Abstract
Lineage-marker depleted (Lin–) murine bone marrow cells expressing stem cell antigen 1 (Sca-1) were sorted on the basis of stem cell factor receptor (c-kit) expression to obtain Lin–Sca+Kit+ or Lin–Sca+Kit– cells. Lin–Sca+Kit– cells have a markedly greater chemotactic response to stromal derived factor-1 (SDF-1). Using a novel fluorescent stain, we show that both populations generate similar levels of a key messenger, phosphatidylinositol 3,4,5 trisphosphate (PIP3), in response to SDF-1. Differences in motile behavior may therefore lie downstream of phosphatidylinositol 3-kinase (PI3-kinase) activation at the level of cytoskeleton regulation. The 2 cell populations were compared using 2-dimensional difference gel electrophoresis (2D-DIGE), with a maleimide CyDye fluorescent protein labeling technique that has enhanced sensitivity for low abundance samples. Comparative proteomic analysis of Cy3- and Cy5-labeled protein samples allows relative quantification of protein spots present in both cell populations; of these, 73% were common. Key protein differences were adseverin and gelsolin, actin micro-filament splicing proteins, regulated by Rac, downstream of PI3-kinase activation. Adseverin was shown to be acetylated, a novel modification for this protein. Differences in major regulators of cell shape and motility between the 2 populations can explain the differential response to SDF-1.
Introduction
Hematopoietic stem cells have the capacity to self-renew and also to differentiate to form a number of mature cell types with distinct morphologies and functional activities. The distinctive features of the stem cell that give it longevity and the ability to balance its self-renewal and differentiation to the required degree in any circumstance remain unclear. These biologic properties are encoded in the transcriptional profile, or transcriptome, of stem cells. The properties of a stem cell can therefore be defined in terms of the gene expression profiles of stem cells. Retention of stem cell characteristics will rely in part on the cohort of genes specifically expressed in these primitive cells, but not in their maturing progeny. This axiom has led to transcriptional analysis of long- and short-term repopulating stem cells and other primitive populations (defined via expression of specific markers) to define the genes expressed for self-renewal and pluripotency.1,2 The generation of these stem cell “blueprints” then offers the opportunity to investigate the functional activity of the proteins produced as a consequence of gene expression and to systematically study gene networks controlling stem cell biology.
The relationship between messenger RNA production and protein synthesis is not, however, strictly correlative.3 In a range of different organisms it has become clear that changes in the expression of a specific protein do not necessarily relate directly to altered transcription. Recent developments in protein separation and relative quantification allow populations of cells to be compared at the proteomic level to complement observations within the transcriptome. Two-dimensional gel electrophoresis (2DE) combined with protein identification by mass spectrometry has already defined differences in hematopoietic cell lines which occur during differentiation or as a result of BCR/ABL oncogene expression.4-7 These approaches relied on intergel sample comparison using silver or Coomassie blue stains, which have a low dynamic range and thus could be subject to the inherent limitations of this classical experimental approach. The development of protein labeling techniques employing fluorescent dyes with distinct excitation/emission spectra but with the same chemical reactivity8,9 enables multiplexing of 2 or more samples so that they can be processed and run on the same gel. The large dynamic range of these dyes (4 orders of magnitude) allows relative quantities of proteins to be accurately determined. The new saturation labeling chemistries give a very high sensitivity of detection (< one ng). These are available as Cy3 and Cy5 for dual-labeling approaches.
These improvements in proteomics technology now make it feasible to obtain relative quantification on protein levels in stem cells. We have initiated such studies by comparison of 2 mouse bone marrow populations: lineage marker negative cells (Lin–), positive for expression of stem cell antigen-1 (Sca-1; Sca+), expressing or not expressing the c-kit cell surface cytokine receptor (Lin–Sca+Kit+ and Lin–Sca+Kit– cells, respectively). Lin–Sca+Kit+ cells are a primitive hematopoietic cell population with long-term marrow reconstitution potential and are predominantly more primitive than their Kit– counterparts. Primitive cells exhibit motile responses.10,11 These cells exhibit differential motility responses to the chemokine, stromal derived factor-1 (SDF-1), with Lin–Sca+Kit– cells showing a greater response than Lin–Sca+Kit+ cells. This initiating study has as its ultimate objective a systematic definition of the component parts of long-term reconstituting cells that define their stem cell nature. We report that definition of comparative protein levels in these 2 cell populations can be achieved using difference gel electrophoresis (DIGE). Sorting on a single parameter, c-kit expression, in Lin–Sca+ cells, major differences in the proteome are apparent. This approach, coupled with mass spectrometry, should facilitate investigation of the molecular basis of their biologic properties.
We have shown that stem cell chemotaxis promoted by SDF-1 and lysophospholipids is governed by the phosphatidylinositol 3-kinase (PI3-kinase)–mediated Vav guanyl nucleotide exchange factor activation of the Rac low-molecular-weight G protein.12 Rac governs hematopoietic stem cell motility13-17 and couples to gelsolin family members (adseverin and gelsolin) to initiate actin splicing and capping.18,19 Gelsolin null cells display decreased motility compared with their wild-type counterparts.20 We initiated a study, using proteomic analysis, aimed at determining aspects that may contribute to the differential motility of Kit+ and Kit– cells.
Materials and methods
Chemicals and reagents
General chemicals were obtained from Sigma-Aldrich (Poole, United Kingdom). Tissue-culture plastic was from Corning Costar (High Wycombe, United Kingdom). Flow cytometry reagents and those for 2D gel electrophoresis were obtained from Becton Dickinson (Oxford, United Kingdom) and from Amersham Biosciences (Chalfont St Giles, United Kingdom) respectively unless otherwise indicated. SDF-1 was from Calbiochem (Nottingham, United Kingdom).
Flow cytometry
Primitive hematopoietic cells were isolated from murine bone marrow from C57Bl/6 mice using techniques previously described.21,22 Briefly, femoral bone marrow was harvested and mononuclear cells prepared by density gradient centrifugation (Optiprep; Axis-Shield; PoC AS, Norway). Lineage-depleted (Lin–) cells were obtained using a cocktail of lineage marker antibodies (Pharmingen, Oxford, United Kingdom). Sheep antirat antibody magnetic beads (Dynal, Wirral, United Kingdom) were employed to deplete cells expressing lineage-specific markers. Lineage-depleted cells were then labeled with fluorescein isothiocyanate–labeled Sca-1 antibody and with phycoerythrin-labeled c-Kit antibody. The lineage-depleted cells were then sorted on a Facs Vantage flow cytometer, to prepare either lineage marker–negative Sca-1 expressing (Sca+) cells or Lin–Sca+ cells expressing c-Kit (Lin–Sca+Kit+ cells) or not (Lin–Sca+Kit–).
Chemotaxis assays
The migration of primary hematopoietic cells in response to agonists was assessed using a Boyden chamber assay system; 96-well transwell plates were used (Neuroprobe, Gaithersburg, MD). These consisted of 2 wells separated by a membrane containing 5-μM diameter pores. Cells (3 × 104 in 30 μL) were placed in the top well and agonists were added to the top and/or bottom wells (bottom well volume, 30 μL) in Iscove medium plus 20% vol/vol batch-tested horse serum. After 6 hours of incubation at 37°C in a 5% CO2 humidified incubator, viable cells in the lower well were counted. Assays of cell migration in the presence of agonists were linear for more than 10 hours.
Assay of phosphatidylinositol 3,4,5 trisphosphate (PIP3) levels
PIP3 levels in enriched primitive cells were analyzed by a PIP3 binding technique using fixed cells. Aliquots of Lin–Sca+Kit+ and Lin–Sca+Kit– cells were incubated with vehicle control or in the presence of 100 ng/mL SDF-1 for 5 minutes. After this time the tubes were placed on ice for 5 minutes to stop the reaction. Slides of 1.8 × 103 cells were prepared using a Shandon cytospin centrifuge and cells fixed in 4% wt/vol paraformaldehyde in phosphate-buffered saline (PBS) for 15 minutes. Cells were labeled using green fluorescent protein (GFP)-tagged pleckstrin homology (PH) domain of murine GRP1 that binds specifically to PIP322 using the following protocol. Cells were washed twice in PBS/0.1 vol/vol Tween 20 (PBST) to permeabilize prior to incubation in blocking solution (3% wt/vol bovine serum albumin in PBST). Following this step, cells were washed once with PBST and incubated with either 1 μg/mL of a dominant-negative form of PH general receptor for phosphoinositides 1 green fluorescent protein (DMN PH GRP1 GFP; this does not bind to PIP3 and gives a measure of nonspecific binding) or PH GRP1 GFP (total binding) for one hour in the dark. Slides were washed 3 times in PBST, nuclei counterstained with propidium iodide, and then samples were mounted with Vectashield mounting medium (Vector Laboratories, Peterborough, United Kingdom). Slides for each condition were prepared in duplicate. Confocal microscopy was performed using a Zeiss LSM 510-Meta microscope (Zeiss, Welwyn Garden City, United Kingdom). All fluorescence data were obtained using the LSM image capture and analysis software using the same camera settings for each data set.
Labeling of cell lysates with Cy3 and Cy5 saturation dyes
In all experiments proteins were labeled with maleimide derivatives of Cy3 and Cy5.8 Cells (0.1 × 106-1.0 × 106) were washed 3 times in PBS pH 7.4 and pelleted at 2000 rpm for 4 minutes. The resultant pellets were snap frozen and stored dry at –80°C until use. When required the cell pellet was resuspended in lysis buffer (9 M urea, 2 M thiourea, 4% wt/vol CHAPS (3-[(3-cholamidopropryl)dimethylammonio]-1-propane sulphonate), 0.5% Triton X-100) to give a concentration of 2.75 μg/μL. Samples were lysed by vortexing at room temperature for 2 minutes using a platform vortex and centrifuged at 13 500 rpm for 10 minutes using a benchtop microfuge. The supernatant was transferred to a fresh tube. Samples were reduced by incubation with 1 mM Tris (tris2-carboxyethyl)phosphine) for one hour at 37°C. Cell lysates were labeled in the ratio 25 μg protein:20 nmol Cy3 or Cy5 protein labeling dye (Amersham Biosciences) in dimethylformamide. After vortexing, samples were incubated for 30 minutes at 37°C. The reaction was quenched by addition of lysis buffer containing 4% vol/vol immobilized pH gradient (IPG) buffers 3-10 nonlinear (NL) (Amersham Biosciences) and 20 mM dithiothreitol. Samples were placed briefly on ice before transferring to storage at –80°C.
Two-dimensional gel electrophoresis
Cy3-labeled samples were mixed with Cy5-labeled protein prior to 2D gel electrophoresis. IPG strips (pH 3-10NL, 24 cm) were rehydrated according to the manufacturer's instructions (Amersham Biosciences) before isoelectric focusing for a total of 72 kVh using a Multiphor II apparatus and the second dimension was a standard sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) protocol using the ISODALT system (Amersham Biosciences). The second dimension was run on 10% wt/vol polyacrylamide gels as previously described.24 Gels were scanned while still between 2 low-fluorescence glass plates, at 532 nm (Cy3) and 633 nm (Cy5) using a Typhoon 8600 fluorescent scanner, and saved in .gel format using ImageQuant software (Amersham Biosciences). The excitation wavelengths for Cy3 and Cy5 are 540 nm and 620 nm, and the emission wavelengths are 590 nm and 680 nm for Cy3 and Cy5, respectively. The gels were removed from the plates and spots for mass spectrometric analysis were excised using a Bio-Rad Proteome Works (Bio-Rad, Hemel Hempstead, United Kingdom) spot picker with a 1.5 mm spot cutter head. Spots for picking were visualized using ultraviolet light and a digital camera with a 520-nm filter. Spot picking coordinates were set using PDQuest Image Analysis software (Bio-Rad).
Gel image analysis
Images were analyzed using Progenesis software (Non-Linear Dynamics, Newcastle, United Kingdom) using the following protocol. Spot detection and background subtraction were performed for the gel image sets imported in .gel format image files. Artefacts (eg, dust particles or streaks detected as protein spots) were removed by manual editing. Where appropriate, spot chains were split into separate entities.
This software automatically created a global reference gel based on the gel containing the most spots, which provides an effective spot index for the analysis. Averaged gels were generated for each of the Lin–Sca+Kit+, Lin–Sca+Kit– gel sets. The averaged gels are a statistical combination of several gels to produce a gel that has mean spot values and associated error terms and provides information about spot variation within the gel set. These gel groups were created for spot pattern comparison. The requirement for inclusion of a spot in the averaged gel was that it be absent from no more than a single gel.
The averaged and reference gels were then recreated to reflect the amended spot detection. Background detection and normalization were performed again to account for any changes in overall volume caused by the manual editing. Once all editing and rematching had been completed, the gels were analyzed for protein spot differences. To compare protein profiles between 2 samples that had been run on the same gel or between samples run on different gels, a normalization procedure was employed to allow for variation in total protein loading onto the gel(s). Total spot volume was calculated and each spot was assigned a normalized spot volume as a proportion of this total value. Normalized spot volumes were compared between Cy3- and Cy5-labeled samples on each gel. Excluding spots that were unmatched to the reference gel did not affect relative quantification of matched spots to a significant extent. Difference thresholds were then applied to identify the proteins with a 1.5-, 2-, or more than 10-fold difference in normalized spot volume.
Mass spectrometry
Spots of interest were excised from the gel, followed by destaining, reduction, alkylation, and digestion with modified porcine trypsin (Promega, Southampton, United Kingdom) as previously described.24 Samples for matrix-assisted laser desorption ionization/time-of-flight (MALDI-ToF) analysis were prepared by mixing a small aliquot of the digest supernatant with an equal volume of a solution of α-cyano-4-hydroxycinnamic acid (10 mg/mL in 1:1 acetonitrile:0.1% vol/vol trifluoroacetic acid). Peptide mass fingerprinting was performed on a reflectron MALDI-ToF mass spectrometer (Micromass, Manchester, United Kingdom). All mass spectra were internally calibrated with trypsin autolysis peaks (mass/charge [m/z] 842.51 or 2211.10). Mascot software (Matrix Science, London, United Kingdom) was employed for protein database searching using monoisotopic mass values for each spectrum. Protein identity was based on at least 5 matching peptides, searching peptide masses allowing for one missed tryptic cleavage with a mass tolerance of 0.2 Da deriving a molecular weight score (MOWSE) with a P value less than .05 within the Mascot software. The monoisotopic mass for the Cy3 tag is 672.3 and for Cy5 684.3. The average masses are 672.8 (Cy3) and 684.8 (Cy5). Allowance was made for these covalent modifications to peptides in all peptide mass fingerprint database searches. Tandem mass spectrometry was performed on a Bruker Ultraflex ToF-ToF instrument25 (Bruker Daltonics, Bremen, Germany).
Transcriptional analysis
The mRNA expression levels for adseverin and gelsolin were assessed using semiquantitative PolyA PCR. The method and its semiquantitative nature have been described previously.26 Primers used were, for gelsolin, caagccatcaatgtcaccaa and agcaattctgtaaagaccctg, and for adseverin, ttgctgtttcccatctcaag and tgctattgagcatgtgctatc. The PCR was repeated 5 times and products visualized using agarose gel electrophoresis. The semiquantitative nature was illustrated by undertaking the PCR on a 10%, 20%, 50%, and 150% dilution series of the cDNA used. Densitometry was undertaken using ImageQuant software (Amersham Biosciences) and the results expressed as a percentage of the starting material.
Results
The differential motility of Lin–Sca+Kit– and Lin–Sca+Kit+ cells in response to SDF-1 is independent of PI3-kinase activation
Primitive hematopoietic cells were enriched from normal murine bone marrow using a 4-stage protocol. Cells were first enriched by density gradient centrifugation and then depleted of cells bearing lineage markers.11,21,22 Finally, cells were sorted for expression of Sca-1 antigen and also either expression or absence of c-kit on the cell surface. These cells show differential motility responses12 : Lin–Sca+Kit+ cells exhibit a reduced chemokinetic response to SDF-1 compared with Lin–Sca+Kit– cells (Table 1). SDF-1 is a chemoattractant for primitive cells,27,28 which promotes migration of hematopoietic progenitor cells via activation of phosphoinositide 3-kinase (Ptd Ins 3-K)29 to generate PIP3, a second messenger that is essential for motile responses in stem cells.12,29
The migration of Lin–Sca+Kit+ and Lin–Sca+Kit– cells in a diffusion chamber assay
Sample . | Kit+ cell migration, % of total . | Kit- cell migration, % of total . |
|---|---|---|
| No addition | 2.2 ± 0.3 | 10.5 ± 0.5 |
| With SDF-1 | 7.9 ± 0.5 | 58.4 ± 0.9 |
Sample . | Kit+ cell migration, % of total . | Kit- cell migration, % of total . |
|---|---|---|
| No addition | 2.2 ± 0.3 | 10.5 ± 0.5 |
| With SDF-1 | 7.9 ± 0.5 | 58.4 ± 0.9 |
SDF-1 was present at 150 ng/mL in the bottom well of a Boyden chamber. The results shown are the mean ± SEM of 3 observations.
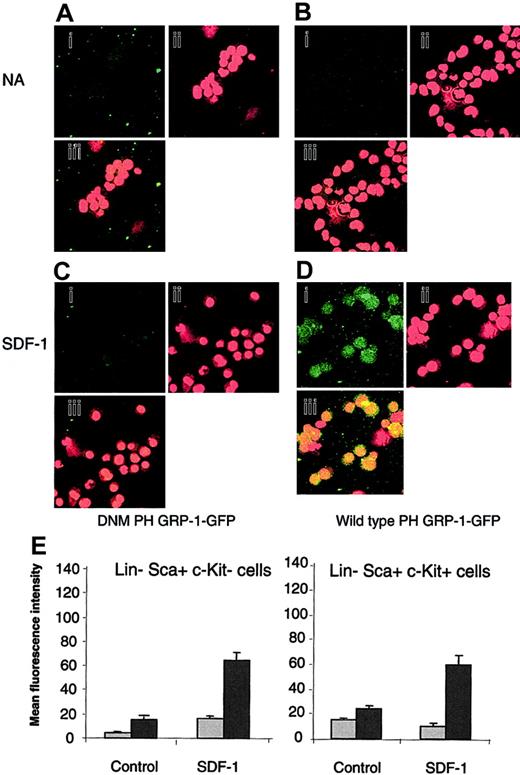
The generation of PIP3 in response to SDF-1 was assessed to determine if the differential response of Lin–Sca+Kit– and Lin–Sca+Kit+ cells to SDF-1 occurs due to differential PIP3 production. A technique based on the selective binding of the PH domain of the GRP1 to PIP3 was used.23 Cells were stained with a fluorescently labeled (GFP tagged) form of the PH domain of GRP1 (PH GRP1 GFP). Background fluorescence was determined using a mutant form of PH GRP1 GFP that is unable to bind PIP3 (DMN PH GRP1 GFP). The data are shown in Figure 1A-D for Lin–Sca+Kit+.
Measurement of PIP3 levels in response to SDF-1 stimulation in Lin–Sca+Kit– and Lin–Sca+Kit+ cells using a selective fluorescence probe. Representative images for unstimulated Lin–Sca+Kit+ (control) cells (A-B) and cells stimulated with SDF-1 for 5 minutes (C-D). Cells were incubated with DNM PH GRP1 GFP (A,C) or wild-type PH-GRP1 GFP (B,D). (A-D) GFP staining (i); propidium iodide (nuclear) staining (ii); and (iii) merged GFP/propidium iodide images of X, Y split images shown in (i) and (ii). Original magnification, ×40. (E) Unstimulated cells (NA) and cells stimulated with SDF-1 for 5 minutes were labeled with either a dominant-negative form of PH GRP1 GFP (DMN PH GRP1 GFP; nonspecific binding; ▦) or wild-type PH GRP1 GFP (total binding; ▪). GFP fluorescence is expressed as mean fluorescence intensity and data are presented as the mean plus or minus SEM from 3 experiments; fluorescence measurements were performed on 12 to 15 cells from cytospin preparations prepared in duplicate.
Measurement of PIP3 levels in response to SDF-1 stimulation in Lin–Sca+Kit– and Lin–Sca+Kit+ cells using a selective fluorescence probe. Representative images for unstimulated Lin–Sca+Kit+ (control) cells (A-B) and cells stimulated with SDF-1 for 5 minutes (C-D). Cells were incubated with DNM PH GRP1 GFP (A,C) or wild-type PH-GRP1 GFP (B,D). (A-D) GFP staining (i); propidium iodide (nuclear) staining (ii); and (iii) merged GFP/propidium iodide images of X, Y split images shown in (i) and (ii). Original magnification, ×40. (E) Unstimulated cells (NA) and cells stimulated with SDF-1 for 5 minutes were labeled with either a dominant-negative form of PH GRP1 GFP (DMN PH GRP1 GFP; nonspecific binding; ▦) or wild-type PH GRP1 GFP (total binding; ▪). GFP fluorescence is expressed as mean fluorescence intensity and data are presented as the mean plus or minus SEM from 3 experiments; fluorescence measurements were performed on 12 to 15 cells from cytospin preparations prepared in duplicate.
Similar results were obtained for Lin–Sca+Kit– cells so that, in contrast to the differential motility responses to SDF-1, both cell populations produce PIP3 upon addition of this chemokine to a similar degree (Figure 1E).
It is important to note that PI3-kinase inhibitors completely abrogate agonist-stimulated motility and chemotaxis in Lin–Sca+Kit– and Lin–Sca+Kit+ cells.12 These data therefore suggest that the mechanism of the differential response is independent of PIP3 generation.
Analysis of protein expression profiles of Lin–Sca+Kit–/+ cells using maleimide Cy dyes
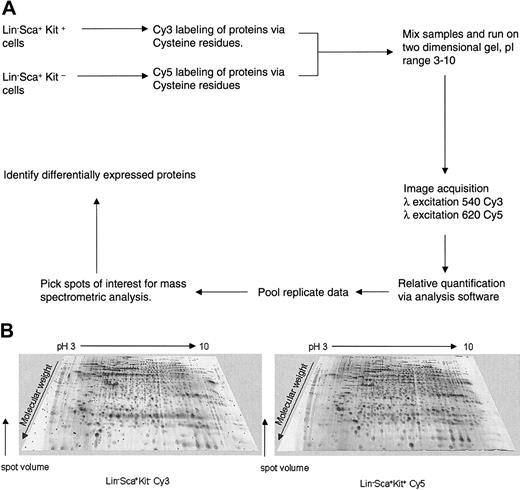
Protein analysis of highly enriched primitive hematopoietic cells can be achieved using the novel approach of 2-dimensional difference gel electrophoresis (2D-DIGE) analysis using maleimide CyDye protein labeling (Amersham Biosciences). This was used to study protein expression profiles of Lin–Sca+Kit– and Lin–Sca+Kit+ cells to identify potential cytoskeletal regulatory components that may explain the different motile behavior of these 2 cell populations. The DIGE technology (Amersham Biosciences) is based on differential protein labeling with fluorescent Cy dyes, which allows sample multiplexing. The workflow of such proteomic analysis is shown in Figure 2A.
2D-DIGE analysis of primitive hematopoietic cells. (A) Workflow for proteomic analysis of Lin–Sca+Kit+ and Lin–Sca+Kit– using Cy dye reagents. The protocol outlined above allows comparison of protein expression profiles, relative protein quantification, and identification of proteins of interest. (B) Representative 2D gel images of Lin–Sca+Kit+ and Lin–Sca+Kit– cells labeled with saturation dyes. The 2 images shown are from a saturation-labeled Cy dye protein gel where the excitation and emission wavelengths were set for Cy3 (left image) or Cy5 (right image). The grayscale 3-dimensional representations show representative gels from Lin–Sca+Kit– and Lin–Sca+Kit+ labeled with Cy3 and Cy5, respectively.
2D-DIGE analysis of primitive hematopoietic cells. (A) Workflow for proteomic analysis of Lin–Sca+Kit+ and Lin–Sca+Kit– using Cy dye reagents. The protocol outlined above allows comparison of protein expression profiles, relative protein quantification, and identification of proteins of interest. (B) Representative 2D gel images of Lin–Sca+Kit+ and Lin–Sca+Kit– cells labeled with saturation dyes. The 2 images shown are from a saturation-labeled Cy dye protein gel where the excitation and emission wavelengths were set for Cy3 (left image) or Cy5 (right image). The grayscale 3-dimensional representations show representative gels from Lin–Sca+Kit– and Lin–Sca+Kit+ labeled with Cy3 and Cy5, respectively.
The first generation of Cy dyes (the “minimal” dyes) label epsilon-amino groups of lysine residues and are N-hydroxysuccinimidyl esters. The lysine content of proteins is relatively high and 5% proteins are typically labeled.9,30 The cysteine-labeling maleimide dyes are a further development of this technique and label cysteine (which is less prevalent than lysine) to saturation to obtain enhanced sensitivity of protein detection. The sensitivity and dynamic range of these dyes make them highly suitable for samples where protein amounts are limiting.8,9
The Cy3 and Cy5 maleimide (saturation) dyes are mass- and charge-matched to minimize differential labeling effects. To determine whether or not preferential labeling with Cy3 over Cy5 or vice versa would lead to artefactual difference generation, experiments were performed on a multipotent hematopoietic progenitor cell line, FDCP-mix. Replicate factor-dependent cell Paterson (FDCP)–mix samples were labeled with either saturation Cy3 or saturation Cy5 dyes; the samples were then run on the same 2D gel. The data revealed no differences in labeling properties of the 2 dyes (results not shown). To confirm Cy3 and Cy5 comparability, FDCP-mix samples were labeled with Cy3 or Cy5 and compared against Cy5- or Cy3-labeled Lin–Sca+Kit+ and Lin–Sca+Kit– cells (results not shown). This revealed that the FDCP-mix cells have an overall similarity in protein expression of 68.2% to Lin–Sca+Kit– and 77.8% to Lin–Sca+Kit+ primitive progenitor cells isolated by flow cytometry from mouse bone marrow.
Having confirmed that the saturation dyes work effectively, aliquots of 25 μg protein (approximately 2.5 × 105 cells) from 2 populations of Lin–Sca+Kit+ and Lin–Sca+Kit– cells were labeled with Cy3 or Cy5 saturation dyes respectively and run on the same 2D gel. Analysis by Progenesis software allowed spots to be detected and ascribed a normalized spot volume, which was then used as the comparator in intragel analyses. To control for dye-labeling artefacts, differences between Lin–Sca+Kit+ and Lin–Sca+Kit– cell samples were only considered significant if they also appeared on reciprocally labeled samples. Representative gels are shown in Figure 2B as 3-dimensional pseudocolor images for Lin–Sca+Kit– (Cy3) and Lin–Sca+Kit+ (Cy5) with spot intensity as the third dimension.
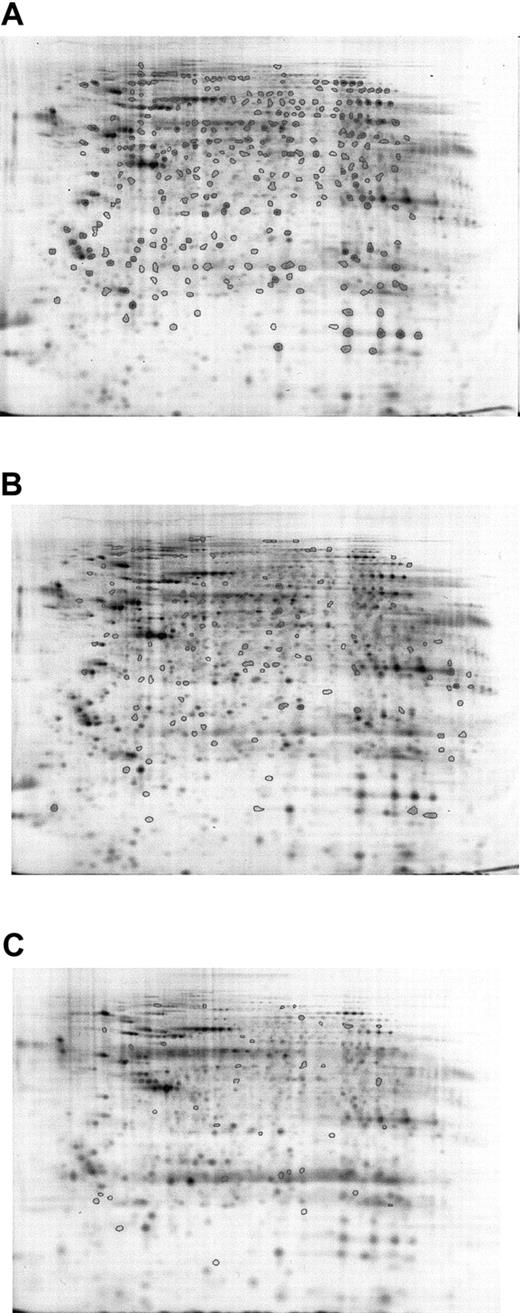
To confirm the results, averaged gels of Cy5- or Cy3-labeled Lin–Sca+Kit+ and Cy5- or Cy3-labeled Lin–Sca+Kit– samples were generated, and all spot differences highlighted in intragel comparisons were cross-checked against these. Five sets of Lin–Sca+Kit– and Lin–Sca+Kit+ cells were compared (4 Cy5-labeled and 1 Cy3-labeled populations for each). This was possible because preliminary experiments indicated no major differences in labeling patterns between the 2 dyes. Results from 5 different sets of Lin–Sca+Kit– and Lin–Sca+Kit+ experiments were pooled to derive data on the significant differences between the 2 populations. A mean value of 550 ± 32 (mean ± SEM; n = 10) protein spots was identified in the samples studied. Analysis of the gel images showed that there are major differences between the 2 cell populations. Progenesis analysis of the data obtained is shown in Figure 3. Spots common to all samples are shown in Figure 3A, those unique to Lin–Sca+Kit– and Lin–Sca+Kit+ are shown in Figure 3B-C, respectively. These can be summarized as 122 spots present in the Lin–Sca+Kit+-averaged gel compared with 41 in the Lin–Sca+Kit–-averaged gel.
Averaged gel comparison of Lin–Sca+Kit+ and Lin–Sca+Kit– cells labeled with saturation dyes. The use of averaged gels to allow sample comparison for Lin–Sca+Kit+ and Lin–Sca+Kit– cell lysates. Data from 5 sets of reciprocally labeled Lin–Sca+Kit–/+ populations were pooled and significant differences in protein expression (normalized volume) calculated using a 2-tailed Student t test. (A) All protein spots common to both gels are shown with enclosed areas. (B) Representative Lin–Sca+Kit– gels showing spots unique to Lin–Sca+Kit–. (C) Spots unique to Lin–Sca+Kit+.
Averaged gel comparison of Lin–Sca+Kit+ and Lin–Sca+Kit– cells labeled with saturation dyes. The use of averaged gels to allow sample comparison for Lin–Sca+Kit+ and Lin–Sca+Kit– cell lysates. Data from 5 sets of reciprocally labeled Lin–Sca+Kit–/+ populations were pooled and significant differences in protein expression (normalized volume) calculated using a 2-tailed Student t test. (A) All protein spots common to both gels are shown with enclosed areas. (B) Representative Lin–Sca+Kit– gels showing spots unique to Lin–Sca+Kit–. (C) Spots unique to Lin–Sca+Kit+.
Fluorescent protein stains also allow relative quantification of spots present in both Lin–Sca+Kit+ and Lin–Sca+Kit– cells. To express these data several thresholds of difference were set, namely 1.5-, 2.0-, and 10-fold between the 2 cell populations (Table 2).
The proteome differences seen using saturation Cy dye analysis of Lin-Sca+Kit+ and Lin-Sca+Kit- cells
Fold difference . | Total number of spots . | No. of spots increased in Kit+ . | No. of spots increased in Kit- . |
|---|---|---|---|
| More than 1.5 | 134 | 73 | 61 |
| More than 2.0 | 72 | 33 | 39 |
| More than 10 | 2 | 0 | 2 |
Fold difference . | Total number of spots . | No. of spots increased in Kit+ . | No. of spots increased in Kit- . |
|---|---|---|---|
| More than 1.5 | 134 | 73 | 61 |
| More than 2.0 | 72 | 33 | 39 |
| More than 10 | 2 | 0 | 2 |
Data for each protein are significant (P < .05) using a 2-tailed Student t test.
We have shown that there are multiple differences in the proteome of Lin–Sca+Kit+ and Lin–Sca+Kit– cells. A difference map is shown in Figure 4. We are in the process of identifying as many proteins as possible using mass spectrometry. The normalized volumes in Lin–Sca+Kit+ and Lin–Sca+Kit– cells are shown in Table 3.
Difference map comparison of Lin–Sca+Kit+ and Lin–Sca+Kit– cells labeled with saturation dyes. Cy5-labeled Lin–Sca+Kit– gel image showing spots increased (green) or decreased (red) relative to Lin–Sca+Kit+ cells.
Difference map comparison of Lin–Sca+Kit+ and Lin–Sca+Kit– cells labeled with saturation dyes. Cy5-labeled Lin–Sca+Kit– gel image showing spots increased (green) or decreased (red) relative to Lin–Sca+Kit+ cells.
Proteins identified by MALDI-ToF mass spectrometry
Spot . | Protein . | Molecular weight . | pl . | Normalized volume Lin-Sca+Kit+ . | Normalized volume Lin-Sca+Kit- . | Function . |
|---|---|---|---|---|---|---|
| 1 | Alcohol dehydrogenase | 42.7 | 6.12 | 0.204 | 0.067 | Metabolic |
| 2 | Actin | 41.7 | 5.29 | 1.501 | 1.697 | Cytoskeletal |
| 3 | Adseverin | 80.2 | 5.64 | 2.99 | 6.7 | Cytoskeletal |
| 4 | Serine protease inhibitor | 55.5 | 7.53 | 0.131 | 0.043 | Metabolic |
| 5 | Enolase | 47 | 6.35 | 0.631 | 0.269 | Metabolic |
| 6 | GAPDH | 47.6 | 8.14 | 1.313 | 0.807 | Metabolic |
| 7 | Pyruvate kinase | 50.7 | 7.42 | 0.39 | 0.169 | Metabolic |
| 8 | Transketolase | 67.6 | 7.23 | 0.389 | 0.205 | Metabolic |
| 9 | Gelsolin | 80.7 | 5.3 | 0.056 | 0.104 | Cytoskeletal |
Spot . | Protein . | Molecular weight . | pl . | Normalized volume Lin-Sca+Kit+ . | Normalized volume Lin-Sca+Kit- . | Function . |
|---|---|---|---|---|---|---|
| 1 | Alcohol dehydrogenase | 42.7 | 6.12 | 0.204 | 0.067 | Metabolic |
| 2 | Actin | 41.7 | 5.29 | 1.501 | 1.697 | Cytoskeletal |
| 3 | Adseverin | 80.2 | 5.64 | 2.99 | 6.7 | Cytoskeletal |
| 4 | Serine protease inhibitor | 55.5 | 7.53 | 0.131 | 0.043 | Metabolic |
| 5 | Enolase | 47 | 6.35 | 0.631 | 0.269 | Metabolic |
| 6 | GAPDH | 47.6 | 8.14 | 1.313 | 0.807 | Metabolic |
| 7 | Pyruvate kinase | 50.7 | 7.42 | 0.39 | 0.169 | Metabolic |
| 8 | Transketolase | 67.6 | 7.23 | 0.389 | 0.205 | Metabolic |
| 9 | Gelsolin | 80.7 | 5.3 | 0.056 | 0.104 | Cytoskeletal |
Identity of spots shown in Figure 4B and normalized volumes calculated relative to spots common to all gels. Values shown for spots 3 and 9 were calculated from the total amount detected in the “chain” of spots (ie, 7 for adseverin [spot 3] and 2 for gelsolin [spot 9]).
GAPDH indicates glyceraldehyde 3 phosphate.
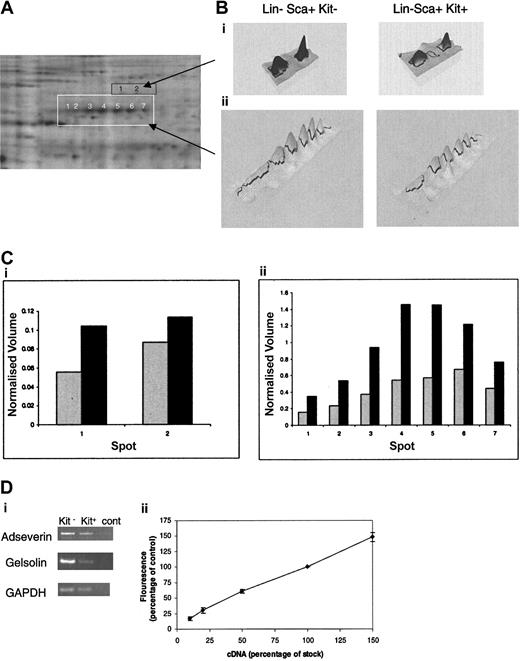
The limited material available precludes identification of proteins corresponding to less-intense spots given the detection limits presently achieved by mass spectrometry. In respect to differences between the Lin–Sca+Kit+ and Lin–Sca+Kit– populations, we are most concerned with changes in cytoskeletal elements and their regulators in these cells for reasons described in the “Introduction.” These are among the most abundant proteins in the cell. One particularly noteworthy feature is the major differences in the expression of a set of proteins of approximate molecular weight 80 kDa (Figure 5A). These proteins form 7 peaks spread across an isoelectric point (pI) range of 5 to 7. This suggests that the proteins are splice variants (with minor changes in mass) and/or posttranslationally modified forms of the same protein. We therefore selected each of the 7 spots and analyzed the tryptic digest using mass spectrometry. Database searching using the mass fingerprints in each case identified adseverin (also known as scinderin or gelsolin-like protein), a member of the gelsolin family that mediates actin capping, nucleation, and filament splicing. The differential levels of expression of these proteins in Lin–Sca+Kit– and Lin–Sca+Kit+ cells are shown in Figure 5B. It is clear that both the total normalized volume for all 7 spots and the relative ratios of each spot compared in Lin–Sca+Kit+ to Lin–Sca+Kit– cells vary markedly. Analyses of spots directly above adseverin (ie, those with a higher molecular weight) yielded mass fingerprints that indicated an identity of gelsolin by database searching; these spot volumes are also increased in Lin–Sca+Kit– cells compared with Lin–Sca+Kit+ cells. The observed increases in protein expression are reflected at the mRNA level with a 1.3- ± 0.1-fold (n = 5, ± SEM) and 4- ± 1.3-fold (n = 3, ± SEM) increase in mRNA for adseverin and gelsolin respectively (Figure 5C).
Differential expression of adseverin and gelsolin spots in Lin–Sca+Kit– and Lin–Sca+Kit+ cells from murine bone marrow. (A) Protein spots yielding tryptic mass fingerprints (using MALDI mass spectrometry) characteristic of adseverin and gelsolin are highlighted in white (for adseverin) and orange (for gelsolin). (B) Areas corresponding to the boxed regions in A are shown in 3D pseudocolor format. (C) Histograms of the normalized volumes of the spot chains Lin–Sca+Kit+ (▦) and Lin–Sca+Kit– (▪) (from left to right: 1-7 for adseverin, shown in white; 1 and 2 for gelsolin, shown in black) for gelsolin (i) and adseverin (ii). Normalized volume was calculated on the basis of spots present in all gels. (Di) The relative expression levels of adseverin and gelsolin mRNA in primitive hematopoietic cells were assessed by semiquantitative PCR using GAPDH as a loading control. Transcripts were amplified and visualized by agarose gel electrophoresis. (Dii) Amalgamated results of RT-PCR reactions from a dilution series of 10%, 20%, 50%, and 150% of cDNA for each primer combination. The data illustrate the semiquantitative nature of the RT-PCR. Error bars indicate SEM of 3 observations.
Differential expression of adseverin and gelsolin spots in Lin–Sca+Kit– and Lin–Sca+Kit+ cells from murine bone marrow. (A) Protein spots yielding tryptic mass fingerprints (using MALDI mass spectrometry) characteristic of adseverin and gelsolin are highlighted in white (for adseverin) and orange (for gelsolin). (B) Areas corresponding to the boxed regions in A are shown in 3D pseudocolor format. (C) Histograms of the normalized volumes of the spot chains Lin–Sca+Kit+ (▦) and Lin–Sca+Kit– (▪) (from left to right: 1-7 for adseverin, shown in white; 1 and 2 for gelsolin, shown in black) for gelsolin (i) and adseverin (ii). Normalized volume was calculated on the basis of spots present in all gels. (Di) The relative expression levels of adseverin and gelsolin mRNA in primitive hematopoietic cells were assessed by semiquantitative PCR using GAPDH as a loading control. Transcripts were amplified and visualized by agarose gel electrophoresis. (Dii) Amalgamated results of RT-PCR reactions from a dilution series of 10%, 20%, 50%, and 150% of cDNA for each primer combination. The data illustrate the semiquantitative nature of the RT-PCR. Error bars indicate SEM of 3 observations.
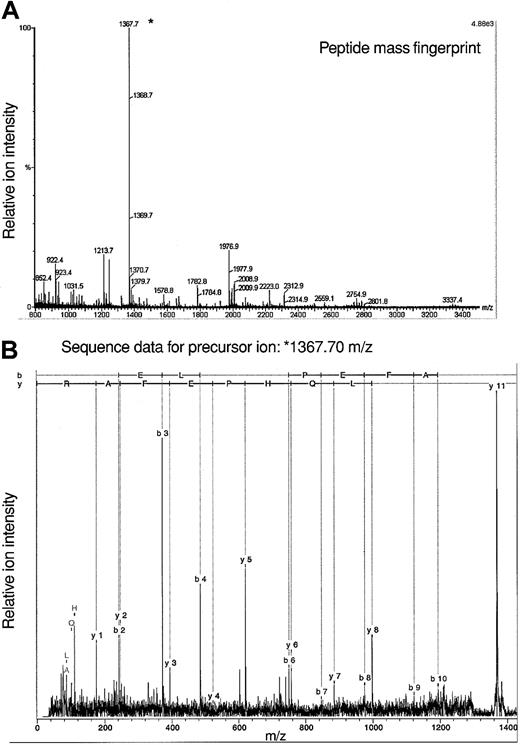
The fact that analyses of protein constituents of 7 spots yielded data consistent with an identification of adseverin leads to the question of how these proteins vary in structure to yield distinct pI. Given the amount of material available we cannot presently answer this directly with Lin–Sca+Kit+ or Lin–Sca+Kit– cells. To gain some idea on this we used adseverin from FDCP-mix cells in a tandem mass spectrometry experiment to identify posttranslational modifications. Using a variety of mass spectrometric techniques, no evidence of protein phosphorylation has been demonstrated (data not shown) although such negative evidence is necessarily inconclusive. We did however observe N-acetylation of adseverin, a posttranslational modification seen in major cytoskeletal proteins and muscle. The acetylated peptide AQELQHPEFAR corresponds to amino acids 2 to 12 of adseverin (Figure 6). It results from removal of the N terminal methionine residue and acetylation of the alanine residue. This commonly occurs in protein where the second amino acid is small and neutral.31 Amino terminal modification is required for the function of alpha-tropomyosin in striated muscle32 and modifies actin function in Drosophila.33
Mass spectrometric analysis of adseverin. (A) Peptide mass fingerprint data for adseverin. The observed peptide MH+ ions characteristic of adseverin are labeled on the spectrum based on a theoretical digest of adseverin allowing for one missed trypsin cleavage and no fixed or variable modifications. (B) Spectrum of product ions derived from a precursor of mass/charge (m/z) 1367.7 Da during tandem mass spectrometric analysis. The b and y ion series were assigned using BioTools software (Bruker, Bremen, Germany).
Mass spectrometric analysis of adseverin. (A) Peptide mass fingerprint data for adseverin. The observed peptide MH+ ions characteristic of adseverin are labeled on the spectrum based on a theoretical digest of adseverin allowing for one missed trypsin cleavage and no fixed or variable modifications. (B) Spectrum of product ions derived from a precursor of mass/charge (m/z) 1367.7 Da during tandem mass spectrometric analysis. The b and y ion series were assigned using BioTools software (Bruker, Bremen, Germany).
The distinct difference in adseverin expression seen between Lin–Sca+Kit+ and Lin–Sca+Kit– cells suggests that there is a potential difference in regulation of actin filaments and thereby motility. Actin filament assembly alone has been suggested to be sufficient to power motility in some systems. Actin levels in the 2 cell populations are equivalent (Table 3).
Discussion
Lin–Sca+Kit+ cells and Lin–Sca+Kit– cells display differential motility and response to agonists that promote motile behavior and chemotaxis. SDF-1 (a chemotactic factor for these cells) and lysophospholipids (agonists that enhance motility) employ a PI3-kinase, Vav-dependent activation of Rho G proteins to promote stem cell motility.12 This has profound implications for engraftment and stem cell homing in vivo. In this respect agents like lysophospholipids that synergistically promote chemotaxis when added with SDF-1 may have an important physiologic role. But this still leaves the fact that the poorly motile Kit+ cells differ in some way in their biochemistry that disallows the more robust response to SDF-1 seen in Kit– cells (Table 1). The most obvious difference is in agonist-stimulated generation of the second messenger critical for motile response, PIP3. Inhibition of PI3-kinase abrogates the motile/chemotactic response of Kit+ and Kit– cells.12 PIP3 levels were assessed using a novel technique never before applied with primitive hematopoietic cells23 and demonstrated not only that it has value in studying signaling in small numbers of cells but also that Kit+ and Kit– cells respond in a similar fashion to SDF-1.
This being the case, the difference in motile behavior may lie elsewhere in the pathway, downstream from receptor-mediated activation of PI3-kinase. Motility is very much driven by proteins and so we decided to investigate the protein content of Kit+ and Kit– cells using the relative quantification offered by the CyDye technique. The amount of material that can be prepared from primary stem and progenitor cells depends on the degree of enrichment required, the speed of the enrichment procedure, and the starting material available. Although this does not limit studies too greatly at the transcriptional level, due to the availability of amplification techniques such as the PCR reaction, it can be crippling to studies on proteins and their posttranslational modification where no such amplification can be achieved. Thus, hematopoietic stem cell biology has been exceptionally well studied and described within the transcriptome arena1,2 while advances at the proteome level remain more problematic. Nonetheless it is clear that there may be marked shifts in the proteome which cannot be approached at all in transcriptomic studies. These include the quantitative definition of subcellular localization, posttranslational modification, protein turnover, and protein-protein interactions. The recent definition of the yeast protein-protein interaction map is a landmark study that suggests how much can be achieved in the future in mapping the natural history of stem cells and other cells.34-36
While these data can be derived perhaps in the future, our immediate goal concerned the study of relatively high-abundance cytosolic proteins regulating motile behavior. With little material available, simply detecting protein can be problematic, but by labeling with Cy dyes via cysteine residues we could visualize proteins present on 2D gels and relatively quantify many proteins from Kit+ and Kit– cells, respectively. We have shown that the task of defining the proteome of a stem cell population can begin using the maleimide Cy3 and Cy5 dyes and this may have value in a number of other scenarios (eg, comparison of transgenic and wild-type hematopoietic cells). The amount of biologic material available determines the limits on protein detection and identification in proteomics approaches. Improvements in the sensitivity of detection by mass spectrometric methods are required to address this issue and indeed these are being realized.37
In general, 2D gel-based approaches are limited by the fact that hydrophobic proteins or proteins with isoelectric points at the extremes of the pH range used are excluded from the 2D gel. Nonetheless, the use of DIGE offers a very broad dynamic range (a criticism of silver-stained gel-based approaches).9 Furthermore, the use of proteome “contigs,” where a series of gels of narrow, overlapping pH range are run, enables the study of a large proportion of the proteome giving relative quantification and protein coverage.38 Gel-free methods such as protein arrays39-41 and multidimensional proteomic identification technology (MUDPIT) offer potential solutions but the latter suffers from a lack of relative quantification.42-44 The term protein array encompasses antibody-based arrays, tissue-section arrays, or arrays of proteins/peptides for screening of protein levels or binding properties. The accompanying review by Cristea et al (beginning on page 3624) covers these in detail.37
MUDPIT can be used to identify very-low-abundance peptides (proteins at 100 copies per cell) by peptide sequencing if sufficient material is available. This is achieved by low flow rate chromatographic separation coupled to electrospray ionization tandem mass spectometry. In theory, this could be adapted to study the stem cell proteome, including poorly soluble proteins43,44 if sufficient material were available. This approach will, however, need adaptation to give comparative analysis between 2 populations. This can be achieved using differential isotope labeling, as in the isotope coded affinity tagging (ICAT) method. The ICAT approach uses isotopically light or deuterated reagents to tag proteins in 2 cell population lysates. The resulting mixtures are mixed and trypsinized, separated by affinity chromatography, and analyzed by mass spectrometry, allowing relative quantification of the 2 isotopic variants of each peptide from each cell population.45,46
In Figure 3 we have shown that a very large number of differences exist between Kit+ and Kit– cells at the proteome level. But these data just scratch the surface of the proteomic definition of stem cells. Our aim was to find differences in cytoskeletal regulators between Kit+ and Kit– cells and this we have done. Multiple gel spots yielding data of the same primary sequence are indicative of differences in posttranslational modification of the protein in each cell type and warrant further investigation. Adseverin and gelsolin are differentially expressed between Lin–Sca+Kit+ and Lin–Sca+Kit– cells. Adseverin is a member of the gelsolin/villin family. Adseverin has a functional domain structure similar to that of gelsolin and can also engage in actin capping, polymerization, and severing actin filaments. Adeverin and gelsolin tend to be expressed at complementary levels in cell types, suggesting a potential common function.47 Rac low-molecular-weight G proteins regulate gelsolin and other members of this family as a means of regulating membrane ruffling and motility.18,19 Gelsolin also has a role in neurite filopodial extension.48 Recently it has been shown that the hematopoietic stem cell can produce magnupodia of considerable length.49 They are also a population that is able to migrate both locally and over extended distances in response to chemotactic stimuli.27,50 Experiments with gelsolin null fibroblasts show clearly the key role this protein family plays in motility and morphologic change. Thus, the enhanced expression of gelsolin/adseverin in Lin–Sca+Kit– cells as opposed to Lin–Sca+Kit+ cells dovetails with the enhanced relative motility seen in the Kit– cells without agonists. This implies that expression of adseverin may be an expression of the potential motile and morphologic changes that a stem cell may engage in. The fact that several different forms of adseverin are present in the same cells argues for a form of posttranslational modification of adseverin. This exemplifies the qualities this form of proteomic study can bring to studies on proteins. Mass spectrometric analysis has revealed adseverin N-acetylation (Figure 6). It may be of relevance that lysine acetylation has been shown to destabilize microtubules via tubulin deacetylation.51 The roles of changes in adseverin and gelsolin in differential chemotactic responses identified by proteomic analysis will be the subject of further exploration.
Prepublished online as Blood First Edition Paper, February 5, 2004; DOI 10.1182/blood-2003-09-3294.
Supported by a Borno State Scholarship from the Federal Republic of Nigeria to H.G.H. and by the Leukaemia Research Fund, United Kingdom.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We wish to thank Rachel Mottram for the flow cytometric enrichment of cells used in this work and Dr Marcus Macht and Dr Arnt Asperger (Bruker Saxonia Analytik GmbH, Bruker Daltonics) for generating tandem mass spectrometry data.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal