Abstract
The bone morphogenetic proteins (BMPs) are required for the development of ventral mesoderm, which contributes to the ventral blood island and primitive (yolk sac stage) hematopoiesis. Primitive erythropoiesis is defective when BMP signaling is blocked during gastrulation of Xenopus embryos. This phenotype might be attributed to changes in mesoderm patterning leading indirectly to altered erythropoiesis. We developed an inducible system in order to block BMP signaling in a controlled fashion at later time points in development. For this purpose, an inhibitory Smad, xSmad6, was fused to the estrogen receptor ligand-binding domain. We show that ER-xSmad6 is inactive when expressed in developing embryos, but its activity is induced by estradiol. When induced early in development, ER-xSmad6 causes a dorsalized phenotype, equivalent to overexpression of native xSmad6. When ER-xSmad6 is induced after gastrulation, there is a specific defect in primitive erythropoiesis without any apparent effect on axial patterning. Our results identify an embryonic signal that is Smad-dependent, is required for maintaining expression of GATA-1, and functions within mesoderm and not the overlying ectoderm. Thus, BMP signaling is necessary both during mesoderm patterning and also following early specification events for proper regulation of the primitive erythroid lineage.
Introduction
The first blood cells develop during embryogenesis on the extra-embryonic yolk sac of birds and mammals, and at analogous sites within the ventral blood island (VBI) of Xenopus, or the ventral intermediate cell mass (ICM) associated with the tail in bony fish. This hematopoiesis is referred to as embryonic or primitive, in order to distinguish it from the subsequent intraembryonic-derived definitive lineage that sustains hematopoiesis throughout later life. Primitive hematopoiesis is devoted largely to erythropoiesis; the primitive erythroid cells have a morphology, gene expression program, and life-span that is distinct from fetal liver or bone marrow–derived definitive erythroid cells. The signaling molecules that regulate primitive hematopoiesis are not known, and at least some of the regulatory factors that are required for definitive erythropoiesis are not essential, including c-myb and runx1.1-5 Studies using embryonic stem (ES) cells6,7 and novel transplantation protocols8-10 indicate that primitive progenitors have an inherent potential to contribute to definitive hematopoiesis. The unique birthplace of primitive progenitor cells must therefore provide a stromal environment conducive for primitive hematopoiesis.
In Xenopus, the first erythrocytes differentiate within the VBI about 2 days after fertilization. Recent fate-mapping studies established that blastomeres with presumptive dorsal or ventral fates both contribute to primitive hematopoiesis,11-13 but all primitive erythroid cells eventually differentiate in the VBI, the most ventral embryonic structure. It is well established that members of the bone morphogenetic protein (BMP) subfamily of transforming growth factor β (TGF-β) growth factors are required during gastrulation for specifying the ventral character of embryonic mesoderm and are therefore candidates for controlling the development of VBI-derived blood progenitors. Indeed, a role for BMPs in regulating hematopoiesis is supported by several lines of investigation. First, the components of BMP signaling (ligands, receptors, and the Smad proteins that mediate signaling) are present and active at the right time and place to regulate the primitive hematopoietic lineage. Second, ectopic BMP signaling activates the primitive erythroid program, while inhibiting the pathway blocks VBI development.14,15 Hedgehog proteins, probably mediated by BMP2 and/or BMP4, are sufficient to reprogram anterior ectoderm and induce primitive erythropoiesis in murine embryonic explants.16 Third, mouse embryos that survive an early embryonic lethality of the targeted BMP4 mutation have severely reduced primitive blood islands,17 and fish embryos mutant for BMP2 or BMP7 lack blood entirely.18 Finally, BMP4 can regulate in vitro hematopoietic development of mouse19,20 or monkey21 ES cells, myeloma cells,22 and purified repopulating progenitors.23,24
Therefore, in addition to their role in patterning the ventral mesoderm (VM), BMPs may act directly on hematopoietic progenitors or stromal elements, in synergy with other embryonic signaling proteins.19,25,26 However, defining precisely the mechanism by which BMPs influence hematopoiesis has been a challenge. The genetic analyses (mutants and knock-outs) are complicated by very early and pleiotropic embryonic requirements for BMP signaling and presumed functional redundancies.27,28 The direct targets of BMP signaling include a class of homeobox-containing genes (Vents) that specify ventral mesoderm, but there is no evidence that these factors regulate later stages of hematopoiesis.29-31 However, direct or indirect BMP target genes also include transcription factors that are critical for hematopoiesis. Expression of GATA-1, GATA-2, LMO-2, SCL, and EKLF are activated by BMP signaling.15,32-34 In order to study the function of BMP signaling beyond stages of ventral specification, it is necessary to bypass the early requirement and provide a conditional block to signaling after specification has occurred. We devised a strategy to accomplish this goal, and we demonstrate the specific requirement of a Smad-dependent signaling pathway for efficient primitive erythroid cell differentiation, independent of any early requirement for cell specification. Our results suggest that GATA-1 is dependent on this signal within the specified mesoderm, and that in its absence decreased levels of GATA-1 correlate with defects in primitive erythroid cell differentiation.
Materials and methods
Plasmid construction
The pCS2-ER-xSmad6 expression plasmid was constructed by first ligating an approximately 1.0–kilobase (kb) EcoRI/BamHI polymerase chain reaction (PCR) fragment generated from pCS2-xSmad6 (a gift from J. Christian) into pBluescript II KS (+/–) to create pKS-xSmad6. The PCR fragment was generated using forward primer (TE647): ATAGAATTCACCATGTTCAGGTCCAGACGC and reverse primer (TE648): ATAGGATCCCCTCTTCCCAGTAGGACCTC. A 945–base pair (bp) ClaI/EcoRI PCR fragment containing the human estrogen receptor ligand-binding domain was generated from pBabepuroNmyc-ER (a gift from A. Iavarone) using forward primer (TE703): ATAATCGATACCATGTCTGCTGGAGACATGAGA and reverse primer (TE704): ATAGAATTCCCCGACTGTGGCAGGGAAACC. This ClaI/EcoRI fragment was ligated into pKS-xSmad6 to create pKS-ER-xSmad6. A ClaI/BstEII fragment from pKS-ER-xSmad6 was then ligated back into pCS2-xSmad6 to generate pCS2ER-xSmad6, which was used to generate mRNA for injection. The pEGFP-ER-xSmad6 expression plasmid was constructed by ligating a BamHI/KpnI fragment from pKS-ER-xSmad6 containing ER-xSmad6 into pEGFP-C1 (Clontech, Palo Alto, CA). The pEGFP-xSmad6 expression plasmid was constructed by ligating an EcoRI/XbaI fragment from pCS2-xSmad6 containing xSmad6 into pEGFP-C1.
Xenopusembryo culture, injection, and explant
Freshly laid Xenopus eggs were obtained by gonadotropin induction, fertilized in vitro using macerated testes, and dejellied in 2% cysteine (pH 7.9). Embryos were staged according to Nieuwkoop and Faber.35 Capped mRNA was prepared by in vitro transcription of NotI linearized pCS2-ER-xSmad6 or pCS2-xSmad6 template using SP6 polymerase from Ambion's mMESSAGE MACHINE SP6 kit (Austin, TX). Purified ER-xSmad6 (0.4-1.0 ng) or xSmad6 (0.25 ng) RNA was injected into both blastomeres of 2-cell stage embryos or into either the 2 dorsal or 2 ventral blastomeres of 4-cell stage embryos. The optimal amount of ER-xSmad6 was determined empirically for each batch of synthesized mRNA. This was defined by preliminary injections, when an induced cohort was strongly dorsalized, while an uninduced cohort was normal. Despite minor batch differences in optimal concentration, once defined, reproducible results were obtained between preparations. All embryos were cultured in 0.1 × modified Barth saline at room temperature. Induction of ER-xSmad6 protein was with 0.1 × MBS containing 1 μM 17β-estradiol (E2; Sigma, St Louis, MO) at specific stages of development, and embryos were cultured in the presence of hormone until harvested, either between stages 22 to 26 for early markers or between stages 35 to 37 for late markers. Benzidine staining of embryos was performed as described.36
For explant assays, uninjected embryos and embryos injected at the 2-cell stage with 2 ng/embryo ER-xSmad6 mRNA were cultured in 0.1 × MBS until stage 10. Animal cap (AC) and ventral mesoderm explants were then cut using a tungsten needle on a bed of 2% agarose in 1 × MBS containing 30 μg/mL kanamycin. Explants were combined in appropriate combinations and cultured together for 10 minutes prior to being transferred as a “sandwich” to a new culture dish. Explant combinations and whole embryo controls were subsequently cultured on a bed of 2% agarose in 1 × MBS in the presence or absence of 1 μM E2 from stage 10.5 until harvesting at stage 35 for Northern blotting analysis.
Tissue culture
HepG2 cells (1.5 × 106 cells per sample) were transfected with 3 μg pEGFP-ER-xSmad6 or 2 μg pEGFP-C1 vector using LIPOFECTAMINE Reagent (Invitrogen, Carlsbad, CA). Prior to addition of DNA, cells were washed 3 times for 5 minutes in Dulbecco modified Eagle medium without phenol red containing 1 × l-glutamine and 1 × nonessential amino acids and then maintained in this medium during transfection. After 5 hours, cells were transferred to the same media containing 10% charcoal-stripped fetal bovine serum. At 4 hours prior to visualization, 10 μM Hoechst 33342 was added to culture medium to stain nuclei. At 24 hours after transfection, pEGFP-ER-xSmad6–transfected cells were cultured either in the presence of 1 μM 17β-estradiol (E2) or the absence of hormone and then photographed using a 12-bit Photometrics cooled CCD camera attached to an Olympus (Melville, NY) IX70 inverted microscope using the × 20 objective.
Western blot analysis
Protein extracts were prepared from uninjected embryos or embryos injected with mRNA for ER-xSmad6 and cultured in the absence of 1 μM E2. Embryos were homogenized in lysis buffer (10 μL per embryo; 20 mM Tris [tris(hydroxymethyl)aminomethane]–-HCl [pH 8.0], 50 mM NaCl, 50 mM NaF, 10 mM β-glycerophosphate, 2 mM EDTA [ethylenediaminetetraacetic acid], 1% nonidet P-40 [NP-40], 1 mM Na3Vo4, 0.75 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitor cocktail [Roche, Mannheim, Germany]) using Kontes pestles in 1.5-mL microfuge tubes. Following homogenization, extracts were centrifuged at 14 000 rpm at 4°C for 10 minutes. The supernatant was assayed for protein concentration using the Biorad Protein Assay system (Hercules, CA).
For Western blotting analysis, 1.2 μg total protein per lane was separated on 7.5% to 8.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels followed by electroblotting to polyvinylidene fluoride filters (Biorad). Blots were incubated in primary antibody (anti-hER [578-595] polyclonal antibody (Sigma) 1:2000 in phosphate-buffered saline, 0.1% Tween-20 (PBST)) for one hour at room temperature, and secondary antibody (antirabbit–horseradish peroxidase [Biorad] 1:8000 in PBST) for one hour at room temperature. Signal was detected using the enhanced chemiluminescence Plus detection system (Amersham, Piscataway, NJ).
Northern blot analysis
Total RNA was isolated from 10 to 15 uninjected stage-35 embryos or ER-xSmad6 mRNA–injected embryos cultured in the presence or absence of 1 μM E2 using TRI REAGENT (Molecular Research Center, Cincinnati, OH). For polyA+ blots, approximately 100 embryos per condition were homogenized. Embryos were homogenized in 1.5-mL microfuge tubes using Kontes pestles. PolyA+ mRNA was harvested from total RNA using PolyATract mRNA Isolation System (Promega, Madison, WI). Then, 10 μg total RNA or 0.8 μg polyA+ mRNA was size fractionated on a 1% agarose gel in 3% formaldehyde, MOPS (3-[N-morpholino]propanesulphonic acid) buffer. Fractionated RNA was transferred to a Nylon filter (Ambion) in 10 × sodium chloride, sodium citrate (SSC). A 600-bp HindIII/XhoI fragment of αT1-globin, 1.9-kb EcoRI fragment of xGATA-1A, 1.3-kb EcoRI fragment of xGATA-2, 1.9-kb EcoRI fragment of xSCL, 2.0-kb EcoRI/NotI fragment of neptune, or a 1.7-kb EcoRI fragment of EF-1α was radiolabeled using High Prime (Roche). Filters were stripped for sequential hybridization by washing twice for 10 minutes each at 95°C in 0.01 × SSC, 0.01% SDS.
Luciferase reporter assay
Xenopus embryos (2- to 4-cell) were injected with combinations of pBRE-Luc (25 pg per blastomere), p3TP-Lux (25 pg per blastomere), Renilla mRNA (0.1 pg per blastomere), xBMP4 mRNA (62.5 pg per blastomere), activin βB mRNA (100 pg per blastomere), xSmad6 mRNA (500 pg per blastomere), ΔBR mRNA (500 pg per blastomere), ER-xSmad6 mRNA (500 pg per blastomere), or EF-1α mRNA (500-600 pg per blastomere) as a control to maintain constant amounts of injected mRNA. Injected embryos were cultured until stage 13/14, when they were harvested in 1 × Passive Lysis Buffer (Promega). Extracts were tested using the Dual Luciferase Reporter Assay Kit (Promega). For each condition, 4 to 6 samples were tested and the assays were performed in triplicate. Luciferase values were averaged and normalized over the average Renilla value.
Results
ER-xSmad6 facilitates a conditional block to BMP-regulated Smad signaling during embryogenesis
BMP signaling is regulated at many levels, but key components are serine/threonine receptor complexes and Smad transcriptional cofactors that transmit the signal from the receptor to the nucleus.37-40 There are 4 subclasses of Smads known: (1) receptor-mediated (R-Smad) 2 and 3 control TGF-β/activin signaling, (2) R-Smads 1, 5, and 8 are activated by BMPs, (3) Smad4 is the co-Smad, (4) while Smads 6 and 7 are negative (inhibitory, I-Smad) factors that function either at the receptor to block R-Smad phosphorylation41 or within the Smad complex to block access to Smad4.42 Smad7 is thought to be a general inhibitor of TGF-β signaling, while Smad6 is more specific to the BMP pathway.43,44 In principle, blocking receptor activity inhibits BMP signaling, and dominant-negative isoforms or antisense RNAs have been used for this purpose. If BMP signaling is blocked during early development the embryos are dorsalized, and since primitive hematopoiesis occurs in ventral tissues, such embryos fail to develop a normal hematopoietic program.45-47 To avoid disrupting mesoderm specification, other approaches have been used, including delayed expression of the dominant-negative receptor using a plasmid promoter15 or titration of the truncated receptor.26 Although the results suggest a hematopoietic role for BMP signaling beyond mesoderm specification, these approaches cannot definitively rule out primary defects in ventral cell-type specification during gastrulation that lead indirectly to the changes seen in hematopoiesis. While still relevant to understanding progenitor cell specification, these approaches are less useful for analyzing signals that act subsequently on the progenitors.
To address this issue, we exploited the natural activity of negative-acting I-Smads to develop an approach for blocking conditionally BMP-dependent Smad signaling. Because xSmad6 is thought to be specific to the BMP pathway, we focused on this gene product. The cDNA encoding xSmad6 was fused to the ligand-binding domain of the human estrogen receptor. We considered that the fusion protein might be sequestered in an inactive state, which would be released upon estrogen induction to function either at the receptor or in the nucleus to block BMP signaling (Figure 1).
Scheme to block BMP signaling conditionally. In the absence of 17β-estradiol (left), ER-xSmad6 is held in an inactive conformation via binding of Hsp90 (square) to the hER ligand-binding domain, allowing signaling (arrows) to occur normally. In the presence of hormone (small circle), ER-xSmad6 is released and blocks signaling downstream of the BMP receptors (right).
Scheme to block BMP signaling conditionally. In the absence of 17β-estradiol (left), ER-xSmad6 is held in an inactive conformation via binding of Hsp90 (square) to the hER ligand-binding domain, allowing signaling (arrows) to occur normally. In the presence of hormone (small circle), ER-xSmad6 is released and blocks signaling downstream of the BMP receptors (right).
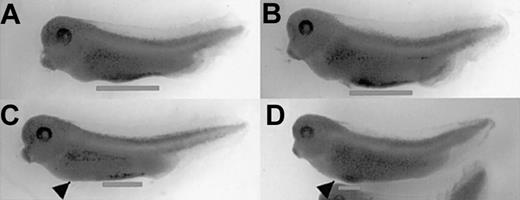
Fusion of ER sequences at the C-terminus inhibited xSmad6 function (not shown). In contrast, the N-terminal ER-xSmad6 fusion protein blocks BMP-regulated Smad signaling, but only in the presence of estrogen (E2). Uninjected embryos cultured alone (Figure 2A) or in the presence of E2 (Figure 2B) appear entirely normal. As shown in Figure 2C, injection of mRNA encoding ER-xSmad6 into fertilized eggs has no effect on development when the embryos are raised in normal medium. However, when injected embryos are cultured in the presence of 1 μM E2, they develop severely dorsalized with a high percentage of twinned dorsal axes (Figure 2D). This phenotype is identical to that of embryos injected with the full-length wild-type xSmad6 mRNA (Figure 2E). Therefore, ER-xSmad6 can be activated conditionally by the addition of E2 and functions like the native protein to block ventral mesoderm development.
Induction of ER-xSmad6 at blastula stages recapitulates overexpression of xSmad6. (A-B) Uninjected embryos develop normally when cultured in the absence (A) or presence (B) of 1 μM 17β-estradiol (E2) from blastula stages. (C) Embryos develop normally when injected at the 2-cell stage with 2 ng/embryo of ER-xSmad6 mRNA and cultured in the absence of E2. (D) Embryos similarly injected with ER-xSmad6 and cultured in the presence of E2 from stage 6 are dorsalized. (E) Embryos are dorsalized when injected at the 2-cell stage with 0.8 ng xSmad6 mRNA. Original magnification, × 20.
Induction of ER-xSmad6 at blastula stages recapitulates overexpression of xSmad6. (A-B) Uninjected embryos develop normally when cultured in the absence (A) or presence (B) of 1 μM 17β-estradiol (E2) from blastula stages. (C) Embryos develop normally when injected at the 2-cell stage with 2 ng/embryo of ER-xSmad6 mRNA and cultured in the absence of E2. (D) Embryos similarly injected with ER-xSmad6 and cultured in the presence of E2 from stage 6 are dorsalized. (E) Embryos are dorsalized when injected at the 2-cell stage with 0.8 ng xSmad6 mRNA. Original magnification, × 20.
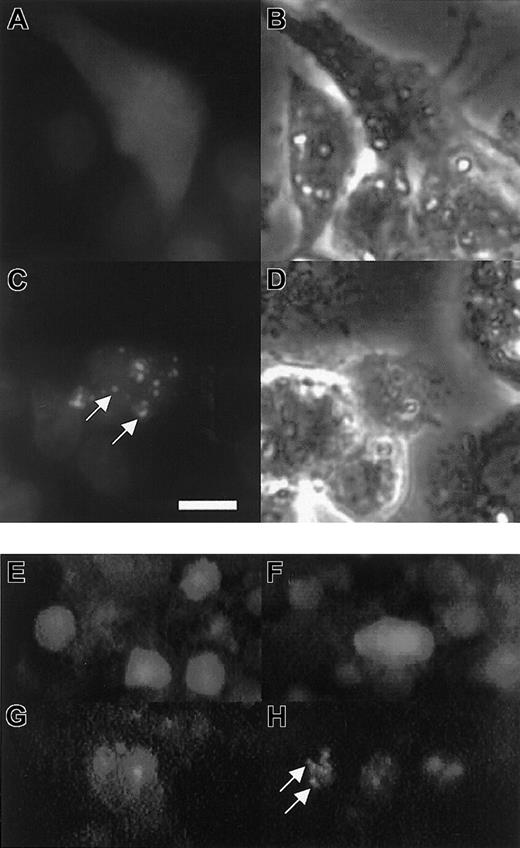
The mechanism by which xSmad6 blocks BMP signaling is not entirely clear but is thought to occur by interfering competitively with R-Smad access to either receptors or Smad4. Alternatively, it is possible that Smad6 interferes in some manner with the nuclear function of R-Smad/Smad4 complexes. The traditional use of ER fusions has been to sequester nuclear transcription factors to the cytoplasm until E2 facilitates release of the protein to act in the nucleus.48-51 Therefore, it was of interest to determine the site of action for the E2-activated ER-xSmad6 fusion protein. For this purpose, the ER-xSmad6 cDNA was fused to the C-terminus of GFP to create a GFP-ER-xSmad6 protein. When expressed in HepG2 cells by transient transfection, GFP-ER-xSmad6 is located diffusely throughout the cytoplasm and excluded from the nucleus (Figure 3A). Within 30 minutes of addition of E2 to the medium, the fusion protein begins to localize to restricted cytoplasmic punctate speckles in many cells (not shown). By 90 minutes after addition of E2, all cells show this cytoplasmic speckling (Figure 3C). Therefore, consistent with some previous analyses of xSmad6 localization,44 the activated ER-xSmad6 fusion protein appears to localize to the plasma membrane, presumably acting at receptor complexes to block signaling. When EGFP plasmid alone was injected into Xenopus embryos and cultured in the absence (Figure 3E) or presence (Figure 3F) of E2, there was no change in localization. When the GFP-ER-xSmad6 fusion protein is expressed in embryos, it dorsalizes embryos conditional to the presence of E2 (not shown) and is localized similarly to speckled cytoplasmic sites (Figure 3H).
Induced ER-xSmad6 localizes to cytoplasmic speckles in cultured cells or embryos. (A-D) HepG2 cells were transfected with 3 μg pEGFP-ER-xSmad6 and shown prior to addition of E2 (A-B) or after addition of E2 for 90 minutes (C-D). In panels A and C, cells are double-labeled with Hoechst 33342 (blue stain) to identify nuclei. Panels B and D are phase images for panels A and C, respectively. (E-H) Embryos were injected at the 2-cell stage with 300 pg pEGFP-C1 DNA (E-F) or 400 pg pEGFP-ER-xSmad6 DNA (G-H). Panels E and G are from embryos cultured in the absence of E2. Panels F and H are from embryos induced with E2 at stage 8. All embryos in panels E-H were photographed between stages 24 and 26. White arrows indicate speckles. (A-D) Bar equals 10 μm. (E-H) Original magnification × 100.
Induced ER-xSmad6 localizes to cytoplasmic speckles in cultured cells or embryos. (A-D) HepG2 cells were transfected with 3 μg pEGFP-ER-xSmad6 and shown prior to addition of E2 (A-B) or after addition of E2 for 90 minutes (C-D). In panels A and C, cells are double-labeled with Hoechst 33342 (blue stain) to identify nuclei. Panels B and D are phase images for panels A and C, respectively. (E-H) Embryos were injected at the 2-cell stage with 300 pg pEGFP-C1 DNA (E-F) or 400 pg pEGFP-ER-xSmad6 DNA (G-H). Panels E and G are from embryos cultured in the absence of E2. Panels F and H are from embryos induced with E2 at stage 8. All embryos in panels E-H were photographed between stages 24 and 26. White arrows indicate speckles. (A-D) Bar equals 10 μm. (E-H) Original magnification × 100.
ER-xSmad6 specifically blocks BMP signaling
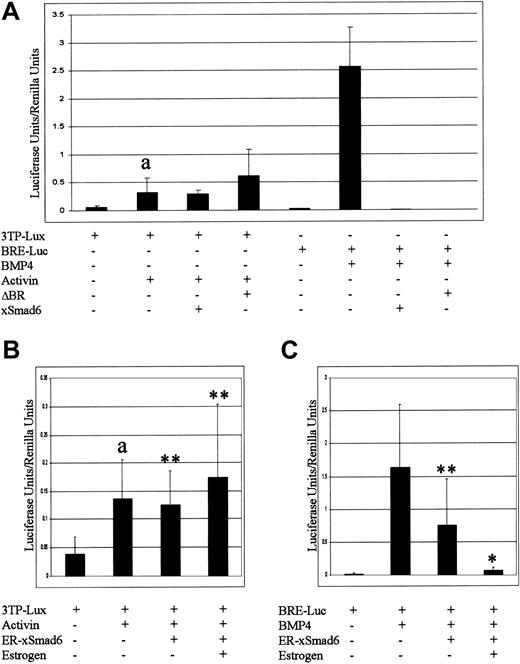
It is thought that xSmad6 mainly inhibits BMP signaling, although inhibition of activin/TGF-β signaling has not been ruled out entirely.42 Because activin/TGF-β signaling may also play roles in hematopoietic development,19,25,52 and to be certain that the addition of ER sequences to Smad6 did not alter its function, we investigated further the specificity of ER-xSmad6. We conducted luciferase reporter assays using a multimerized BMP response element from the xVent2 promoter (BRE-luc)53 or the TGF-β responsive reporter, p3TP-Lux.54 BRE-luc and p3TP-Lux serve to assay active signaling by Smads 1/5/8 or Smads 2/3, respectively. Either xSmad6 or the dominant-negative BMP type I receptor ΔBR strongly inhibits BMP4-mediated activation of the BRE-luc reporter, whereas neither of these proteins inhibits activin-mediated activation of the p3TP-Lux reporter (Figure 4A). ER-xSmad6 was similarly unable to reduce activin-mediated p3TP-Lux activation (Figure 4B). In comparison, ectopic expression of ER-xSmad6, in the absence of E2, inhibits BMP-mediated activation of the reporter approximately 50% (Figure 4C). In this assay the ER-xSmad6 protein is therefore slightly “leaky.” However, this activity may not be physiologically relevant, since these embryos develop normally. In the presence of E2, ER-xSmad6 completely eliminates BMP reporter activity. Therefore, ER-xSmad6 is as effective at blocking BMP signaling as native xSmad6 and does not inhibit activin/TGF-β signaling.
ER-xSmad6 specifically blocks BMP signaling. (A) The luciferase reporter constructs p3TP-Lux or pBRE-Luc were injected alone or coinjected into 2-cell stage embryos with mRNA encoding BMP4, activin βB, ΔBR, or xSmad6 (“Materials and methods”). a indicates p3TP-Lux alone is significantly different from p3TP-Lux + activin (P < .001). (B-C) ER-xSmad6 (1 ng) was coinjected at the 2-cell stage with either p3TP-Lux and activin βB (B) or pBRE-Luc and xBMP4 (C) and cultured in the presence or absence of E2 from the time of injection. In panel B, a indicates p3TP-Lux alone is significantly different from p3TP-Lux + activin (P < .001) and ** indicates the values are not significant compared with p3TP-Lux + activin. In panel C, ** indicates the value is not significant compared with pBRE-Luc + xBMP4, whereas * indicates the value is significant (P < .001). (A-C) All values reported as luciferase value normalized to control Renilla value. Error bars represent one standard deviation above the mean.
ER-xSmad6 specifically blocks BMP signaling. (A) The luciferase reporter constructs p3TP-Lux or pBRE-Luc were injected alone or coinjected into 2-cell stage embryos with mRNA encoding BMP4, activin βB, ΔBR, or xSmad6 (“Materials and methods”). a indicates p3TP-Lux alone is significantly different from p3TP-Lux + activin (P < .001). (B-C) ER-xSmad6 (1 ng) was coinjected at the 2-cell stage with either p3TP-Lux and activin βB (B) or pBRE-Luc and xBMP4 (C) and cultured in the presence or absence of E2 from the time of injection. In panel B, a indicates p3TP-Lux alone is significantly different from p3TP-Lux + activin (P < .001) and ** indicates the values are not significant compared with p3TP-Lux + activin. In panel C, ** indicates the value is not significant compared with pBRE-Luc + xBMP4, whereas * indicates the value is significant (P < .001). (A-C) All values reported as luciferase value normalized to control Renilla value. Error bars represent one standard deviation above the mean.
ER-xSmad6 can be used to evaluate postspecification requirements of BMP signaling
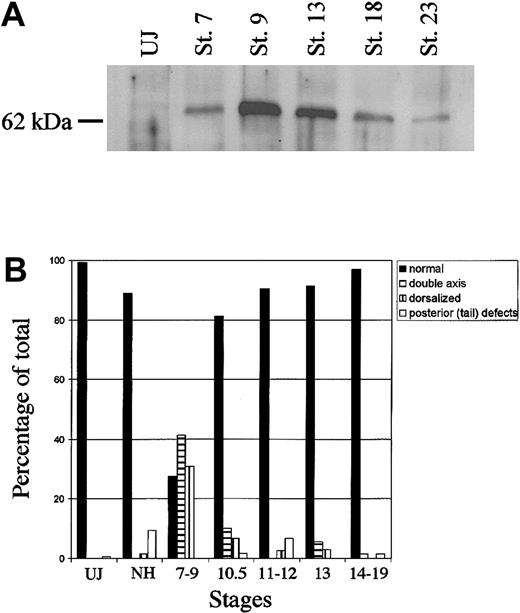
The stability of ectopic ER-xSmad6 was evaluated by Western blotting experiments. The protein derived from mRNA injected into fertilized eggs is stable through gastrula and neurula stages, but eventually declines by around stage 23 of tailbud tadpoles (Figure 5A). However, induction of the fusion protein with E2 did not provoke significant morphologic defects as long as the E2 was added to the medium at or after stage 10.5 (midgastrulation). Beyond this point, 80% to 95% of the embryos developed with a normal body axis, compared with only 25% of the embryos induced with E2 between stages 7 to 9 (Figure 5B). This provides a window of development (stages 13-15) to test the effect of blocking BMP signaling between the end of gastrulation and tailbud stages.
ER-xSmad6 remains stable through neurulation but does not disrupt morphology unless induced prior to stage 11. (A) Western blot analysis of ER-xSmad6 stability. Embryos were injected at the 2-cell stage with 1 ng/embryo of ER-xSmad6 mRNA and cultured in the absence of E2. Protein was harvested at the stages indicated (UJ = uninjected; St. = stage). The Western blot was probed with a polyclonal antibody against the F domain of the human estrogen receptor. (B) Graphic summary of the morphologic phenotypes derived from induction of ER-xSmad6 at various stages throughout development after injection into ventral blastomeres at the 4-cell stage. n = 125 (uninjected), 63 (no hormone), 58 (stages 7-9), 59 (stage 10.5), 74 (stages 11.5-12), 35 (stage 13), and 68 (stages 14-19)
ER-xSmad6 remains stable through neurulation but does not disrupt morphology unless induced prior to stage 11. (A) Western blot analysis of ER-xSmad6 stability. Embryos were injected at the 2-cell stage with 1 ng/embryo of ER-xSmad6 mRNA and cultured in the absence of E2. Protein was harvested at the stages indicated (UJ = uninjected; St. = stage). The Western blot was probed with a polyclonal antibody against the F domain of the human estrogen receptor. (B) Graphic summary of the morphologic phenotypes derived from induction of ER-xSmad6 at various stages throughout development after injection into ventral blastomeres at the 4-cell stage. n = 125 (uninjected), 63 (no hormone), 58 (stages 7-9), 59 (stage 10.5), 74 (stages 11.5-12), 35 (stage 13), and 68 (stages 14-19)
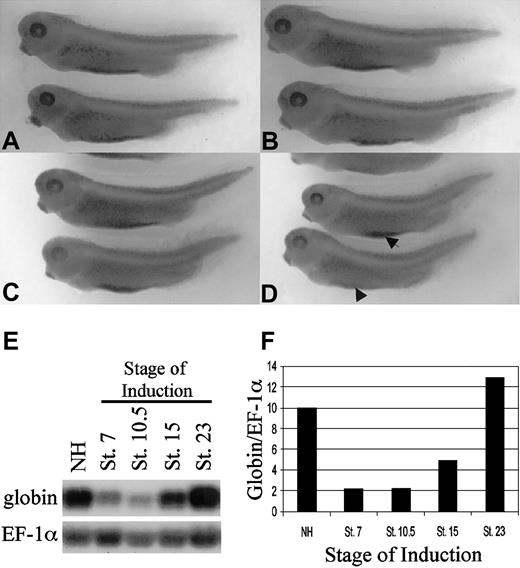
As an initial test for alterations in blood island development, embryos were injected into both ventral blastomeres at the 4-cell stage with the ER-xSmad6 mRNA and cultured in normal medium or medium that was supplemented with E2 at stage 13. All embryos were fixed at stage 35/36 and analyzed for normal blood island development by assaying reactivity with benzidine, which stains hemoglobin. Mock-injected embryos cultured without (Figure 6A) or with (Figure 6B) addition of E2 showed normal patterns of benzidine staining. As shown in Figure 6C, injected embryos cultured in the absence of E2 show no reduction of blood island development, whereas activation of ER-xSmad6 after gastrulation (stage 13) results in defects of primitive erythropoiesis, illustrated by significantly decreased VBI staining (Figure 6D). The embryos induced at stage 13 appear otherwise entirely normal, with a complete dorsal-ventral axis and no evidence of dorsalized structures. Injection of the ER ligand-binding domain alone had no effect on the gross morphology or VBI development of the embryos (not shown). This result demonstrates a Smad6-sensitive signaling pathway required after gastrulation stages for normal VBI development.
ER-xSmad6 inhibits primitive blood island development during neurulation. (A-B) Water-injected embryos cultured in the absence (A) or presence (B) of E2 develop normal ventral blood islands as assayed by benzidine staining at stage 35. (C-D) Embryos injected with 1 ng/embryo in the ventral blastomeres of 4-cell stage embryos and cultured in the absence of E2 (C) develop normal ventral blood islands. Embryos similarly injected and cultured in the presence of E2 from stage 13 (D) develop with reduced (arrow) or lacking ventral blood islands (arrowhead). (E) Northern blotting analysis using 10 μg per lane of total RNA harvested from pools of embryos injected at the 2-cell stage with 0.8 ng/embryo of ER-xSmad6 and cultured in E2 from the indicated stages (NH = no hormone). The filter was first probed with αT1-globin, then stripped and reprobed with EF-1α as a loading control. (F) Graphic representation of normalized values from panel E. Original magnification for A-D, × 20.
ER-xSmad6 inhibits primitive blood island development during neurulation. (A-B) Water-injected embryos cultured in the absence (A) or presence (B) of E2 develop normal ventral blood islands as assayed by benzidine staining at stage 35. (C-D) Embryos injected with 1 ng/embryo in the ventral blastomeres of 4-cell stage embryos and cultured in the absence of E2 (C) develop normal ventral blood islands. Embryos similarly injected and cultured in the presence of E2 from stage 13 (D) develop with reduced (arrow) or lacking ventral blood islands (arrowhead). (E) Northern blotting analysis using 10 μg per lane of total RNA harvested from pools of embryos injected at the 2-cell stage with 0.8 ng/embryo of ER-xSmad6 and cultured in E2 from the indicated stages (NH = no hormone). The filter was first probed with αT1-globin, then stripped and reprobed with EF-1α as a loading control. (F) Graphic representation of normalized values from panel E. Original magnification for A-D, × 20.
Late induction of ER-xSmad6 decreases expression levels for globin and GATA-1
The defect in benzidine staining could be due to a block in erythropoiesis at any of several levels. To determine if the defect is related to the erythroid transcriptional program, embryos expressing ER-xSmad6 were incubated with E2 starting at various stages; RNA was harvested from all the embryos at stage 35/36 and analyzed by Northern blotting experiments for globin transcripts. As shown in Figure 6E, activation of ER-Smad6 prior to gastrulation (stage 7) or during gastrulation (stage 10.5) nearly eliminates globin mRNA, compared with embryos that were not exposed to E2. The results at these stages are consistent with the failure to specify hematopoietic mesoderm. However, when exposure to E2 was delayed until stage 15 (neurulation), the resulting embryos still show a significant relative decrease in globin transcript levels. If E2 is not added until stage 23, the embryos express normal levels of globin RNA, but this may simply reflect the fact that ER-xSmad6 protein levels decline by this stage (Figure 5A). Levels of globin transcript in the injected, untreated controls (NH) are equivalent to noninjected embryos (not shown).
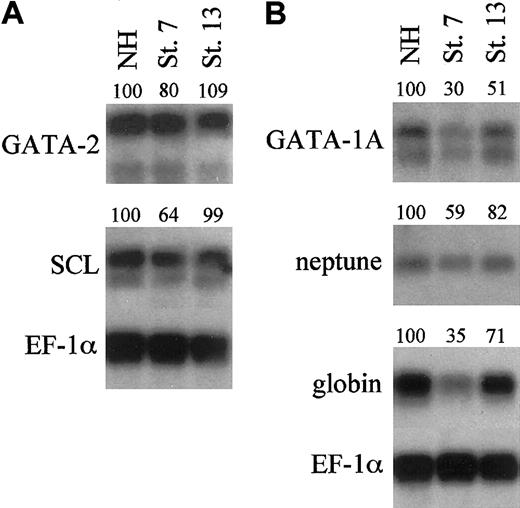
To determine if this defect in erythropoiesis acts at the level of regulatory genes, the transcript levels for known hematopoietic transcription factors were analyzed quantitatively in polyA+ Northern blotting experiments. Embryos were treated with E2 at stage 7 or 13 and harvested at either stage 22 or stage 35/36 to analyze early or late markers of primitive hematopoiesis, respectively. As expected, transcript levels for GATA-2 and SCL are decreased when embryos are dorsalized by induction of ER-xSmad6 at stage 7 (Figure 7A). Interestingly, when injected embryos were induced at the onset of neurulation (stage 13), transcript levels of GATA-2 and SCL at stage 22 are equivalent to untreated controls. This suggests that these early specification genes (which are expressed already during gastrula stages) do not require subsequent BMP signals to maintain steady-state transcript levels at least up to stage 22. Transcript levels for globin, neptune, and GATA-1 were also reduced in the stage-7 induced embryos (Figure 7B), as expected. When embryos were treated with E2 at stage 13, GATA-1, Neptune, and globin transcript levels were decreased by 20% to 50% at stage 35. GATA-1 mRNA levels were too low to assess quantitatively at stage 22, but reverse transcriptase (RT)–PCR analysis indicated that, similar to GATA-2 and SCL, these were normal during those early stages of VBI development, as long as E2 addition was delayed to stage 13 (not shown). These data suggest that BMP signaling continues to function at relatively late stages of blood island development for efficient differentiation of the primitive erythroid lineage.
Blocking BMP signaling during neurulation reduces differentiation markers of primitive hematopoiesis. Northern blotting analysis using 0.8 μg per lane of polyA+ mRNA harvested from uninjected embryos or embryos injected at the 2-cell stage with 2 ng/embryo of ER-xSmad6 mRNA and induced with E2 at the stages indicated (NH = no hormone). mRNA was harvested from stage 22 embryos (A) to test early markers of primitive hematopoiesis (GATA-2 and SCL) or from stage 35/36 embryos (B) to test late markers (GATA-1A, neptune, and αT1-globin). Numbers above lanes indicate percent of signal relative to the sample that is uninduced (NH).
Blocking BMP signaling during neurulation reduces differentiation markers of primitive hematopoiesis. Northern blotting analysis using 0.8 μg per lane of polyA+ mRNA harvested from uninjected embryos or embryos injected at the 2-cell stage with 2 ng/embryo of ER-xSmad6 mRNA and induced with E2 at the stages indicated (NH = no hormone). mRNA was harvested from stage 22 embryos (A) to test early markers of primitive hematopoiesis (GATA-2 and SCL) or from stage 35/36 embryos (B) to test late markers (GATA-1A, neptune, and αT1-globin). Numbers above lanes indicate percent of signal relative to the sample that is uninduced (NH).
Erythroid cells derived from either dorsal or ventral blastomeres require BMP signaling during and after gastrulation
Recent studies revealed that the VBI is composed of distinct anterior and posterior domains that are derived from early dorsal- or ventral-fated blastomeres.13 To determine if the anterior VBI (aVBI) is dependent on BMP signaling, embryos were injected at the 4-cell stage into the 2 prospective dorsal blastomeres and induced with E2 at stage 10.5. Similar experiments using ΔBR had shown that this has no effect on dorsal/ventral patterning,55 and indeed no morphologic defects occurred. However, benzidine staining shows that the aVBI fails to differentiate primitive erythroid cells (Figure 8A,C; Table 1). We next tested if targeting ER-xSmad6 RNA to the 2 ventral-fated blastomeres at the 4-cell stage would be sufficient to inhibit VBI development in the posterior compartment. Embryos treated with E2 at stage 10.5 did have a significant block to erythropoiesis, determined by staining for hemoglobin with benzidine. This block was restricted to the posterior VBI (pVBI) (Figure 8B,D; Table 1), and the embryos retained a small segment of differentiated anterior blood island. Therefore, the entire blood island is dependent on BMP signaling for erythroid differentiation, regardless of the embryonic origin of the progenitors.
Both anterior and posterior VBI are dependent on BMP signaling for erythropoiesis. (A-D) Embryos were injected into dorsal (A,C) or ventral (B,D) blastomeres at the 4-cell stage with 0.8 ng/embryo of ER-xSmad6 mRNA and cultured in the absence (A-B) or presence (C-D) of E2 from stage 10.5. Embryos were fixed and stained with benzidine (blue stain) at stage 35. Arrowheads indicate approximate location of liver primordium and lines indicate extent of ventral blood island staining. Original magnification, × 20.
Both anterior and posterior VBI are dependent on BMP signaling for erythropoiesis. (A-D) Embryos were injected into dorsal (A,C) or ventral (B,D) blastomeres at the 4-cell stage with 0.8 ng/embryo of ER-xSmad6 mRNA and cultured in the absence (A-B) or presence (C-D) of E2 from stage 10.5. Embryos were fixed and stained with benzidine (blue stain) at stage 35. Arrowheads indicate approximate location of liver primordium and lines indicate extent of ventral blood island staining. Original magnification, × 20.
ER-xSmad6 blocks conditionally both aVBI and pVBI development
. | . | Ventral blood island phenotype, % embryos . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| Injection site (n) . | Induced at stage 10.5 . | Normal VBI . | Anterior VBI only . | Posterior VBI only . | No VBI . | |||
| Dorsal (11) | No | 100 | 0 | 0 | 0 | |||
| Dorsal (16) | Yes | 13 | 0 | 56 | 31 | |||
| Ventral (40) | No | 63 | 17 | 5 | 15 | |||
| Ventral (30) | Yes | 27 | 67 | 6 | 0 | |||
. | . | Ventral blood island phenotype, % embryos . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| Injection site (n) . | Induced at stage 10.5 . | Normal VBI . | Anterior VBI only . | Posterior VBI only . | No VBI . | |||
| Dorsal (11) | No | 100 | 0 | 0 | 0 | |||
| Dorsal (16) | Yes | 13 | 0 | 56 | 31 | |||
| Ventral (40) | No | 63 | 17 | 5 | 15 | |||
| Ventral (30) | Yes | 27 | 67 | 6 | 0 | |||
Embryos were injected with ER-xSmad6 mRNA at the 4-cell stage into either the 2 presumptive dorsal or ventral blastomeres (n = number of embryos) and either cultured alone or induced at stage 10.5 with E2. Some leakiness of the fusion protein is evident in the ventral-targeted injections cultured in the absence of E2, but there is a clear trend toward blocking of either anterior or posterior domains by activated ER-xSmad6.
Overexpression of xSmad1, but not xSmad2, rescues the ER-xSmad6 phenotype
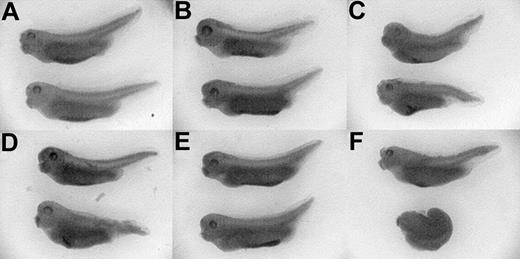
BMP signaling is mediated by 3 receptor-activated Smads, (1/5/8), whereas activin/TGF-β signaling is mediated by Smads 2/3. To confirm that ER-xSmad6 interferes with blood development by antagonizing BMP-dependent Smads, rescue experiments were performed using specific R-Smads (Figure 9). When control xEF-1α mRNA was coinjected with ER-xSmad6 mRNA into the ventral blastomeres of 4-cell embryos, the embryos had absent or reduced posterior blood islands after addition of E2 at stage 10.5, whereas the injected, untreated embryos had normal (anterior and posterior) ventral blood islands (Figure 9A,D). When xSmad1 mRNA was coinjected with ER-xSmad6 mRNA, the induced embryos developed normal VBIs (Figure 9E). When xSmad2 mRNA was coinjected with ER-xSmad6 mRNA, the untreated embryos developed only anterior blood islands (indicating that Smad2 inhibits VBI development), and the E2-induced embryos were either fully dorsalized or developed only anterior blood islands (Figure 9C,F). These data support the conclusion that ER-xSmad6 is a specific inhibitor of BMP-dependent Smad signaling and indicate that the postmesoderm specification requirement for BMP signaling in the primitive hematopoietic compartment is Smad-dependent.
xSmad1, but not xSmad2, can rescue the ER-xSmad6–induced block of primitive erythropoiesis. Embryos were coinjected at the 4-cell stage in the ventral blastomeres with 1 ng/embryo ER-xSmad6 mRNA and either 0.6 ng/embryo of EF-1α (A,D), xSmad1 (B,E), or xSmad2 (C,F) mRNA. Half of each cohort of injected embryos was cultured in the presence of E2 from stage 10.5 (D-F). All embryos were fixed and stained with benzidine at stage 35. Original magnification, × 20.
xSmad1, but not xSmad2, can rescue the ER-xSmad6–induced block of primitive erythropoiesis. Embryos were coinjected at the 4-cell stage in the ventral blastomeres with 1 ng/embryo ER-xSmad6 mRNA and either 0.6 ng/embryo of EF-1α (A,D), xSmad1 (B,E), or xSmad2 (C,F) mRNA. Half of each cohort of injected embryos was cultured in the presence of E2 from stage 10.5 (D-F). All embryos were fixed and stained with benzidine at stage 35. Original magnification, × 20.
BMP signaling is differentially required in mesoderm and overlying ectoderm
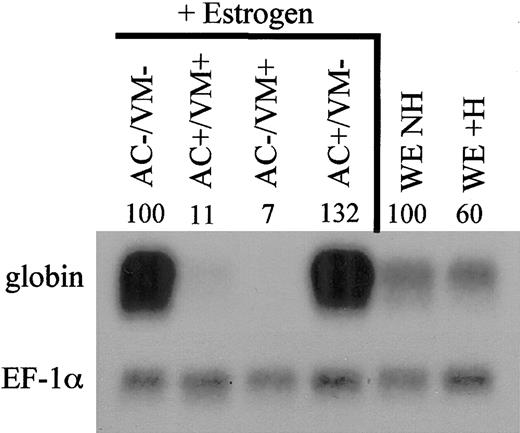
Previous studies suggested that ectoderm overlying ventral mesoderm (VM) during gastrulation provides a stimulatory factor required for erythropoiesis.56 In explant assays, stage-10 ventral mesoderm cultured alone fails to develop erythroid cells, while globin expression is induced if the explant is cocultured with stage-10 (but not stage-7) animal cap (AC) presumptive ectoderm.33,76 BMP4 is expressed in both the mesoderm and ectoderm, and forced expression of BMP4 confers the ability of stage-7 AC ectoderm to induce globin in VM explants.33 By attempting to target the dominant-negative BMP receptor to these tissues, evidence was presented that BMP signaling in ectoderm is required for erythroid differentiation in the adjacent mesoderm.76 Using ER-xSmad6, we are now able to investigate the temporal requirement of Smad signaling in defined embryonic tissues. For this purpose, VM and AC explants were isolated from embryos derived from ER-xSmad6 injected or uninjected eggs (Figure 10). When uninjected AC and VM explants are cocultured in the presence of E2 from the equivalent of stage 13, globin is readily induced. If AC and VM explants both preloaded with ER-xSmad6 are cocultured in the presence of E2 from stage 13, globin expression is blocked. In other samples, equivalent cultures were prepared using explant pieces that differentially express ER-xSmad6. When the AC explant (but not the VM explant) expressed ER-xSmad6, globin expression was not effected (or even increased) compared with the control explants in which neither piece expressed the fusion protein. In marked contrast, globin expression was inhibited when the VM explant (but not the AC explant) was blocked for Smad signaling by ER-xSmad6 at stage 13. These data clearly indicate that during VBI development, at least 2 distinct signals are required for efficient primitive erythropoiesis. One is provided by ectoderm during gastrulation, while BMP-dependent Smad signaling is required after gastrulation, but instead within ventral mesoderm.
BMP signaling in ventral mesoderm is required for erythroid development. Animal cap and ventral mesoderm explants were isolated at stage 10 from uninjected embryos, or embryos injected at the 2-cell stage with 2 ng/embryo of ER-xSmad6 mRNA, and cocultured in the combinations as indicated (WE = whole embryo, NH = no hormone, and + or – indicates presence or absence, respectively, of ER-xSmad6). E2 was added to all explant cocultures at the equivalent of stage 13. Total RNA was harvested from explants and control whole embryos at stage 35 and samples were analyzed by Northern blotting (2 μg per lane). The filter was first probed with αT1-globin, then stripped and reprobed with EF-1α as a loading control. Numbers above lanes indicate percent of signal relative to uninjected control.
BMP signaling in ventral mesoderm is required for erythroid development. Animal cap and ventral mesoderm explants were isolated at stage 10 from uninjected embryos, or embryos injected at the 2-cell stage with 2 ng/embryo of ER-xSmad6 mRNA, and cocultured in the combinations as indicated (WE = whole embryo, NH = no hormone, and + or – indicates presence or absence, respectively, of ER-xSmad6). E2 was added to all explant cocultures at the equivalent of stage 13. Total RNA was harvested from explants and control whole embryos at stage 35 and samples were analyzed by Northern blotting (2 μg per lane). The filter was first probed with αT1-globin, then stripped and reprobed with EF-1α as a loading control. Numbers above lanes indicate percent of signal relative to uninjected control.
Discussion
An inducible inhibitor of BMP-dependent Smad signaling demonstrates a role for active signaling in mesoderm after progenitor specification
BMP signaling is implicated in regulating primitive erythropoiesis based on the ability of BMP4 to induce the erythroid program in uncommitted AC cells that serve as a model stem cell system in Xenopus.15,33 Similar ectopic expression studies show that a conserved pathway functions in murine ES cells.19,32 This is supported by genetic studies since BMP-4–/– embryos that survive long enough to be analyzed lack yolk sac blood islands.17 Zebrafish mutants defective in BMP signaling components also lack primitive blood.57 However, it has been less clear if BMP function is restricted to the early specification of hematopoietic progenitors, since the approaches used (null mutations or forced expression of dominant-negative receptors) inherently lead to mesoderm patterning defects that preclude a clear examination of BMP signaling at later developmental stages.
In order to address the question of whether BMP-dependent Smad signaling is required after hematopoietic progenitors are specified, we generated ER-xSmad6, a cell-autonomous inducible inhibitor of BMP-regulated Smads. Induction of this protein during neurulation, after the erythroid regulatory program has already been initiated in the presumptive VBI, is sufficient to cause quantitative defects in primitive erythropoiesis. Morphologic analysis showed that induction of ER-xSmad6 before gastrulation causes a dorsalized phenotype, consistent with the phenotype of embryos where BMP signaling is blocked in a nonconditional manner.45,47,55,58 BMP signaling at this time is required for specification of the embryonic hematopoietic progenitors in ventral mesoderm.57 BMPs are also required at this time for ectoderm development, which in the absence of BMP signals forms a default neural fate.59 Indeed BMP4,36,60 active BMP signaling,61 and expression of GATA-262,63 all occur in ventral mesoderm and ectoderm during gastrula stages. The results of Maeno and colleagues33,56,76 indicate that BMP signaling in the ventral ectoderm is also important at these stages for hematopoiesis in the underlying ventral mesoderm.
In contrast, induction of ER-xSmad6 at the onset (stage 13) or during neurulation does not affect morphologic development of the embryos, yet causes a reduction in primitive erythropoiesis. We showed that this inhibitor blocks a BMP-regulated Smad-dependent pathway since xSmad1 rescues the defect, while xSmad2 does not. BMP signaling remains active in the presumptive ventral blood island during neurula stages at least up to stage 20, demonstrated by immunostaining for activated xSmad1.64 The erythroid regulatory program is initiated by stage 11, based on the expression of critical regulatory transcription factors including GATA-2, LMO-2, SCL, and GATA-1.34,62,65,66 The approach of using an inducible inhibitor permits the normal specification of progenitors to occur during gastrulation and demonstrates that continued BMP signaling is required for efficient primitive erythropoiesis. When ER-xSmad6 is activated during these later stages (stage 13), the expression levels of GATA-2 and SCL are not reduced during VBI development. This indicates that the initial induction events were sufficient to activate these genes associated with early progenitor state, and BMP signaling is no longer required after gastrulation for their maintenance. However, the eventual reduction in globin, neptune, and GATA-1 transcript levels demonstrate that continued BMP signaling is still required for the efficient differentiation of primitive erythroid cells in the VBI. Our explant assays support the idea that this signal must occur in mesoderm and is no longer required by stage 13 in ectoderm, although this does not distinguish whether the signals act on the progenitors themselves or act indirectly through stromal elements of the mesoderm.
Consistent with the explant studies of Maeno and colleagues,33,56,76 Walters et al26 confirmed that BMP signaling is required in ectoderm for embryonic erythropoiesis, since expression of a dominant-negative receptor in blastomeres that give rise to anterior ectoderm derivatives (and not VBI cells) is sufficient to block globin expression. This occurs even though hematopoietic specification is not affected in mesoderm, based on normal expression of SCL. Inhibition of BMP signaling in the marginal zone also leads to defects in erythropoiesis, but this functions at the earlier level of specification, since SCL is reduced in these embryos.67 By using targeted expression of an inducible xSmad6, our data can be placed in the context of this work to now define 3 distinct roles for BMP signaling. Initially, BMP signals are needed in mesoderm for specification of hematopoietic progenitors and also in ectoderm (negatively regulated by CaM KIV26 ) for expression of a signal that is required not for specification but for erythroid differentiation. Using the ER-xSmad6 conditional block, we now identify a third function for BMP signaling following specification that works in mesoderm (not ectoderm) and is also required for normal levels of erythroid differentiation.
BMP signaling is required throughout the VBI
Tracey et al12 first proposed, based on the expression pattern of xAML1, that the VBI is composed of both anterior and posterior halves, derived from presumptive dorsal or ventral blastomeres, respectively. Fate-mapping studies have since confirmed that anterior and posterior halves of the VBI are derived from different blastomeres of the 32-cell stage embryo.13,68 We show that both halves of the ventral blood island require BMP signaling for their development. This has also been demonstrated independently with respect to earlier requirements during specification.67 However, we note that the temporal requirement for BMP signaling may differ between the 2 halves of the blood island. By stage 15, erythroid differentiation occurs normally in the aVBI, while cells of the pVBI remain sensitive to the blocking activity of ER-xSmad6 (data not shown). This suggests that the aVBI is influenced by a distinct signaling source. A likely candidate is the developing fetal liver, which lies at the anterior boundary of the developing VBI. BMPs may provide a specific cue for the pVBI cells which lie caudal, or permit survival of these progenitors until additional differentiation cues function; it has long been noted that the VBI differentiates in a rostral-caudal wave.69,70
The requirement for BMPs in mesoderm after specification is most likely a survival cue that requires expression of GATA-1. When signaling is blocked, expression of GATA-1 is not maintained. GATA-1 is required for the survival of mature erythrocytes and in the absence of GATA-1 erythroid cells undergo apoptosis.71 Regulatory pathways downstream of BMP signaling that function in the ectoderm (early) or mesoderm (early or later) for specification and differentiation of primitive erythrocytes are yet to be defined. Candidate factors include Mix.1,72 GATA-2,33 and target genes of Smad5,73,74 all of which are also expressed in definitive progenitor cells. Therefore, defining the key signaling pathways downstream of BMPs that control primitive hematopoiesis may be relevant as well to adult-stage bone marrow–derived definitive hematopoiesis.75
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2003-04-1094.
Supported by grant HL56182 from the National Institutes of Health (NIH) (T.E.), the Irma T. Hirschl Trust (T.E.), and NIH training grant T32GM 07491 (M.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ashwini Ghatpande for technical assistance; Joan Massague for the pBRE-Luc reporter; Erwin Böttinger for the p3TP-Lux reporter; Jan Christian for the xSmad6 cDNA; Gerry Thompson for the xSmad1 cDNA; Doug Melton for the xSmad2 cDNA; Len Zon for the xSCL, neptune, and xGATA-2 cDNAs; Ali Hemmati-Brivanlou for the activin βB cDNA; Antonio Iavarone for the hER ligand-binding domain; and Chris Wright for the xBMP4 cDNA. We thank the Albert Einstein College of Medicine Analytical Imaging Facility for technical assistance. We thank Charlie Hall for statistical assistance. We also would like to thank Brian Zafonte, Yuko Miyanaga, and Tal Oren for their helpful criticism of this manuscript.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal