Abstract
CDKN2B (INK4B), which encodes the cyclin-dependent kinase inhibitor p15INK4b, is up-regulated by many cytokines found in hematopoietic environments in vivo. In human acute myeloid leukemias (AMLs), it is inactivated with high frequency. To gain insight into the regulatory pathways leading to the normal activation of p15Ink4b expression, we examined interferon β (IFNβ)–induced transcription. Using reporter gene assays in murine myeloid cells M1, we determined that a 328-bp fragment, located 117 to 443 bp upstream of the translation initiation site, was sufficient to activate transcription. Both the interferon consensus sequence-binding protein/interferon regulatory factor 8 (ICSBP/IRF-8) and PU.1 were able to increase transcription from this region. It was determined that both ICSBP and PU.1 must bind to DNA to form a stable PU.1/ICSBP binding complex. Interestingly, introduction of the ICSBP into ICSBP-null Tot2 cells led to a significant increase in p15Ink4b RNA expression. This regulation of the Ink4b promoter is apparently myeloid specific because both ICSBP and PU.1 are myeloid commitment factors. Importantly, this provides a mechanism to explain in part the tumor suppressor activity of ICSBP, since ICSBP-deficient mice develop a chronic myelogenous leukemia (CML)–like disease and a high percentage of human AML and CML lack ICSBP transcripts.
Introduction
The INK4 family of tumor suppressors play an important role in the development of neoplastic diseases.1,2 In particular, one of its members, p15INK4b, is inactivated with high frequency in hematopoietic neoplasms. This protein, like other family members, is a cyclin-dependent kinase inhibitor (CDKI), acting at the early G1 phase of the cell cycle. It binds to CDK4 and CDK6 inhibiting the interaction of the kinases with cyclin Ds. The CDKN2B (INK4B) gene, which encodes this protein, is inactivated by hypermethylation in approximately 80% of all acute myeloid leukemias (AMLs) and a high proportion of acute lymphocytic leukemias (ALLs) and myelodysplastic syndromes (MDSs). In addition, it is deleted in approximately 50% of all ALLs.1 Ink4b-deficient mice develop a lymphoproliferative disease and a low incidence of angiosarcomas and have an increased susceptibility to retrovirus-induced myeloid leukemia.3,4
According to recent studies, p15INK4b functions in pathways that involve extrinsic signaling. For example, its expression is up-regulated in myeloid cells at the RNA level in response to interleukin-6 (IL-6)5-7 in lymphoid cells in response to type 1 interferon (IFNα)8 and in epithelial cells treated with transforming growth factor β (TGFβ).9 In addition, as demonstrated here, it is induced in myeloid cells treated with type 1 IFN.
Since much of the research on p15INK4b strongly indicates that it is involved in the physiologic control of hematopoietic cell growth, it is important to understand how its expression is regulated at the transcription level through cytokines. To date, little is known about how the Ink4b promoter is regulated in myeloid cells, although studies have looked at its response in human and mouse epithelial cells following treatment with TGFβ. It was reported, for example, that specificity protein 1 (Sp1) is responsible, in part, for the transcriptional activation of the human INK4b gene in response to TGFβ.9 This result was confirmed by Malumbres and coworkers10 in murine fibroblasts. The INK4b gene is characterized as a member of the family of TATA-less promoters whose transcription is induced at the transcriptional initiator (Inr) region that plays an important role in both the positive and negative regulation of the promoter as in TGFβ-treated epithelial cells.11,12
In the present study, we specifically examined the transcriptional response of the Ink4b gene to treatment with type 1 IFN in murine myeloid cells. We chose to look at the IFN response for 2 reasons. First, type 1 IFN is used as a treatment to inhibit progression of human chronic myelogenous leukemia (CML) and little is known about its growth inhibitory mechanisms. Second, autocrine expression of IFNβ is induced during myeloid cell differentiation and implicated in growth arrest.13,14 In this work, we demonstrate that the transcription factors PU.1 and interferon consensus sequence-binding protein (ICSBP) are involved in the IFN induction of p15Ink4b transcription. PU.1 is well known to play an important role during myeloid cell differentiation into granulocytes and macrophages.15,16 The observation that ICSBP may play a role in this regulation is especially intriguing because it has been shown that ICSBP-deficient mice develop a CML-like disease and a high percentage of human myeloid leukemias lack normal levels of ICSBP RNA.17-19 Furthermore, CMLs with decreased expression of ICSBP are less responsive to IFN treatment.20
Materials and methods
Cells
The murine myeloid cell line M121 was maintained in RPMI 1640 medium supplemented with 10% heat-inactivated horse serum. For induction of growth arrest or differentiation, cells were seeded at a concentration of 2 × 105/mL and cultivated in medium containing 344 U/mL IFNβ (PBL Biochemical Laboratories, New Brunswick, NJ), 100 U/mL IFNγ (Pepro-Tech, Rocky Hill, NJ), 50 ng/mL lipopolysaccharide (LPS; Sigma, St Louis, MO), or IL-6. The IL-6 stocks were prepared as described recently.22 Tot2 cells were described previously.23
Plasmids and primers
The mammalian expression vectors pcDNA HA and pcDNA HA PU.1, encoding the murine full-length, wild-type PU.1 protein, were obtained from Harrinder Singh and described in Eisenbeis et al.24 The plasmids pcXN2, pcXN2 ICSBP, and pcDNA ICSBP expressing murine full-length, wild-type ICSBP were described in Minucci et al25 and Thornton et al.26 The ICSBP DNA-binding mutant pcDNA ICSBP K79E and the ICSBP interactivation domain mutant pcDNA ICSBP R289E were described in Tsujimura et al27 and Tamura et al.23 The plasmid pmp15 containing the genomic murine Ink4b DNA was a generous gift of Marcos Malumbres and is described in Malumbres et al.28 The plasmid was used as a template to determine the sequence of the Ink4b upstream region together with the following primers: -576/-595 bp, 5′gctagttcatctctaggcgg3′; -1075/-1094 bp, 5′cctgtcattaaaaccctctc3′; and -1630/-1649 bp, 5′ccagcaggttgatattgatg3′. Reporter vectors were constructed using the pGL3 luciferase reporter vectors pGL3-Basic and pGL3-Promoter (Promega, Madison, WI). Promoter regions were amplified using pmp15 as a template and the following primers: -1891/-1872 bp, 5′cagccaccggtctacctcaa3′; -1277/-1261 bp, 5′cagccgttcccacacag3′; -1006/-985bp, 5′gctctttattacatatgataac3′; -688/-669 bp, 5′ggatccttgggatgtgttat3′; -444/-424 bp, 5′ctaaataaagacctctgctcc3′; -293/-271 bp, 5′ctatttgtctcatgacgtcacca3′; -213/-193 bp, 5′tgcagaacgctgcagctcagt3′ as forward primers and -117/-134 bp, 5′ggaacgctcgagcgctag3′; and -193/-213 bp, 5′actgagctgcagcgttctgca3′ as reverse primers. The bp position is shown in reference to the translation start point as described in Malumbres et al28 (GenBank accession no. U66084). The resulting amplification products were cloned in the appropriate luciferase vector.
Mutations of potential PU.1 and Sp1 binding sites were introduced by site-directed mutagenesis using the QuickChange Multi Site–Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The 2 proximal PU.1 sites at position -364 bp and -357 bp were mutated with 5′ccggccacggtaagttaac-gactcttaagcttaaagttctgcgcagg3′ and the downstream PU.1 site at -320 bp with 5′ggcgtctaagatcggccgactctctcaagataccacccc3′. Mutated bases are shown in bold. The nonmutated sequence is depicted in Figure 3.
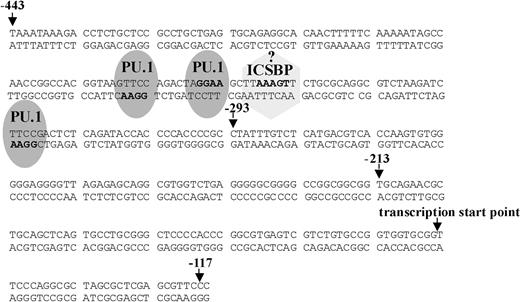
Potential transcription factor binding sites in the Ink4b promoter sequence. Shown is the sequence from -443 to -117 bp of the Ink4b promoter. Marked are potential PU.1 and ICSBP binding sites, identified according to their reported core-binding motifs (in boldface).
Potential transcription factor binding sites in the Ink4b promoter sequence. Shown is the sequence from -443 to -117 bp of the Ink4b promoter. Marked are potential PU.1 and ICSBP binding sites, identified according to their reported core-binding motifs (in boldface).
Preparation of RNA, Northern blot analysis, and semiquantitative RT-PCR
Total RNA was prepared and analyzed by Northern analysis as described.7 The following cDNA fragments were used as probes: Ink4b, 1.3-kb EcoRI fragment29 ; PU.1, 0.9-kb HindIII/ApaI fragment30 ; ICSBP, 1.9-kb EcoRI fragment31 ; β-actin.32 Semiquantitative reverse transcriptase–polymerase chain reaction (RT-PCR) was carried out using the Titan One-Tube RT-PCR System (Roche Applied Science, Indianapolis, IN) following the manufacturer's recommendations. Five nanograms of total RNA was used as a template with the following primers: Ink4b (sense 5′gttgggcggcagcagtgac3′, antisense 5′caatctccagtggcag3′), β-actin (sense 5′tgtgatggtgggaatgggtcag3′, antisense 5′tttgatgtcacgcacgatttcc3′).
Preparation of protein lysates and Western blot analysis
Preparation of nuclear extracts
Nuclear extracts were prepared as reported in Marecki et al34 using a modified procedure originally described by Schreiber et al.35 The murine myeloid cell line M1 was plated at a concentration of 2 × 105/mL and stimulated for 16 hours with IFNβ (344 U/mL). The cells (107) were washed once with Mg2+- and Ca2+-free phosphate-buffered saline (PBS; Invitrogen, Carlsbad, CA) and resuspended in 400 μL ice-cold buffer I (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH 7.0-7.6; 10 mM KCl; 0.1 mM EDTA [ethylenediaminetetraacetic acid]; 0.1 mM EGTA [ethyleneglycoltetraacetic acid]; 1 mM dithiothreitol [DTT]; 10 mM iodoacetamide; 1 mM phenylmethylsulfonyl fluoride [PMSF]; and Complete protease inhibitor cocktail according to the manufacturer's instructions [Boehringer-Mannheim, Mannheim, Germany]). Following incubation on ice for 15 minutes, 25 μL of 10% Nonidet P-40 (NP40) was added to the samples and they were vortexed. The cytoplasmic fraction was separated from the nuclei by centrifugation for 1 minute at 5000g. The nuclear pellets were resuspended in 300 μL ice-cold buffer II (20 mM HEPES, pH 7.0-7.6; 0.4 M NaCl; 1 mM EDTA; 1 mM EGTA; 1 mM DTT; 10 mM iodoacetamide; 1 mM PMSF; and Complete protease inhibitor cocktail according to the manufacturer's instructions [Boehringer-Mannheim]) and rocked on a shaking platform for 15 minutes at 4.0° C, followed by a 5-minute spin at 18 000g at 4.0° C. The lysates were stored at -70° C and protein concentration was determined using the BCA Protein Assay Kit (PIERCE, Rockford, IL).
Electrophoretic mobility shift assays (EMSA)
Complementary oligonucleotides spanning the region from -311 bp to -336 bp (30-bp probe), -311 bp to -362 bp (60-bp probe), or -375 bp to -336 bp (40-bp probe) of the Ink4b promoter region containing the predicted PU.1 and ICSBP binding sites (Figure 3) were annealed and labeled by filling in 3′ recessed ends with DNA polymeraseI Klenow fragment (New England BioLabs, Beverly, MA) and [α-32P]dCTP (Amersham Biosciences, Arlington Heights, IL). Oligonucleotides harboring mutations in the predicted PU.1 and ICSBP sites were used for a closer analysis of the PU.1/ICSBP binding complex by changing the ggaa PU.1 core-binding motive to ttaa and the partial ICSBP core-binding motive aaagt to cacga. For gel shift assays, 3 μg nuclear extract was incubated for 60 minutes on ice with 0.1 ng labeled DNA probe in 20 μL binding buffer containing 1.0 mM EDTA, 10 mM Tris-HCl (pH 8.0), 25 mM glycerol, 0.5 mM DTT, and 1.0 μg poly dI-dC. Unlabeled probe was added up to a 50-fold molar excess in competition experiments and 2 μg antiserum (PU.1, Ets-2, TEL, ICSBP, interferon regulatory factor 1 [IRF-1]) was added in supershift experiments. Following incubation, each reaction was separated on a 7% nondenaturating low-ionic strength polyacrylamide gel for samples containing the 30-bp and 40-bp probes and on a 6% gel for samples containing the 60-bp probes. Gels were dried and exposed to X-ray films.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) analysis was performed as described previously36 with some modifications. Proteins were cross-linked to the genomic DNA in M1 cells by addition of formaldehyde (1% wt/vol) directly to culture medium and incubated for 30 minutes at room temperature. Glycine (125 mM) was added to the reaction and incubated for an additional 5 minutes to stop the reaction. The cells were washed twice with ice-cold PBS, resuspended in nuclei-releasing buffer (5 mM piperazine diethanesulfonic acid [PIPES], pH 8.0; 85 mM KCl; 0.5% NP40), swelled on ice for 10 minutes, and homogenized in a Dounce homogenizer to facilitate the release of the nuclei. Nuclei were pelleted at 2700g, resuspended in nuclei lysis buffer (50 mM Tris-HCl, pH 8.1; 10 mM EDTA; 1% sodium dodecyl sulfate [SDS]) supplemented with proteases inhibitors (Complete; Roche Applied Science), and sonicated twice for 30 seconds. After centrifugation at 2700g the supernatant was incubated with Staph A cells for 15 minutes at 4° C. The Staph A cells were removed from the chromatin solution by centrifugation. Precleared chromatin from 2 × 107 cells was incubated overnight at 4° C either with 2 μgaffinity-purified rabbit polyclonal antibody to PU.1 (Santa Cruz Biotechnology), or 3 μL rabbit polyclonal serum to murine ICSBP37 or control immunoglobulin G (IgG) antibody. A control reaction with no antibody was also included in the experiment. The immune complexes were precipitated by a 15-minute incubation at 4° C with preblocked Staph A cells and collected by centrifugation. The precipitates were washed 5 times with immunoprecipitation buffer (100 mM Tris-HCl, pH 9.0; 500 mM LiCl, 1% NP40, 1% deoxycholic acid), resuspended in 300 μL elution buffer (50 mM NaHCO3, 1% SDS), and vortexed for 20 minutes at room temperature. Released Staph A cells were separated from the supernatant by centrifugation. Supernatant containing immunoprecipitated, cross-linked chromatin was transferred to a new tube supplemented with NaCl and RNase A (final concentration 300 mM and 30 μg/μL, respectively). To reverse formaldehyde cross-linking, the samples were incubated 5 hours at 67° C and the genomic DNA was ethanol precipitated. The pellets were resuspended in proteinase K buffer (10 mM Tris-HCl, pH 7.5; 5 mM EDTA, and 0.25% SDS) and treated with proteinase K (200 μg/μL) for 2 hours at 45° C. Immunoprecipitated DNA was further cleaned by phenol extraction and ethanol precipitation with glycogen. The pellet was dissolved in Tris-EDTA buffer and used in PCR analysis. Ink4b and c-myb promoter DNAs were detected by PCR analysis using HotStarTaq Master Mix Kit (Qiagen, Valencia, CA) and specific primers: p15diag+, 5′ggcctgggctaaataaagacct3′; and p15diag-, 5′ttcgccggccgtgagattgctaca3′ for Ink4b; and Myb-pr1 as 5′gcacaagttcctgagaactg3′, and Myb-pr1 as 5′aatacagtcagccctgtgg3′ for c-myb. After 35 cycles of amplification, PCR products were separated on a 1.5% agarose gel containing ethidium bromide.
DNA transfections and reporter gene analyses
Transfection of M1 cells was carried out by electroporation using 5 × 106 cells in 0.5 mL RPMI medium containing 0.5 μg pRL-TK (Promega), a Renilla luciferase vector to control for transfection efficiency, 3.0 μg of a luciferase reporter construct, and 20 μg of PU.1 and/or ICSBP expression plasmid or the corresponding empty vector. Purified DNA with an endotoxin level of less than 0.04 endotoxin units (EU)/μg of DNA was purchased from Qbiogene (Carlsbad, CA) or Lofstrand Labs Limited (Gaithersburg, MD). Electroporation was performed at 275 V with a pulse length of 50 milliseconds in an Electro Square Porator ECM 830 (BTX, San Diego, CA). The cells were plated at a density of 2 × 105/mL and, if indicated in the text, treated with IFNβ (344 U/mL) starting at 4 hours following transfection. They were harvested after 24 hours and lysed in 40 μL of 1 × Reporter Lysis buffer (Promega). Thirty microliters of lysates were tested for reporter gene activity using the dual luciferase assay system (Promega) and a Turner TD-20e luminometer (Turner Designs, Sunnyvale, CA). Each experiment was carried out in triplicate and was repeated several times using different plasmid preparations. If not indicated differently in the text, data are expressed as average values ± standard error.
Results
The tumor suppressor p15Ink4b is up-regulated in M1 cells in response to IFNβ treatment
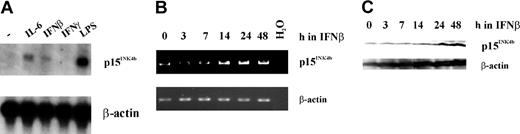
Differentiation toward the macrophage lineage and/or growth arrest can be induced by a variety of different stimuli and cytokines in the murine myeloid cell line, M1.38 It was previously shown that IL-6 causes up-regulation of p15Ink4b mRNA,6,7 and we presume its up-regulation promotes the growth arrest associated with IL-6–induced differentiation. We were interested in determining if IFNβ, a strong inducer of growth arrest in the absence of differentiation, would induce p15Ink4b expression in M1 cells. The responsiveness of the Ink4b promoter in M1 cells to cytokines was demonstrated here following treatment for 24 hours with IFNβ as well as IL-6, LPS, and IFNγ. As shown in the northern analysis in Figure 1A, p15Ink4b mRNA expression was up-regulated following treatment with IFNβ, IL-6, and LPS but not IFNγ. In an additional experiment, we treated M1 cells for 0, 3, 7, 14, 24, and 48 hours with IFNβ and analyzed them for p15Ink4b expression by RT-PCR and Western blot. We observed up-regulation of the tumor suppressor at the RNA level after 14 hours and at the protein level after 24 hours. Because activation of transcription took several hours (Figure 1B-C), it is very likely due to an indirect mechanism, for example through activation of another gene.
Expression of p15Ink4b in response to treatment with growth arrest stimuli. (A) M1 cells were cultivated in the presence of IL-6, IFNβ, IFNγ, and LPS, all of which are known to induce growth arrest. Twenty-four hours following treatment with these agents, cells were harvested and total RNA was isolated. Northern analysis was carried out using a p15Ink4b cDNA probe. To control for sample loading, the same blot was rehybridized with a β-actin probe. (B) RT-PCR analysis of p15Ink4b expression in M1 cells cultivated in the presence of IFNβ for 0, 3, 7, 14, 24, and 48 hours. To control for sample loading, β-actin cDNA was amplified from the same RNA samples. (C) Western blot analysis of M1 cells that were treated with IFNβ for 0, 3, 7, 14, 24, and 48 hours, using a monoclonal antibody against p15Ink4b and an antibody against β-actin.
Expression of p15Ink4b in response to treatment with growth arrest stimuli. (A) M1 cells were cultivated in the presence of IL-6, IFNβ, IFNγ, and LPS, all of which are known to induce growth arrest. Twenty-four hours following treatment with these agents, cells were harvested and total RNA was isolated. Northern analysis was carried out using a p15Ink4b cDNA probe. To control for sample loading, the same blot was rehybridized with a β-actin probe. (B) RT-PCR analysis of p15Ink4b expression in M1 cells cultivated in the presence of IFNβ for 0, 3, 7, 14, 24, and 48 hours. To control for sample loading, β-actin cDNA was amplified from the same RNA samples. (C) Western blot analysis of M1 cells that were treated with IFNβ for 0, 3, 7, 14, 24, and 48 hours, using a monoclonal antibody against p15Ink4b and an antibody against β-actin.
Deletion analysis of the Ink4b promoter region
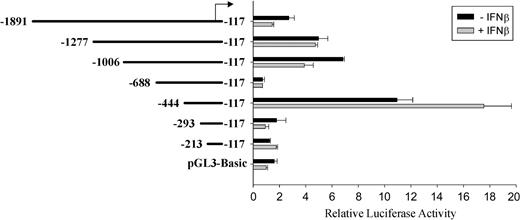
Since we were interested in the ability of IFNβ to activate transcription, we determined the specific region of the Ink4b promoter involved in this activity. Fragments of different lengths were evaluated for their ability to be transcribed in M1 cells, with or without IFNβ treatment, using a luciferase reporter construct, pGL3-Basic. A fragment from -117bpto -294 bp (in reference to the translation initiation site) did not have promoter activity when compared with the control pGL3-Basic vector (Figure 2). However, extension of the promoter region another 151 bp resulted in more than a 5-fold increase in activity in the absence of IFNβ. Very low promoter activity was observed for fragment -688/-117 bp, indicating the presence of a negative regulatory element between -688 bp and -444 bp, although this possibility has not been further investigated. Higher reporter gene activity was present in IFNβ-treated cultures (-444/-117; Figure 2).
Deletion analysis of the Ink4b promoter region with and without IFNβ. Fragments of the Ink4b promoter region were cloned into the reporter plasmid pGL3-Basic. M1 cells, transiently transfected with these constructs, were cultivated in the presence or absence of IFNβ and analyzed after 24 hours for luciferase activity. Shown are relative values normalized for Renilla luciferase activity. Error bars depict standard error.
Deletion analysis of the Ink4b promoter region with and without IFNβ. Fragments of the Ink4b promoter region were cloned into the reporter plasmid pGL3-Basic. M1 cells, transiently transfected with these constructs, were cultivated in the presence or absence of IFNβ and analyzed after 24 hours for luciferase activity. Shown are relative values normalized for Renilla luciferase activity. Error bars depict standard error.
PU.1 and ICSBP increase Ink4b promoter activity
The -444/-117-bp promoter fragment was studied further because it showed the highest promoter activity in both the absence and presence of IFNβ stimulation. Interestingly, we did not find an obvious potential binding site for interferon regulatory factor (IRF) in this region. We did, however, find 3 potential binding sites for PU.1 based on the known core-binding sequence (Figure 3). The transcription factor PU.1 is a member of the Ets family of transcription factors and one of the key players involved in maturation of myeloid cells into granulocytes and macrophages.14,39 Recently, it was reported that PU.1 can enhance transcription by interacting with IRFs.40 One IRF, shown to interact with PU.1 in myeloid cells, irrespective of its own binding to DNA, is interferon consensus binding protein ICSBP (IRF-8). It has been reported to be lymphoid and myeloid cell specific, and ICSBP-deficient mice were found to possess an increased number of myeloid progenitor cells and to develop a CML-like disease.17,18 We, therefore, decided to test ICSBP's ability to transactivate the Ink4b promoter in conjunction with PU.1 in response to IFNβ.
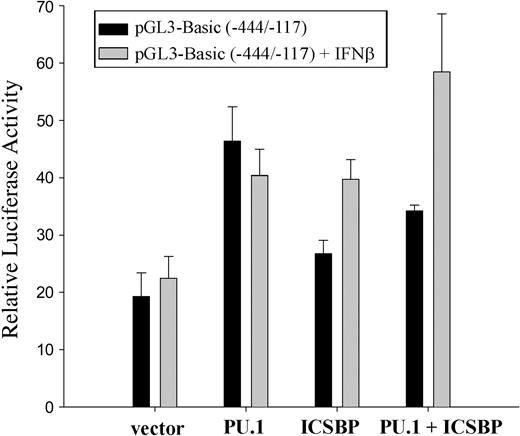
M1 cells were transfected with the luciferase vector construct pGL3-Basic (-444/-117) and cotransfected with plasmids expressing PU.1, ICSBP, or both. The cells were cultivated in the presence or absence of IFNβ and analyzed for reporter gene activity (Figure 4). In IFNβ-treated samples, transfection of PU.1 or ICSBP individually resulted in an approximate 2-fold increase in pGL3-Basic (-444/-117) activity. Together the transcription factors led to a 3- to 4-fold increase in promoter activity.
Ink4b promoter activity is positively regulated by PU.1 and ICSBP and can be further increased by addition of IFNβ. M1 cells, cultivated in the presence or absence of IFNβ, were transiently cotransfected with the -444/-117 bp promoter construct as well as expression plasmids for PU.1 and/or ICSBP or the corresponding empty vector and analyzed for promoter activity. Shown are relative values normalized for Renilla luciferase activity. Error bars depict standard error.
Ink4b promoter activity is positively regulated by PU.1 and ICSBP and can be further increased by addition of IFNβ. M1 cells, cultivated in the presence or absence of IFNβ, were transiently cotransfected with the -444/-117 bp promoter construct as well as expression plasmids for PU.1 and/or ICSBP or the corresponding empty vector and analyzed for promoter activity. Shown are relative values normalized for Renilla luciferase activity. Error bars depict standard error.
Although PU.1 and ICSBP consistently induced transcription from the Ink4b promoter, we observed variability in the degree of PU.1/ICSBP-induced activation and this appeared to be due to a response to cell density– and/or stress-related activation following electroporation. In any case, we consistently observed at least a 2- to 3-fold increase in activation following transfection of ICSBP and PU.1. This range of increase is consistent with that observed by others.41
IFNβ increased ICSBP's, but not PU.1's, potential to stimulate 15Ink4b expression. Interestingly, it increased the activity of the combined factors (Figure 4). The ability of the 2 transcription factors to activate p15Ink4b expression in response to IFNβ is presumed to be regulated at the posttranslational level because M1 cells treated with IFNβ for up to 48 hours were not demonstrated to have significant changes in expression levels for ICSBP and PU.1 (data not shown).
Localization of the PU.1 binding site in the Ink4b promoter region involved in the transcriptional response to IFNβ
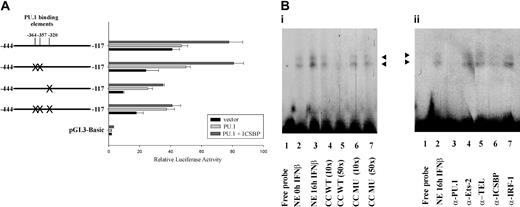
Experiments were performed to determine which of the 3 potential PU.1 binding sites are responsible for the stimulatory effect of PU.1 on p15Ink4b expression (Figure 3). The PU.1 sites were mutated in pGL3-Basic (-444/-117; Figure 5A) and the resulting vectors were analyzed for reporter gene activity. Mutation of the 2 upstream PU.1 binding sites (-364 bp and -357 bp) did not show any inhibitory effect on promoter activity compared with the nonmutated, original sequence. However, mutation of the PU.1 binding site at -320 bp lead to a significant drop in promoter activity. The same effect was observed with the construct harboring the deletion of all 3 potential PU.1 binding sites. Thus, the PU.1 binding site closest to the transcription start point (-320 bp) alone is responsible for the positive regulatory effect of PU.1 in this promoter region.
The potential PU.1 binding site at -320 bp mediates PU.1's stimulatory effect on Ink4b promoter activity. (A) M1 cells were transiently transfected with pGL3-Basic -444/-117 or constructs harboring site-directed mutagenesis of potential PU.1 binding sites. The cells were cotransfected with expression plasmids for PU.1, PU.1 and ICSBP, or the corresponding empty vectors. The samples were cultivated in the presence of IFNβ and analyzed for luciferase activity. Shown are absolute values normalized for Renilla luciferase activity. Error bars depict standard error. (Bi) EMSA analysis was performed using nuclear extracts (NEs) from M1 cells and a radiolabeled oligonucleotide Probe A spanning the region from -337 bp to -312 bp of the Ink4b promoter region. Two specific complexes (arrowheads) were resolved. No nuclear extract was added in lane 1. Nuclear extract from cells that were cultivated in the absence of IFNβ was added in lane 2. For all of the other lanes, nuclear extracts from cells cultivated in the presence of IFNβ for 16 hours were used. For lanes 4 to 7 an unlabeled wild-type (WT) oligonucleotide or an oligonucleotide carrying a mutation (MU) in the PU.1 binding site at -320 bp was added as a cold competitor (CC) at 10- or 50-fold excess compared with the labeled oligonucleotide. (Bii) EMSA analysis was performed as described for panel Bi. To determine which proteins are present in the observed complexes, the binding reactions were performed in the presence of antibodies against PU.1 (lane 3), Ets-2 (lane 4), TEL (lane 5), ICSBP (lane 6), or IRF-1 (lane 7). Arrowheads indicate protein/DNA complexes proposed to contain PU.1 and ISCBP.
The potential PU.1 binding site at -320 bp mediates PU.1's stimulatory effect on Ink4b promoter activity. (A) M1 cells were transiently transfected with pGL3-Basic -444/-117 or constructs harboring site-directed mutagenesis of potential PU.1 binding sites. The cells were cotransfected with expression plasmids for PU.1, PU.1 and ICSBP, or the corresponding empty vectors. The samples were cultivated in the presence of IFNβ and analyzed for luciferase activity. Shown are absolute values normalized for Renilla luciferase activity. Error bars depict standard error. (Bi) EMSA analysis was performed using nuclear extracts (NEs) from M1 cells and a radiolabeled oligonucleotide Probe A spanning the region from -337 bp to -312 bp of the Ink4b promoter region. Two specific complexes (arrowheads) were resolved. No nuclear extract was added in lane 1. Nuclear extract from cells that were cultivated in the absence of IFNβ was added in lane 2. For all of the other lanes, nuclear extracts from cells cultivated in the presence of IFNβ for 16 hours were used. For lanes 4 to 7 an unlabeled wild-type (WT) oligonucleotide or an oligonucleotide carrying a mutation (MU) in the PU.1 binding site at -320 bp was added as a cold competitor (CC) at 10- or 50-fold excess compared with the labeled oligonucleotide. (Bii) EMSA analysis was performed as described for panel Bi. To determine which proteins are present in the observed complexes, the binding reactions were performed in the presence of antibodies against PU.1 (lane 3), Ets-2 (lane 4), TEL (lane 5), ICSBP (lane 6), or IRF-1 (lane 7). Arrowheads indicate protein/DNA complexes proposed to contain PU.1 and ISCBP.
In conjunction with our data in Figure 5A that strongly indicated that the 320-bp PU.1 potential binding site is involved in IFNβ-induced transcription, we wished to demonstrate that PU.1 binds to this site in M1 cells by EMSA. Following IFNβ treatment for 0 hours or 16 hours, nuclear lysates were prepared and used in EMSA with radiolabeled oligonucleotides panning the promoter region from -337 bp to -312 bp (Probe A) or from -374 bp to -335 bp (Probe B). Probe A contains the PU.1 site at -320 bp, identified before as being responsible for mediating PU.1 transcriptional activity. Using this probe, we observed 2 specific protein/DNA complexes, which significantly diminished following addition of the unlabeled oligonucleotide as a cold competitor (Figure 5Bi). Unlabeled oligonucleotide, carrying a mutation in the PU.1 site at -320 bp, was not able to compete for these protein/DNA complexes. Incubation of M1 cells with IFNβ lead to a strong increase in formation of the 2 complexes compared with the nonstimulated control, indicating that both complexes were formed in response to cytokine stimulation. Interestingly, addition of antibodies to PU.1 or ICSBP lead to a specific decrease in intensity of both protein/DNA complexes (Figure 5Bii). Control antibodies directed to Ets-2, TEL, or IRF-1 did not have any effect. This result not only indicated that PU.1 can bind to the potential PU.1 binding site at -320 bp and is present in each of the 2 detected specific complexes but also demonstrated that ICSBP is present in the 2 DNA/protein complexes. Inhibition of complex formation, rather than a supershift, was observed and is consistent with previously described results using the antibodies against PU.1 and ICSBP from Santa Cruz Biotechnology.42 Probe B included the other 2 potential PU.1 sites (-364 and -357) and the ICSBP site. Since it was not possible to make a good probe in which the 2 potential PU.1 sites were separated (because of their close proximity to each other), we examined them together. In an EMSA experiment using this probe, no specific bands were present that could compete with the unlabeled wild-type probe and, although there was a band present, it was not supershifted or blocked by anti-PU.1 antibody (data not shown). Therefore, we concluded that the potential PU.1 sites at -364 and -357 are not bound by PU.1 in M1 cells.
Mechanism of ICSBP's interaction with PU.1 at the Ink4b promoter
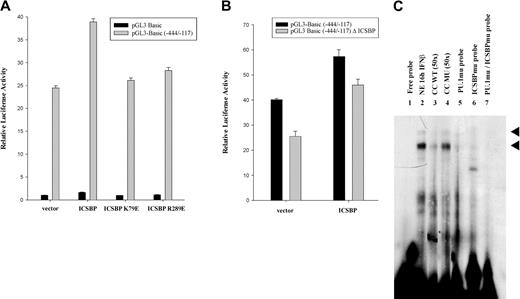
Regulation of promoter activity by PU.1 has been described by others to occur either by (1) binding of the PU.1 and an IRF to a PU.1/IRF composite element (adjacent sites) to form a complex or (2) tethering of an IRF to DNA through interaction with PU.1 in the absence of direct IRF binding.40 Since we did not find a PU.1/IRF composite element or complete ISRE sequence motif (binding site for IRFs) in the -444 bp to -117 bp promoter region, an indirect mode of binding initially seemed more likely. It was important, however, to clarify whether ICSBP is simply tethered to the DNA through PU.1 or could actually bind to the DNA itself. Therefore, we performed reporter gene assays using 2 ICSBP mutants described before by Tsujimura et al27 and Tamura et al23 to assist in the determination of the correct mechanism of this ICSBP promoter interaction. ICSBP K79E carries a mutation in the DNA binding domain that inhibits its binding to target elements. ICSBP R289E carries a mutation in the IRF association domain believed to be indispensable for interaction with PU.1.43 M1 cells were transfected with pGL3-Basic (-444/-117) and cotransfected with plasmids expressing ICSBP, ICSBP K79E, or ICSBP R289E. The cells were cultivated in the presence of IFNβ and analyzed for reporter gene activity (Figure 6A). In contrast to samples transfected with wild-type ICSBP, cotransfection of either one of the ICSBP mutants resulted in a negligible increase in promoter activity with an overall transactivation rate comparable with that of the negative control cotransfected empty vector. This result suggested that in order for ICSBP to activate transcription, both DNA binding and interaction with another protein is required. Surprisingly, cotransfection of PU.1 together with the ICSBP mutants abrogated the observed differences on reporter gene activity between the wild-type and the mutated ISCBPs (data not shown). We suggest that this is due to a large quantity of PU.1 in the cells following cotransfection with the PU.1-expressing vector that is able to counteract the destabilizing effect of the ICSBP mutants on PU.1/ICSBP transcription complex formation.
Analysis of ICSBP's binding to DNA and protein-protein interaction. (A) M1 cells cultivated in the presence of IFNβ were transiently transfected with either the pGL3-Basic vector or the pGL3-Basic -444/-117 promoter construct and cotransfected with expression plasmids for wild-type ICSBP, the DNA binding domain (DBD) mutant ICSBP K79E, the interaction domain (IAD) mutant ICSBP R289E, or the corresponding empty vector and analyzed for luciferase activity 24 hours following transfection. Shown are relative values normalized for Renilla luciferase activity. Error bars depict standard error. (B) M1 cells cultivated in the presence of IFNβ were transiently transfected with the pGL3-Basic -444/-117 promoter construct or a construct harboring a mutation in a potential ICSBP binding site at position -347 bp, pGL3 Basic (-444/-117) Δ ICSBP. An expression plasmid for ICSBP or the corresponding empty vector was cotransfected .The samples were analyzed for luciferase activity 24 hours following transfection. Shown are relative values normalized for Renilla luciferase activity. Error bars depict standard error. (C) EMSA analysis was performed using nuclear extracts (NEs) from M1 cells and radiolabeled oligonucleotides spanning the Ink4b promoter region from -364 bp to -312 bp (Probe C). The -320 bp PU.1 binding site, as well as the potential ICSBP binding site at -360 bp, are present in this region. For lanes 2 to 7, nuclear extract from cells that were cultivated in the presence of IFNβ for 16 hours was incubated with the following oligonucleotide probes: wild-type (WT; lanes 1-4), a probe carrying a mutation in the PU.1 site (PU.1mu probe; lane 5), a probe carrying a mutation in the ICSBP binding site (ICSBPmu probe; lane 6), and a probe carrying mutations in both the PU.1 and ICSBP binding site (PU.1mu/ICSBPmu probe; lane 7). Two specific complexes were resolved. Cold competitor oligonucleotide (CC) at 50-fold excess compared with the labeled oligonucleotide was included for lanes 3 and 4, using either unlabeled wild-type (WT) probe (lane 3) or a probe carrying a mutation (MU) in the PU.1 binding site at -320 bp as well as in the potential ICSBP binding site at -360 bp (lane 4). Arrowheads indicate protein/DNA complexes proposed to contain PU.1 and ISCBP.
Analysis of ICSBP's binding to DNA and protein-protein interaction. (A) M1 cells cultivated in the presence of IFNβ were transiently transfected with either the pGL3-Basic vector or the pGL3-Basic -444/-117 promoter construct and cotransfected with expression plasmids for wild-type ICSBP, the DNA binding domain (DBD) mutant ICSBP K79E, the interaction domain (IAD) mutant ICSBP R289E, or the corresponding empty vector and analyzed for luciferase activity 24 hours following transfection. Shown are relative values normalized for Renilla luciferase activity. Error bars depict standard error. (B) M1 cells cultivated in the presence of IFNβ were transiently transfected with the pGL3-Basic -444/-117 promoter construct or a construct harboring a mutation in a potential ICSBP binding site at position -347 bp, pGL3 Basic (-444/-117) Δ ICSBP. An expression plasmid for ICSBP or the corresponding empty vector was cotransfected .The samples were analyzed for luciferase activity 24 hours following transfection. Shown are relative values normalized for Renilla luciferase activity. Error bars depict standard error. (C) EMSA analysis was performed using nuclear extracts (NEs) from M1 cells and radiolabeled oligonucleotides spanning the Ink4b promoter region from -364 bp to -312 bp (Probe C). The -320 bp PU.1 binding site, as well as the potential ICSBP binding site at -360 bp, are present in this region. For lanes 2 to 7, nuclear extract from cells that were cultivated in the presence of IFNβ for 16 hours was incubated with the following oligonucleotide probes: wild-type (WT; lanes 1-4), a probe carrying a mutation in the PU.1 site (PU.1mu probe; lane 5), a probe carrying a mutation in the ICSBP binding site (ICSBPmu probe; lane 6), and a probe carrying mutations in both the PU.1 and ICSBP binding site (PU.1mu/ICSBPmu probe; lane 7). Two specific complexes were resolved. Cold competitor oligonucleotide (CC) at 50-fold excess compared with the labeled oligonucleotide was included for lanes 3 and 4, using either unlabeled wild-type (WT) probe (lane 3) or a probe carrying a mutation (MU) in the PU.1 binding site at -320 bp as well as in the potential ICSBP binding site at -360 bp (lane 4). Arrowheads indicate protein/DNA complexes proposed to contain PU.1 and ISCBP.
As mentioned above, there is no consensus ISRE binding motif in the -444/-117-bp Ink4b promoter region and no composite element at the PU.1 site at -320. However, since our data with the ICSBP mutants suggested that ICSBP binds to DNA, we examined the DNA sequence more carefully for a motif that might at least have part of the consensus binding site for ISCBP.40 We found a sequence at -350 bp that matched the core of the ICSBP binding motif (Figure 3). In order to determine if this partial ICSBP binding site is able to facilitate ICSBP activation of the Ink4b promoter, we mutated the ICSBP core-binding sequence in pGL3-Basic (-444/-117) and subsequently analyzed it for reporter gene activity (Figure 6B). The degree of transactivation of mutated reporter construct pGL3-Basic (-444/-117) Δ ICSBP in the presence of IFNβ was significantly lower than that of the nonmutated construct both in the presence and absence of ectopically expressed ICSBP. We, therefore, hypothesize that binding of ICSBP to this partial ISRE binding motif at -350 bp stabilizes the ICSBP/PU.1 binding complex and facilitates transcription.
To establish if binding of ICSBP to the proposed binding site 30 bp upstream of the -320 PU.1 binding site stabilizes the ICSBP/PU.1 binding to DNA, we carried out an EMSA with a radiolabeled oligonucleotide that spanned from -363 bp to -312 bp (Probe C) and that included potential binding sites for both transcription factors. Nuclear lysates were harvested from M1 cells treated for 16 hours with IFNβ and were used in a binding assay. As shown in Figure 6C, one strong and one weak band were observed. The 2 protein/DNA complexes significantly diminished following competition with unlabeled oligonucleotide (Figure 6C lane 3), but did not diminish with unlabeled oligonucleotide carrying either a mutation in the -320-bp PU.1 binding site or the -350-bp ICSBP binding site (Figure 6C lane 4). Radiolabeled probes with a mutation in the -320-bp PU.1 binding site or in the potential -350-bp ICSBP binding site were not able to form complexes seen with the wild-type probe (Figure 6C lanes 5-6). A radiolabeled probe carrying mutations in both binding sites completely abolished complex formation (Figure 6C lane 7). These observations further support the idea that both transcription factors, PU.1 and ICSBP, bind to the DNA producing a stable DNA binding complex. This hypothesis is further strengthened by the fact that one of the PU.1/ICSBP complexes we detected in Figure 6C is significantly stronger than either of those we detected using the 30-bp probe (Figure 5B) containing only the PU.1 site.
PU.1 and ICSBP are both bound to the endogenous Ink4b promoter in M1 cells
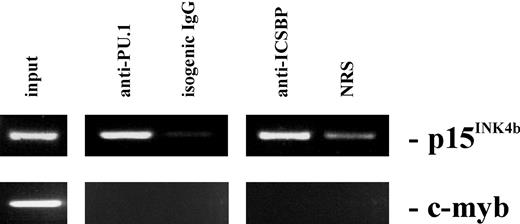
To see if PU.1 and ICSBP proteins bind to the Ink4b promoter in vivo, we performed a ChIP analysis. M1 cells were treated with IFNβ for 16 hours and the proteins were cross-linked to DNA using formaldehyde. Following immunoprecipitation with anti-PU.1, anti-ICSBP, or control antibody, cross-linking was reversed and the DNA was amplified with Ink4b-specific primers. As a negative control, specific primers that amplify part of the c-Myb promoter were also used. Although both primer pairs amplified a DNA product of the predicted size from the total input DNA (Figure 7), only the Ink4b-specific primers produced a highly visual product from anti-PU.1–precipitated or anti-ICSBP–precipitated DNA. These data indicate that both PU.1 and ICSBP proteins bind the endogenous Ink4b promoter in vivo in M1 cells treated with IFNβ.
PU.1 and ICSBP are both present at the Ink4b promoter region. Chromatin immunoprecipitation (ChIP) analysis was performed with M1 cells cultivated in the presence of IFNβ for 16 hours. Cross-linked protein-DNA complexes were precipitated by addition of antibodies against PU.1 or ICSBP to the cell lysates and analyzed by PCR for the presence of the Ink4b promoter region or for the c-myb promoter region as a negative control. As an additional negative control, normal rabbit serum or isogenic IgGs was used for precipitation.
PU.1 and ICSBP are both present at the Ink4b promoter region. Chromatin immunoprecipitation (ChIP) analysis was performed with M1 cells cultivated in the presence of IFNβ for 16 hours. Cross-linked protein-DNA complexes were precipitated by addition of antibodies against PU.1 or ICSBP to the cell lysates and analyzed by PCR for the presence of the Ink4b promoter region or for the c-myb promoter region as a negative control. As an additional negative control, normal rabbit serum or isogenic IgGs was used for precipitation.
Expression of ICSBP in an ICSBP-/- myeloid progenitor cell line induces the expression of p15Ink4b
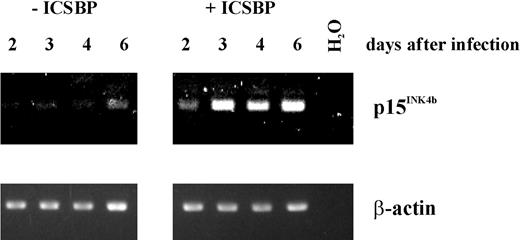
Tamura and coworkers23 reported the establishment of an ICSBP-/- immature blast cell line, Tot2, from bone marrow cells of ICSBP knockout mice. Transduction of these cells with an ICSBP-expressing vector was shown to induce differentiation toward the macrophage lineage in association with growth arrest. We were interested in whether p15Ink4b expression is induced following expression of ICSBP and, therefore, might play a role in promoting growth arrest. We analyzed the cells using RT-PCR for p15Ink4b mRNA expression 2, 3, 4, and 6 days following transduction with the ICSBP-expressing vector and compared this expression with that in cells transfected with the empty vector (Figure 8). As early as 2 days following transduction of ICSBP, a significant level of RT-PCR product could be detected that continued to rise during the subsequent 4 days. In contrast to the ICSBP-expressing cells, there was only basal expression of the CDK inhibitor RNA in the control cells with exception of day 6 (where we observed a slight increase in p15Ink4b expression).
Expression of ICSBP in ICSBP-/- cells activates the Ink4b promoter. Tot2 cells were cultivated for 2, 3, 4, or 6 days following transduction with an ICSBP-expressing vector. The cells were harvested and total RNA was isolated. The p15Ink4b cDNA was amplified by RT-PCR using 50 ng of total RNA. To control for sample loading, β-actin cDNA was amplified from the same RNA samples.
Expression of ICSBP in ICSBP-/- cells activates the Ink4b promoter. Tot2 cells were cultivated for 2, 3, 4, or 6 days following transduction with an ICSBP-expressing vector. The cells were harvested and total RNA was isolated. The p15Ink4b cDNA was amplified by RT-PCR using 50 ng of total RNA. To control for sample loading, β-actin cDNA was amplified from the same RNA samples.
Discussion
This study presents a model to explain how the expression of the tumor suppressor p15Ink4b in myeloid cells is controlled in response to treatment with the growth inhibitory cytokine IFNβ. Data here shows that the transcription factors PU.1 and ICSBP bind to DNA and regulate transcription from the promoter in myeloid cells in response to this type I IFN. The role of ICSBP in regulating the promoter was further substantiated in an experiment where its ectopic expression in an ICSBP-null hematopoietic cell line resulted in a dramatic increase in p15Ink4b RNA. The regulation of the Ink4b promoter by ICSBP and PU.1 is interesting for 2 reasons. First, both p15Ink4b and ICSBP are proposed tumor suppressors. The finding that ICSBP positively regulates the expression of another tumor suppressor provides an explanation for its own tumor suppressive function. Second, both PU.1 and ICSBP are myeloid commitment factors, suggesting that the observed regulation of p15Ink4b expression by these factors is lineage specific.
The fact that the ICSBP product regulates the Ink4b promoter shows that the 2 genes are in the same pathway of growth control. It had been previously found that ICSBP-deficient mice develop a CML-like disease and that 66% of all AML and 79% of all CML patients have reduced levels of ICSBP or no expression at all.17,19 Functions of ICSBP that would explain this tumor suppressor role have been elusive until now. Therefore, our observation that ICSBP regulates p15Ink4b expression provides, at least in part, an explanation for ICSBP's role in growth inhibition. It was also demonstrated recently that ICSBP inhibits growth of Bcr/Abl-transformed cells by activating several genes that interfere with the c-Myc pathway.44
PU.1 has been shown to be important not only for lineage commitment of hematopoietic progenitors into myeloid cells and B cells but also for terminal differentiation of myeloid cells into macrophages.45-47 ICSBP is also a myeloid commitment factor.17 For example, ICSBP-/- mice harbor an increased number of myeloid progenitor cells compared with normal mice. These myeloid cells have impaired macrophage differentiation and mature mainly into granulocytes. Although, we did not observe a composite element for ICSBP and PU.1 in the Ink4b promoter, similar to that found in the gp91phox promoter and in other myeloid specific promoters,40 we found a consensus binding site for PU.1 and a partial consensus binding site for ICSBP that was 30 bp upstream. We provide evidence that both binding sites are used to form a stable complex that is active in transcription of the Ink4b promoter. Even though ICSBP and PU.1 are present in cells prior to treatment with IFNβ, we postulate that posttranslation modification of one or both factors is required for promoter activation. In fact, binding of ICSBP is probably so weak that it is not bound without interaction with PU.1. Following treatment with IFNβ, modification of the ICSBP and/or PU.1, most likely through phosphorylation, would be predicted to promote interaction of ICSBP with PU.1 and subsequent binding of ICSBP to DNA. The resulting formation of a stable PU.1/ICSBP complex would increase transactivation of p15Ink4b expression. It has been shown for PU.1, in other experimental systems, that phosphorylation of Ser148 in the PEST (proline-glutamic acid-serine-threonine) domain of the protein is critical for complex formation.34,48,49 For ICSBP, it has been reported that tyrosine phosphorylation is essential for efficient protein-protein interaction.50
The Myc-interacting zinc finger protein 1 (Miz-1) has been shown recently to be instrumental in regulating p15Ink4b expression in TGFβ-treated fibroblasts by binding to the Inr.11,12 Although we cannot rule out that Miz-1 may play a role in inducing p15Ink4b expression in cells of the myeloid compartment, its presence is definitively not required for PU.1's or for ICSBP's ability to increase expression. In Del-444/-274, the Inr region was removed and the remaining sequence was cloned in front of an SV40 promoter. Using this construct, we were able to observe an increase in promoter activity in response to cotransfection of PU.1 and ICSBP.
Administration of type I interferons is an important treatment for CML patients in chronic phase. These agents are also widely used in the treatment of myeloproliferative disorders.51 The study presented here suggests a mechanism to explain how IFN may inhibit growth. The INK4b gene is not usually inactivated in CMLs as it is in AML. Therefore, the fact that IFN is effective for treatment of CML, but not AML, could be correlated with the INK4b promoter status. It would be interesting to determine if p15INK4b expression is up-regulated in CML in response to the IFN treatment.
Prepublished online as Blood First Edition Paper, February 19, 2004; DOI 10.1182/blood-2003-01-0285.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Marcos Malumbres for kindly providing us with the plasmid pmp15, harboring the genomic Ink4b promoter sequence, which enabled us to do this study. Additionally, we thank Harinder Singh for providing us with the PU.1 expression plasmids, John Brady for sharing his expertise in EMSA technology, Carol Stocking for invaluable and stimulating discussions, and Jan Markus for critical reading of the manuscript.
J.B. is on leave from the Cancer Research Institute, Slovak Academy of Sciences, Bratislava, Slovak Republic.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal