenesis.28 It is hence possible that a single cryptic primary change may cause hyperdiploidy in childhood ALL as a secondary effect. Alternatively, the trisomic chromosomes may harbor cryptic rearrangements, and the extra copies of mutated genes may be the pathogenetically important outcome of the gains, in line with previous findings of mutations in KIT in acute myeloid leukemia with +4 and in MET in hereditary papillary kidney carcinoma with trisomy 7.29,30 However, to explain the presence of hyperdiploidy in this way, hidden abnormalities would have to be present on each duplicated chromosome. Furthermore, equal allele dosages were detected for tetrasomy 21 in most of the ALL patients in this study, strongly suggesting that there was no selection for a specific mutated gene located on this chromosome. Although the trisomies of hyperdiploid childhood ALL may not be primary, they may confer a positive selection for the leukemic blast because of dosage effects. In fact, support for this possibility comes from a recent microarray study showing that 70% of the class-defining genes of the hyperdiploid subgroup were localized to chromosomes 21 and X,31 2 of the most frequently gained chromosomes.6,12
In addition to gains of extra copies, gene expression patterns can also be influenced by imprinting, an epigenetic event causing the expression of a gene to be dependent on whether the allele is maternally or paternally inherited.32 Such genes are known to be involved in the pathogenesis of many congenital disorders, such as Prader-Willi and Angelman syndromes, in which the underlying imprinting may be unmasked by, for example, a deletion or a UPD.33 It has been suggested that imprinting plays an important role in several neoplasms; it is implicated because of a preferential parental origin of chromosomal aberrations, such as loss of the maternally derived copy of chromosome band 11p15 in Wilms tumors and of the paternally inherited 19q in oligodendrogliomas,34,35 or because of the presence of a UPD for a chromosome that is usually monosomic in a certain neoplastic disorder, such as UPD for chromosome 3 in uveal melanoma.36 In childhood ALL, LOH in the 9p region surrounding the CDKN2A tumor-suppressor gene has been reported in 35% to 60% of patients,37,38 and a preferential loss of the maternally inherited allele has been suggested.23,24 In this study, 2 microsatellite markers, situated approximately 3 and 6 Mb centromeric to CDKN2A (D9S1679 and D9S746, respectively; http://www.ensembl.org),23 were used to identify 9p deletions. Patient 10 displayed LOH for both these loci, with loss of the paternal alleles (Figure 1B), whereas patient 8 had LOH for D9S1679, with the maternal allele deleted, and a homozygous deletion of D9S746. Hence, the present results do not agree with previous studies showing loss of the maternal 9p alleles. However, including our patients, 14 of 17 investigated ALL patients23,24 have displayed loss of the maternal homologue.
Given that imprinting is known to be important in many neoplastic disorders, Haas19 speculated that this phenomenon may also be involved in hyperdiploid ALL. In the present study, this hypothesis has, for the first time, been tested experimentally, using microsatellite markers to ascertain the parental origin of the trisomic chromosomes in hyperdiploid childhood ALL. No evidence for preferential duplications of the maternally or paternally inherited copies of chromosomes X, 4, 6, 9, 10, 17, 18, and 21 was found (Table 3). It may be important that there was a trend toward significance for chromosomes 8 and 14. In all 4 patients with +8, the extra copies were derived from the paternal homologue (P = .125; Table 3). Although the sample size is too small to draw any conclusions based on this study alone, a differentially methylated region in 8q21 has been observed in uniparental tissues—complete hydatidiform moles, ovarian teratomas, and sperms39 —suggesting that imprinted genes may reside in this chromosome. Furthermore, of the 8 patients with +14, 7 displayed duplication of the copy inherited from the mother (P = .0703; Table 3). This finding is worthy of note because chromosome 14 is known to harbor several imprinted genes.40 The duplicated X chromosome was of paternal origin in both patients with trisomy (patients 1 and 4; Tables 1, 3). The extra X was methylated in one and unmethylated in the other, suggesting that the activation status of the duplicated chromosome has no or little impact in hyperdiploid ALL, a finding similar to what has previously been reported in non-Hodgkin lymphoma with trisomy X.41
In conclusion, the results of the present study suggest that hyperdiploid childhood ALL originates in a single aberrant mitosis, though a sequential gain of chromosomes other than 21 in consecutive cell divisions remains a possibility. Furthermore, we found no support for the hypothesis that imprinting effects related to the origin of the trisomic/tetrasomic chromosomes are of major pathogenetic importance, with the possible exception of the observed parental skewness of +8 and +14.
Prepublished online as Blood First Edition Paper, June 26, 2003; DOI 10.1182/blood-2003-05-1444.
Supported by grants from the Swedish Cancer Society and the Swedish Children's Cancer Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal