Abstract
The hematopoietic microenvironment, approximated in vitro by long-term marrow cultures (LTCs), consists of both nonhematopoietic-derived stromal elements and hematopoietic-derived monocyte/macrophages. To better understand the consequences of monocyte-stroma interactions, we compared gene expression profiles of CD14+ peripheral blood monocytes and HS-27a stromal cells cultured alone and together in cocultures. Results from 7 separate experiments revealed 22 genes were significantly up- or down-regulated in the cocultures, with osteopontin (OPN) up-regulated more than 15-fold. The microarray OPN data were confirmed by Northern blot, real-time polymerase chain reaction (PCR), and by detection of OPN protein. High levels of OPN gene expression were also detected in 2- to 3-week-old primary LTCs. Using Transwells we determined that stromal cells were secreting a factor that up-regulated OPN gene expression in CD14+ cells. When CD34+ cells were cultured in the presence of purified OPN, tyrosine phosphorylation of a 34-kDa molecule was increased 2- to 3-fold, an effect that was diminished in the presence of an OPN neutralizing monoclonal antibody. In addition, Notch1 gene expression was decreased 5-fold in OPN-treated CD34+ cells. We conclude that interactions between stroma and monocytes can result in activities that limit the role of Notch signaling in hematopoietic regulation. (Blood. 2004;103:4496-4502)
Introduction
The hematopoietic microenvironment (ME), defined largely by function, is a complex mixture of cells and factors critical for the regulation of hematopoiesis. The ME is assayed in vitro using primary long-term marrow cultures (LTCs) that consist of nonhematopoietic-derived stromal components as well as hematopoietic components, including monocytes and macrophages. A critical in vivo role for the macrophage in the marrow ME has long been inferred from histologic studies, best exemplified by the nurse cell of the erythropoietic island. In primary LTCs, up to 40% of all cells can be macrophages even after 12 weeks of culture.1 These cells are known to secrete many cytokines such as interleukin-1 (IL-1), interleukin-6 (IL-6), granulocyte colony-stimulating factor (G-CSF), macrophage colony-stimulating factor (M-CSF), and tumor necrosis factor (TNF), all of which have direct effects on hematopoietic cells. Monocytes/macrophages are also capable of stimulating stromal cells to produce cytokines.2-5 Because macrophages also produce transforming growth factor-β (TGFβ) and platelet-derived growth factor (PDGF), both of which can influence the mitotic rate of fibroblasts, the macrophage may also be capable of regulating stromal cell growth.6-10
Gaining a more precise understanding of monocyte-stroma interactions within the ME is complicated by the heterogeneous nature of the stromal compartment. In an effort to functionally dissect the ME, several groups have established cloned stromal cell lines that identify individual ME components.11-15 We previously reported on immortalized human marrow stromal cell lines that represent functionally distinct components of the ME.16 One of these, designated HS-27a, supports cobblestone area formation of early hematopoietic progenitors and expresses a Notch ligand, Jagged-1, as well as other cell surface and matrix molecules.17
In the present study, we investigated the changes in gene expression caused by coculturing HS-27a with CD14+ peripheral blood monocytes. In some experiments CD34+ cells were also added to the cocultures. A comprehensive study of gene expression in cocultures was undertaken using DNA microarray technology. We found that HS-27a provided a signal to CD14+ cells that stimulated the expression of osteopontin (OPN). Although some additional chemokine genes were also induced (eg, CXCL7 and CXCL1), the stromal-derived factor did not result in general activation of monocytes, because gene expression of other activities normally associated with monocyte activation by lipopolysaccharide (LPS)18 (eg, interleukin-1, CXCL2, and prostaglandin endoperoxide synthase-2) was not induced. Stroma-stimulated CD14+ cell-conditioned media as well as purified OPN were found to down-regulate gene expression of Notch1 in CD34+ cells. Given the role of Notch signaling in hematopoietic cell fate determination,19,20 a similar interaction in vivo between a subset of stromal cells and monocytes could affect the responsiveness of CD34+ cells to differentiation signals.
Materials and methods
Cell culture
Marrow stromal cell lines HS-27a, HS-5, and human foreskin fibroblasts (HFFs) were grown in RPMI 1640 medium (Gibco, Carlsbad, CA) supplemented with L-glutamine (0.4 mg/mL), sodium pyruvate (1 mM), and 10% fetal calf serum (FCS) at 37°C under 95% air, 5% CO2. After reaching confluency, cells were trypsinized and transferred at a cell density of 40 × 103/cm2 to multiwell plates for coculture studies. At this density, cultures were about 80% confluent and reached complete confluency at 3 days.
Bone marrow aspirates and peripheral blood were obtained from healthy donors after written informed consent using forms approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center (FHCRC). Primary long-term cultures (LTCs) from marrow of healthy donors were prepared under conditions previously described.21
Normal peripheral blood mononuclear cells (PBMCs) were isolated over a Ficoll step gradient, hemolyzed, and washed 3 times in Hanks balanced salt solution (HBSS). CD14+ cells were isolated by AutoMacs (Miltenyi Biotec, Auburn, CA) after incubation with a CD14 monoclonal antibody (clone Tük4; Dako, Carpinteria, CA) and antimouse immunoglobulin G2a+b (IgG2a+b)-conjugated magnetic microbeads (Miltenyi Biotec). In brief, 10 × 106 to 50 × 106 PBMCs were suspended in 0.1 to 0.5 mL phosphate-buffered saline (PBS) containing EDTA (ethylenediaminetetraacetic acid) (1.488 g/L) and 0.5% bovine serum albumin (BSA). The CD14 antibody was added at 5 μL/107 cells and incubated at 4°C for 10 minutes. The cells were washed, and the magnetic microbeads were added at 7 μL/107 cells. After incubating at 4°C for 10 minutes, the cells were washed 3 times and positively selected with an AutoMacs magnetic separation apparatus (Miltenyi Biotec). This magnetic separation was repeated twice. Purity of CD14+ cells from healthy donors was more than 95%.
CD34-enriched cells from G-CSF-mobilized blood mononuclear cells were obtained through the Cell Processing Shared Resource of the FHCRC, as previously described.22 After thawing, more than 90% of the cells were found to be alive and expressing the CD34 antigen by flow analysis.
Transwell cultures
In some experiments, CD14+ cells were cultured in a Transwell insert (Millicell-CM, 0.4-μm mesh, 30-mm diameter; Millipore, Bedford, MA). Nonadherent cells were harvested and adherent cells on the insert released by immersion in 0.05% trypsin-EDTA solution (Gibco, Rockville, MD). Both adherent and nonadherent cells were combined, and total RNA was isolated as described below.
In some experiments, 0.2 × 106 CD34+ cells were seeded in a Transwell insert (Millicell-CM, 0.4 μm, 30-mm diameter; Millipore, Billerica, MA) and cultured for 4 days in Iscove medium (Gibco, Carlsbad, CA) containing a growth factor cocktail of 100 ng/mL stem cell factor (SCF), 5 ng/mL IL-3, 5 ng/mL G-CSF, 5 ng/mL IL-6, 0.4 mg/mL L-glutamine, 1 mM sodium pyruvate, and 10% FCS.
Conditioned media
HS-27a cells (3 × 106 cells) were grown in supplemented RPMI 1640 medium containing 10% FCS in a T-75 flask. Conditioned medium was harvested after 5 days and clarified by centrifugation. The conditioned medium was diluted with more than 1 volume of supplemented RPMI 1640 medium before use.
DNA microarray sample preparation
HS-27a cells (3 × 106) were cultured for 3 days in a T-75 flask in the presence or absence of 1.5 × 106 CD14+ cells. Nonadherent cells were removed by washing 3 times with HBSS, and total RNA was extracted from the adherent cells using RNeasy spin columns (Qiagen, Valencia, CA). Seven RNA preparations for each experimental group were prepared separately over a period of 7 months. RNA from HS-27a cultured alone was used as a control. The RNA (30 μg) was annealed with 5 μg oligo dT12-18 (Pharmacia, Piscataway, NJ) and reverse-transcribed into cDNA with Superscript II reverse transcriptase (Gibco, Rockville, MD) for 2 hours at 42°C in the presence of 0.5 mM deoxyguanosine triphosphate (dGTP), 0.5 mM deoxycytidine triphosphate (dCTP), 0.5 mM deoxyadenosine triphosphate (dATP), 0.3 mM deoxythymidine triphosphate (dTTP), and 0.2 mM amino-allyl deoxyuridine triphosphate (dUTP; Sigma, St Louis, MO). After hydrolysis of RNA in 0.2 M NaOH, followed by neutralization of the reaction with Tris (tris(hydroxymethyl)aminomethane)-HCl, pH 7.4, Tris was removed from the reaction with a Microcon-30 concentrator (Amicon, Beverly, MA). The cDNA from cocultures and HS-27a-alone cultures was covalently coupled separately with cyanine 3 (Cy3; Amersham, Piscataway, NJ) and Cy5 monoreactive fluors, respectively, in 50 mM sodium bicarbonate, pH 9.0, followed by quenching with 2.7 M hydroxylamine. The Cy5- and Cy3-labeled cDNAs were combined and purified with QIAquick polymerase chain reaction (PCR) purification kit (Qiagen) and suspended in 36 μL3 × SSC and 0.8 mg/mL polyA (Roche, Indianapolis, IN).
Microarray hybridization and data analysis
Fluor-labeled cDNA was hybridized to human spotted microarrays constructed using a set of more than 17 000 sequence-verified clones from the Research Genetics IMAGE (Integrated Molecular Analysis of Genomes and their Expression) clone set, available through Invitrogen (http://clones.invitrogen.com/). The cDNA inserts were amplified from replica plate cultures by PCR, purified, and verified as unique before being mechanically spotted onto polylysine-coated glass microscope slides using an OmniGrid high-precision robotic gridder (GeneMachines, San Carlo, CA). Microarray construction and hybridization protocols were modified from those described elsewhere.23 The hybridization was conducted in the DNA Array Facility at the Fred Hutchinson Cancer Research Center. Fluorescent array images were collected for both Cy3 and Cy5 by using a GenePix 4000A fluorescent scanner (Axon Instruments, Foster City, CA), and image intensity data were extracted and analyzed by GENEPIX PRO 3.0 microarray analysis software. Gene expression was shown as a ratio of fluorescence intensity of Cy3-labeled HS-27a+monocyte coculture cDNA to Cy5-labeled cDNA from the HS-27a-alone culture. Intensity-dependent normalization of all samples was done with the scatter plot smoother “Lowess” implemented in the statistical software package R (as described by Yang et al24 ). This regression analysis-based normalization method performs locally linear fits, which are not affected by a small percentage of differentially expressed genes. The fraction of the data used for smoothing at each point (f value) was 50%. GeneSpring 5.0 software (Silicon Genetics, Redwood City, CA) was used both for the Lowess normalization procedure and the preliminary analysis of variance (ANOVA) determination of genes differentially expressed between CD14+ cells and HS-27a. Statistical analysis of differences between the cultures was carried out by CyberT software25 available at http://visitor.ics.uci.edu/genex/cybert/help/. Genes with spots flagged by the GenePix program for low signal or high noise in more than 5 of the 7 samples analyzed were not included in the analysis. The prenormalization dataset has been submitted to the GEO database (http://www.ncbi.nlm.nih.gov/geo/) as series GSE831. Raw data, Lowess normalized data, summary tables, and Minimal Information About a Microarray Experiment (MIAME), as recommended by the Microarray Gene Expression Data society, are available as Supplemental Materials (see the Supplemental Materials link at the top of the online article on the Blood website).
Syber Green real-time polymerase chain reaction (PCR) analysis
Expression levels of RNA transcripts were quantitated by real-time PCR. Total RNA was purified using RNeasy spin columns (Qiagen) and then DNase-treated at 37°C for 30 minutes using 1000 units of RQ1 RNase-free DNase (Promega, Madison, WI). Samples were then reverse-transcribed into cDNA with an oligo dT12-18 primer. Complementary DNAs were mixed with SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) and primers, and real-time PCR was performed using ABI Prism 7700 Sequence Detector (Perkin Elmer, Boston, MA). PCR conditions were 15 seconds at 95°C and 2 minutes at 68°C with 40 cycles for real-time PCR. Forward and reverse oligonucleotide primers are as follows, in 5′ to 3′ orientation: ATCGCTCTGAAATTAGCCTACTGCC and TAAGGCTGGCAAATACACCAACACC for glyceraldehyde 3-phosphate dehydrogenase (G3PDH); GGAAGGACTCATGACCACAGTCC and TCGCTGTTGAAGTCAGAGGAGACC for GAPDH; AGGCTGATTCTGGAAGTTCTGAGG and ACTCCTCGCTTTCCATGTGTGAGG for OPN; GGACATCACGGATCATATGGACC and GAGGGTTGTATTGGTTCGGCACC for Notch1; TTGGAGCTCCGTATGATGACTTGG and GGATCTCCACTGAGGCAGTTATGG for integrin alpha 6 (ITGA6). The data were represented by the average ± SD for at least 2 samples.
Northern blot
Northern blot analysis was performed using a 32P-labeled DNA probe of OPN (DNA clone no. 835575R; American Type Culture Collection, Manassas, VA), which has the same DNA sequence as a DNA spot for OPN in the spotted microarray (Research Genetics clone ID 378461). Five micrograms of total RNA were prepared from CD14+ cells cultured alone, HS-27a cultured alone, and adherent cells from the cocultures using RNeasy spin columns (Qiagen). Formaldehyde RNA agarose gels, capillary blotting, and hybridization were conducted as previously described.26,27 The blot was exposed to a phosphor screen that was quantitatively scanned by a Typhoon scanner (Molecular Dynamics, Santa Cruz, CA), and analysis was done using ImageQuant software, also from Molecular Dynamics.
Detection of tyrosine phosphorylation
Tyrosine phosphorylation of proteins in CD34+ cells was detected by flow-based and Western blot analyses. CD34+ cells (0.3 × 106 cells) were suspended in 0.1 mL PBS containing 0.5% BSA (PBS-BSA) and incubated at 22°C for 2 minutes with 1 μg/mL purified milk OPN (Kamiya Biomedical, Seattle, WA) in the presence or absence of 10.8 μg/mL of a neutralizing antibody (MPIIIB10; Developmental Studies Hybridoma Bank, Iowa City, IA). The cells were fixed and permeabilized using a Fix and Perm Cell Permeabilization kit (Caltag Laboratories, An Der Grub, Austria) according to the manufacturer's protocol. Tyrosine-phosphorylated proteins were detected using fluorescein isothiocyanate (FITC)-labeled antiphosphotyrosine antibody (4G10; Upstate Biotechnology, Lake Placid, NY). FITC-labeled isotype antibody was used as a negative control. The fluorescence intensity of the cells was analyzed by flow cytometry (FACSCalibur; Becton Dickinson, San Jose, CA).
In some experiments, tyrosine-phosphorylated proteins were detected by Western blot as previously described.27 CD34+ cells (1 × 106 to 2 × 106 cells) were suspended in 0.1 mL PBS-BSA and incubated at 22°C for 2 minutes with 1 μg/mL OPN in the presence or absence of the neutralizing antibody or isotype control at 10.8 μg/mL. OPN and the antibodies were preincubated at 4°C for 30 minutes before applying to the cells. The cells were quickly spun down, supernatant was completely aspirated, and 0.1 mL boiling Laemmli sample buffer containing 5% β-mercaptoethanol was added to the cell pellets. The cell extracts were briefly sonicated to reduce viscosity and heat-denatured at 100°C for 5 minutes. Forty microliters of the extracts were applied to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a precast 10% polyacrylamide gel (Invitrogen, Carlsbad, CA) followed by immunoblot with PY-20 antiphosphotyrosine antibody (Transduction Laboratories, Lexington, KY) as described elsewhere.27 Bench Mark protein ladder (Gibco, Rockville, MD) was used to calculate molecular weight of the phosphorylated proteins.
Cytokine, endotoxin, and mycoplasma analysis
Culture supernatants were harvested from cell cultures and frozen at -80°C until analyzed. Cytokine enzyme-linked immunosorbent assays (ELISAs) of OPN were performed using a TiterZyme EIA kit (Assay Design, Ann Arbor, MI). The data were represented by the average ± SD for more than 2 samples.
Endotoxin and mycoplasma levels of media, cytokine, and reagents were tested using a Limulus Amebocyte Lysate test kit (Charles River Laboratories, Wilmington, MA) by a kinetic-turbidmetric method and DAPI (4,6-diamidino-2-phenylindole)-based cytochemical detection, respectively. Both assays were performed by the Biologics Production Shared Resource at FHCRC.
Results
Microarray comparison of gene expression in cocultures of HS-27a and CD14+ cells
After 3 × 106 HS-27a cells were cocultured with 1.5 × 106 CD14+ monocytes for 3 days, adherent cells were harvested and total RNA was isolated. Microarray experiments were conducted using monocytes from 7 healthy donors over a period of more than 7 months. Because it is not possible to remove all CD14+ cells from the adherent layer of stromal cells, the contribution of CD14+ cells (5%-10% of total adherent cells) to the expressed gene profiles had to be evaluated. Previously generated expression profiles of CD14+ cells alone (L.G. and B.T.-S., unpublished data, June 2001) and HS-27a alone27 relative to a universal control RNA were used to obtain information on genes with large expression differences between these 2 cell types. Statistical analysis (ANOVA, P < .05, n = 3, with Benjamini and Hochberg false discovery rate correction) showed 3268 genes differentially expressed. Of those, 411 gene sequences were more than 5-fold higher in HS-27a than in CD14+ cells, predominantly those coding for collagens and proteoglycans, as well as matrix metalloproteinases and their regulators. Additionally, 444 sequences were more than 5-fold higher in CD14+ cells than in HS-27a, with the largest differences in major histocompatibility complex (MHC) class II expression and that of chemokines, cytokines, and their receptors. Any sequences found in this comparison of freshly isolated CD14+ cells and HS-27a to be more than 2-fold higher in expression in CD14+ cells than in HS-27a were virtually subtracted from the analysis of the coculture data.
Table 1 provides a list of those transcripts significantly up- or down-regulated in adherent cells of the cocultures. Statistical analysis was performed based on t test using point estimates generated by a Bayesian framework (CyberT). The complete data set and gene identifications are available as Supplemental Materials and on the NCBI Gene Expression Omnibus Web site (www.ncbi.nlm.nih.gov/geo). The most highly up-regulated gene in the coculture was OPN, also known as secreted phosphoprotein 1 or early T-lymphocyte antigen 1. Other differentially expressed genes included chemokines (up-regulated) and immunomodulatory protein PSG1 (down-regulated). The 15-fold increase in OPN mRNA was of interest given the many different cellular signaling events attributed to this protein.28,29
Genes significantly overexpressed or underexpressed in HS-27a/CD14+ monocyte cocultures compared with either population cultured alone
Clone ID . | Accession . | Cluster ID . | Name . | Annotation . | Ratio* . | P† . |
|---|---|---|---|---|---|---|
| 378461 | AA775616 | Hs.313 | SPP1 | Osteopontin | 15.4 | 1.63 × 10−6 |
| 418422 | W92812 | Hs.2164 | PPBP | CXCL7 | 13.7 | 2.69 × 10−9 |
| 840493 | AA485893 | Hs.78224 | RNASE1 | Ribonuclease, RNase A family | 7.6 | 4.36 × 10−8 |
| 773330 | AA425450 | — | — | ESTs | 4.5 | 1.13 × 10−10 |
| 841046 | AA486747 | Hs.8904 | Z391G | Ig superfamily protein | 4.4 | 2.03 × 10−9 |
| 502244 | AA133297 | — | — | ESTs | 3.8 | 2.30 × 10−6 |
| 470393 | AA031514 | Hs.2256 | MMP7 | Matrix metalloproteinase 7 | 3.2 | 1.28 × 10−6 |
| 292833 | N69227 | Hs.94953 | C1QG | Complement component 1 | 3.1 | 5.93 × 10−8 |
| 247587 | N58136 | — | — | ESTs | 2.4 | 1.65 × 10−6 |
| 73268 | T56021 | Hs.5057 | CPD | Carboxypeptidase D | 2.0 | 1.89 × 10−6 |
| 50182 | H17883 | — | — | ESTs | 1.8 | 9.72 × 10−7 |
| 813614 | AA447684 | Hs.1076 | SPRR1B | Small proline-rich protein 1B (cornifin) | 1.8 | 7.66 × 10−7 |
| 784360 | AA447196 | Hs.12451 | EML1 | Microtubule-associated protein | 1.7 | 1.13 × 10−6 |
| 324437 | W46900 | Hs.789 | CXCL1 | CXCL1 (GRO1 oncogene) | 1.6 | 7.09 × 10−7 |
| 739625 | AA479623 | Hs.227489 | SAST | Syntrophin-associated serine/threonine kinase | 1.6 | 5.21 × 10−7 |
| 382654 | AA069517 | — | — | ESTs | 1.6 | 3.22 × 10−6 |
| 786069 | AA448660 | Hs.49349 | BACE | Beta-site APP-cleaving enzyme | 0.7 | 2.53 × 10−6 |
| 130057 | R20779 | Hs.155223 | STC2 | Stanniocalcin 2 | 0.6 | 2.26 × 10−6 |
| 825785 | AA505117 | — | — | ESTs | 0.6 | 2.53 × 10−6 |
| 325641 | W51985 | Hs.251850 | PSG1 | Pregnancy-specific beta-1-glycoprotein | 0.5 | 7.40 × 10−7 |
| 811785 | AA463463 | — | — | ESTs | 0.5 | 7.94 × 10−7 |
| 199635 | R96522 | — | — | ESTs | 0.5 | 3.71 × 10−7 |
Clone ID . | Accession . | Cluster ID . | Name . | Annotation . | Ratio* . | P† . |
|---|---|---|---|---|---|---|
| 378461 | AA775616 | Hs.313 | SPP1 | Osteopontin | 15.4 | 1.63 × 10−6 |
| 418422 | W92812 | Hs.2164 | PPBP | CXCL7 | 13.7 | 2.69 × 10−9 |
| 840493 | AA485893 | Hs.78224 | RNASE1 | Ribonuclease, RNase A family | 7.6 | 4.36 × 10−8 |
| 773330 | AA425450 | — | — | ESTs | 4.5 | 1.13 × 10−10 |
| 841046 | AA486747 | Hs.8904 | Z391G | Ig superfamily protein | 4.4 | 2.03 × 10−9 |
| 502244 | AA133297 | — | — | ESTs | 3.8 | 2.30 × 10−6 |
| 470393 | AA031514 | Hs.2256 | MMP7 | Matrix metalloproteinase 7 | 3.2 | 1.28 × 10−6 |
| 292833 | N69227 | Hs.94953 | C1QG | Complement component 1 | 3.1 | 5.93 × 10−8 |
| 247587 | N58136 | — | — | ESTs | 2.4 | 1.65 × 10−6 |
| 73268 | T56021 | Hs.5057 | CPD | Carboxypeptidase D | 2.0 | 1.89 × 10−6 |
| 50182 | H17883 | — | — | ESTs | 1.8 | 9.72 × 10−7 |
| 813614 | AA447684 | Hs.1076 | SPRR1B | Small proline-rich protein 1B (cornifin) | 1.8 | 7.66 × 10−7 |
| 784360 | AA447196 | Hs.12451 | EML1 | Microtubule-associated protein | 1.7 | 1.13 × 10−6 |
| 324437 | W46900 | Hs.789 | CXCL1 | CXCL1 (GRO1 oncogene) | 1.6 | 7.09 × 10−7 |
| 739625 | AA479623 | Hs.227489 | SAST | Syntrophin-associated serine/threonine kinase | 1.6 | 5.21 × 10−7 |
| 382654 | AA069517 | — | — | ESTs | 1.6 | 3.22 × 10−6 |
| 786069 | AA448660 | Hs.49349 | BACE | Beta-site APP-cleaving enzyme | 0.7 | 2.53 × 10−6 |
| 130057 | R20779 | Hs.155223 | STC2 | Stanniocalcin 2 | 0.6 | 2.26 × 10−6 |
| 825785 | AA505117 | — | — | ESTs | 0.6 | 2.53 × 10−6 |
| 325641 | W51985 | Hs.251850 | PSG1 | Pregnancy-specific beta-1-glycoprotein | 0.5 | 7.40 × 10−7 |
| 811785 | AA463463 | — | — | ESTs | 0.5 | 7.94 × 10−7 |
| 199635 | R96522 | — | — | ESTs | 0.5 | 3.71 × 10−7 |
Identifiers and names are derived from information available in public databases, primarily the National Center for Biotechnology Information (NCBI) UniGene database and GenBank. Cultures were washed thoroughly to remove monocytes, and flow cytometry of the harvested cells showed 5% to 10% monocytes remaining in the cultures at 3 days.
— indicates that cluster ID or name is not available; ESTs, expressed sequence tags; and APP, amyloid precursor protein.
Linearized ratio of gene expression of cells in coculture to gene expression of HS-27a cells cultured alone. (Known monocyte-specific genes, such as HLA-DR, have been excluded.)
P value is based on a t test using point estimates by a Bayesian framework (CyberT) on 7 independent experiments.
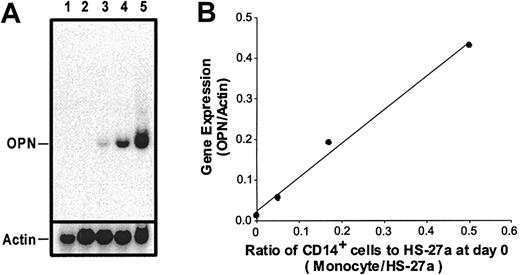
The microarray data on OPN was confirmed by Northern blot analysis, as shown in Figure 1. When HS-27a cells were cocultured with different numbers of CD14+ cells, OPN gene expression in the adherent cells increased proportionally to the CD14+ cell number (Figure 1A, lanes 3 through 5). Quantitation by phosphor image analysis of the Northern blot showed a linear relationship between numbers of CD14+ cells in coculture and OPN gene expression (Figure 1B).
Northern blot analysis of OPN gene expression. (A) Northern blot of peripheral blood monocytes (lane 1) and 3-day cultures of HS-27a alone (lane 2) and adherent cells from cocultures of HS-27a and monocytes with different monocyte-HS-27a ratios (lanes 3-5). The monocyte-HS-27a ratios at day 0 in lanes 3, 4, and 5 are 1:20, 1:6, and 1:2, respectively. Total RNA (5 μg per lane) was separated on an RNA agarose gel and blotted onto a nylon membrane. The membrane was probed with a 32P-labeled OPN probe. The bound probe was stripped off, and the membrane was reprobed with a 32P-labeled actin probe. (B) Quantitation of the Northern blot by phosphor image analysis showed a strong correlation between the ratio of monocytes to HS-27a and OPN gene expression (R2 = 0.99). The data indicate a strong signal for OPN in RNA isolated from cocultures but not HS-27a or CD14+ cells cultured alone.
Northern blot analysis of OPN gene expression. (A) Northern blot of peripheral blood monocytes (lane 1) and 3-day cultures of HS-27a alone (lane 2) and adherent cells from cocultures of HS-27a and monocytes with different monocyte-HS-27a ratios (lanes 3-5). The monocyte-HS-27a ratios at day 0 in lanes 3, 4, and 5 are 1:20, 1:6, and 1:2, respectively. Total RNA (5 μg per lane) was separated on an RNA agarose gel and blotted onto a nylon membrane. The membrane was probed with a 32P-labeled OPN probe. The bound probe was stripped off, and the membrane was reprobed with a 32P-labeled actin probe. (B) Quantitation of the Northern blot by phosphor image analysis showed a strong correlation between the ratio of monocytes to HS-27a and OPN gene expression (R2 = 0.99). The data indicate a strong signal for OPN in RNA isolated from cocultures but not HS-27a or CD14+ cells cultured alone.
HS-27a induces OPN expression and secretion in CD14+ cells
To determine whether the CD14+ cells or HS-27a cells were responsible for the increased transcription of OPN seen in cocultures, gene expression in the adherent cells (HS-27a plus adherent CD14+ cells) and nonadherent cells (CD14+ cells only) was compared by real-time PCR assay (Figure 2A). Nonadherent CD14+ cells had more than 7-fold higher OPN gene expression than adherent cells, suggesting that the CD14+ cells are responsible for the increased gene expression of OPN. The increased OPN gene expression was time dependent, as seen in Figure 2B. OPN gene expression was only minimally increased in CD14+ cells cultured alone (Figure 2B, ▪).
Gene expression and secretion of OPN. (A) HS-27a cells were cultured alone or cocultured with CD14+ cells at a HS-27a/CD14 ratio of 2:1 for 3 days. Adherent and nonadherent cells were harvested separately from the coculture. Total RNA was isolated and reverse-transcribed into cDNA. OPN gene expression was determined by a Syber Green real-time PCR assay and normalized to G3PDH gene expression. (B) CD14+ cells (0.2 × 106 cells) were cultured in the presence (○) or absence (▪) of HS-27a (0.4 × 106 cells) for 3 days. Nonadherent cells were harvested daily, and OPN gene expression was determined as described for panelA. (C) CD14+ cells from 13 different healthy donors were cultured alone (CD14 cells alone) or cocultured with HS-27a (CD14 cells in coculture) for 3 days. The nonadherent monocytes were harvested, and OPN gene expression was determined by a Syber Green assay as above. (D) CD14+ cells (0-0.2 × 106 cells) were cultured alone (CD14 alone) or cocultured with 0.4 × 106 cells of HS-27a (coculture) for 3 days. ELISA was used to determine the concentration of OPN in conditioned media. Each symbol represents the average ± SD for 3 samples.
Gene expression and secretion of OPN. (A) HS-27a cells were cultured alone or cocultured with CD14+ cells at a HS-27a/CD14 ratio of 2:1 for 3 days. Adherent and nonadherent cells were harvested separately from the coculture. Total RNA was isolated and reverse-transcribed into cDNA. OPN gene expression was determined by a Syber Green real-time PCR assay and normalized to G3PDH gene expression. (B) CD14+ cells (0.2 × 106 cells) were cultured in the presence (○) or absence (▪) of HS-27a (0.4 × 106 cells) for 3 days. Nonadherent cells were harvested daily, and OPN gene expression was determined as described for panelA. (C) CD14+ cells from 13 different healthy donors were cultured alone (CD14 cells alone) or cocultured with HS-27a (CD14 cells in coculture) for 3 days. The nonadherent monocytes were harvested, and OPN gene expression was determined by a Syber Green assay as above. (D) CD14+ cells (0-0.2 × 106 cells) were cultured alone (CD14 alone) or cocultured with 0.4 × 106 cells of HS-27a (coculture) for 3 days. ELISA was used to determine the concentration of OPN in conditioned media. Each symbol represents the average ± SD for 3 samples.
CD14+ cells from 13 healthy donors were cultured alone or in coculture with HS-27a cells for 3 days to determine the variability among individuals in the induction of OPN gene expression (Figure 2C). CD14+ cells from all donors increased OPN gene expression in cocultures (mean = 19-fold higher than CD14+ cells alone; range, 3-fold to 89-fold: P = .0007 by paired Student t test). OPN protein secretion was also found to be increased in the coculture (Figure 2D), in parallel with the number of CD14+ cells added. OPN secretion was also increased in CD14+ cells cultured alone, but to a much lesser degree. OPN secretion into media clearly correlated with gene expression.
All media and cells used in this study were tested for endotoxin and mycoplasma and found to be negative, thereby excluding the possibility of endotoxin- and mycoplasma-induced activation of monocytes.
HS-27a secretes a signal that stimulates OPN gene expression and secretion in CD14+ cells
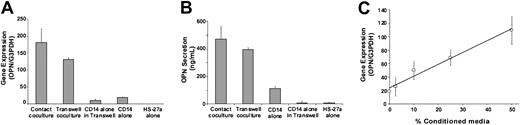
To determine whether a soluble factor(s) induces OPN gene expression in monocytes, additional cocultures were set up in standard coculture format, which allows cell contact, or in Transwells, with monocytes in the upper chamber and HS-27a cells in the lower chamber. Cultures were maintained for 3 days, cells were harvested, total RNA was isolated, and OPN gene expression was measured (Figure 3A). Culture media were also harvested, and OPN protein secretion was measured by ELISA (Figure 3B). Monocytes in Transwells showed almost the same level of OPN gene expression and secretion as seen in standard cocultures. This indicates that the monocytes are expressing OPN and that the HS-27a cells secrete a signal that stimulates this effect. When HS-27a-conditioned media were added to monocytes cultured alone, OPN gene expression was increased in a concentration-dependent manner (Figure 3C), confirming the presence of the soluble factor(s) from HS-27a. The conditioned media was fractionated by ultrafiltration filter devices (Centriprep YM-10; Millipore, Billerica, MA). The OPN-stimulating activity was recovered in a filtrate containing molecules smaller than 10 kDa. Precise identification of the factor(s) is currently under investigation.
OPN gene expression and protein secretion from monocytes cocultured with HS-27a. (A) CD14+ cells were cocultured in contact with HS-27a (contact coculture) or separated from HS-27a by a Transwell insert (transwell coculture). Controls included CD14 alone in a Transwell insert, CD14 alone, and HS-27a alone. Cells were cultured for 3 days. Both adherent and nonadherent monocytes were harvested and combined, except in the contact culture where only the nonadherent cells were harvested. Total RNA was harvested, and OPN gene expression was determined by Syber Green PCR assay and normalized to G3PDH gene expression. (B) Conditioned media were also harvested after 3 days, and concentrations of OPN protein were determined by ELISA. (C) CD14+ cells were cultured in multiwell culture plates in the absence or presence of conditioned media from HS-27a at different concentrations for 3 days. Adherent and nonadherent CD14+ cells were combined, and total RNA was isolated. OPN gene expression was determined as described above. The data represent the average ± SD for at least 2 samples.
OPN gene expression and protein secretion from monocytes cocultured with HS-27a. (A) CD14+ cells were cocultured in contact with HS-27a (contact coculture) or separated from HS-27a by a Transwell insert (transwell coculture). Controls included CD14 alone in a Transwell insert, CD14 alone, and HS-27a alone. Cells were cultured for 3 days. Both adherent and nonadherent monocytes were harvested and combined, except in the contact culture where only the nonadherent cells were harvested. Total RNA was harvested, and OPN gene expression was determined by Syber Green PCR assay and normalized to G3PDH gene expression. (B) Conditioned media were also harvested after 3 days, and concentrations of OPN protein were determined by ELISA. (C) CD14+ cells were cultured in multiwell culture plates in the absence or presence of conditioned media from HS-27a at different concentrations for 3 days. Adherent and nonadherent CD14+ cells were combined, and total RNA was isolated. OPN gene expression was determined as described above. The data represent the average ± SD for at least 2 samples.
Not all stromal cells induce OPN gene expression in CD14+ monocytes
We next investigated the basal and inducible levels of OPN expression in normal fresh bone marrow, primary LTCs, and peripheral blood mononuclear cells (PBMCs). Little or no expression was detected in PBMCs or the uncultured marrow cells, but a high level of OPN gene expression was found in LTCs, which contain both stroma and macrophages. Only mild induction of OPN gene expression was found in monocytes cocultured with human foreskin fibroblasts (HFFs), whereas cocultures with another stromal cell line, HS-5, failed to induce OPN (Figure 4).
OPN gene expression in PBMC, primary stroma, and stromal cell lines. (A) Total RNA was immediately isolated from peripheral blood mononuclear cells (PBMCs) and the buffy coat layer of normal bone marrow cells (BM). Remaining cells from the buffy coat were cultured for 2 to 3 weeks to establish primary LTCs, which contain both stroma and macrophages, at which time total RNA from the LTCs was isolated. OPN gene expression was determined by a Syber Green assay. (B) CD14+ cells (0.2 × 106 cells) were cultured alone or cocultured with 0.4 × 106 cells of HS-27a, HS-5, or human foreskin fibroblasts (HFFs) for 3 days. Total RNA of nonadherent CD14+ cells was harvested, and OPN gene expression was measured. The data represent the average ± SD for at least 2 samples.
OPN gene expression in PBMC, primary stroma, and stromal cell lines. (A) Total RNA was immediately isolated from peripheral blood mononuclear cells (PBMCs) and the buffy coat layer of normal bone marrow cells (BM). Remaining cells from the buffy coat were cultured for 2 to 3 weeks to establish primary LTCs, which contain both stroma and macrophages, at which time total RNA from the LTCs was isolated. OPN gene expression was determined by a Syber Green assay. (B) CD14+ cells (0.2 × 106 cells) were cultured alone or cocultured with 0.4 × 106 cells of HS-27a, HS-5, or human foreskin fibroblasts (HFFs) for 3 days. Total RNA of nonadherent CD14+ cells was harvested, and OPN gene expression was measured. The data represent the average ± SD for at least 2 samples.
Signal transduction in CD34+ cells exposed to purified OPN
Given that CD34+ cells express the 2 OPN receptors CD44 and integrin alpha 4,30-34 we next studied the effect of OPN on CD34+ cells. In these experiments CD34+ cells were obtained from apheresis products from G-CSF-mobilized healthy donors. Short-term signal transduction was detected by measuring changes in tyrosine phosphorylation in whole cells by immune fluorescence and flow cytometry.35 We exposed CD34+ cells to purified OPN for 2 minutes. The data shown in Figure 5A indicate that the amount of phosphotyrosine in the CD34+ cells increased after exposure and that this effect was reduced in the presence of an anti-OPN neutralizing antibody. Increased tyrosine phosphorylation could be detected at 2 minutes but decreased after 15 minutes of exposure (data not shown).
Signal transduction in OPN-treated CD34+ cells. (A) Purified CD34+ cells were incubated with 1 μg/mL OPN in the presence or absence of a neutralizing antibody against OPN for 2 minutes. The cells were fixed and permeabilized, followed by incubation with FITC-conjugated antiphosphotyrosine antibody (4G10) and analysis by flow cytometry. Cells were gated for forward and side scatter as shown in the inserted panel. Solid and dashed lines in the histogram show the degree of phosphorylation of cellular proteins in the absence and presence of the neutralizing antibody, respectively. Black filled area shows untreated control cells. (B) Immunoblot analysis of tyrosine-phosphorylated proteins in CD34+ cells. Total protein extracts of untreated or OPN-treated CD34+ cells were analyzed for tyrosine phosphorylation by Western blot. CD34+ cells were cultured for 2 minutes in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 1 μg/mL OPN. Total protein extracts were prepared, run on SDS-PAGE under reducing conditions, and transferred onto a nitrocellulose membrane. The membrane was stained with Ponceau S (Sigma, St. Louis, MO) to show protein load and incubated with PY20 antiphosphotyrosine antibody. An arrow indicates the position of the tyrosine-phosphorylated protein (34 kDa) that has increased phosphorylation after exposure to OPN. (C) CD34+ cells were cultured for 2 minutes in the absence (none) or presence of 1 μg/mL OPN (+OPN), OPN plus 13.5 μg/mL of a neutralizing antibody (+OPN+Ab), or OPN plus 13.5 μg/mL mouse IgG1 (+OPN+isotype). Total protein extracts were prepared, and tyrosinephosphorylated proteins were analyzed by immunoblots. The band intensity of the tyrosine-phosphorylated 34-kDa protein was measured by phosphor image analysis. Comparable results were obtained from 3 different cell preparations.
Signal transduction in OPN-treated CD34+ cells. (A) Purified CD34+ cells were incubated with 1 μg/mL OPN in the presence or absence of a neutralizing antibody against OPN for 2 minutes. The cells were fixed and permeabilized, followed by incubation with FITC-conjugated antiphosphotyrosine antibody (4G10) and analysis by flow cytometry. Cells were gated for forward and side scatter as shown in the inserted panel. Solid and dashed lines in the histogram show the degree of phosphorylation of cellular proteins in the absence and presence of the neutralizing antibody, respectively. Black filled area shows untreated control cells. (B) Immunoblot analysis of tyrosine-phosphorylated proteins in CD34+ cells. Total protein extracts of untreated or OPN-treated CD34+ cells were analyzed for tyrosine phosphorylation by Western blot. CD34+ cells were cultured for 2 minutes in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 1 μg/mL OPN. Total protein extracts were prepared, run on SDS-PAGE under reducing conditions, and transferred onto a nitrocellulose membrane. The membrane was stained with Ponceau S (Sigma, St. Louis, MO) to show protein load and incubated with PY20 antiphosphotyrosine antibody. An arrow indicates the position of the tyrosine-phosphorylated protein (34 kDa) that has increased phosphorylation after exposure to OPN. (C) CD34+ cells were cultured for 2 minutes in the absence (none) or presence of 1 μg/mL OPN (+OPN), OPN plus 13.5 μg/mL of a neutralizing antibody (+OPN+Ab), or OPN plus 13.5 μg/mL mouse IgG1 (+OPN+isotype). Total protein extracts were prepared, and tyrosinephosphorylated proteins were analyzed by immunoblots. The band intensity of the tyrosine-phosphorylated 34-kDa protein was measured by phosphor image analysis. Comparable results were obtained from 3 different cell preparations.
Western blotting with a different antiphosphotyrosine antibody (PY20) was used to identify a specific protein(s) that is phosphorylated on tyrosine residues after OPN signaling (Figure 5B). Tyrosine phosphorylation of a 34-kDa protein was increased after 2 minutes of stimulation with 1 μg/mL OPN, and this increased phosphorylation was diminished in the presence of the OPN neutralizing antibody but not an isotype control (Figure 5C). Additional studies are needed to identify the phosphorylated protein and its signaling pathway in CD34+ cells.
Coculture with HS-27a plus monocytes decreases Notch1 gene expression in CD34+ cells
Using real-time PCR analysis, we found that Notch1 gene expression was almost completely abolished in CD34+ cells cocultured with HS-27a plus monocytes (Figure 6A). The concentration of OPN protein in the conditioned media was inversely correlated with Notch1 gene expression (Figure 6B), which suggests that OPN could be one of the effector molecules. In support of this, exposure of CD34+ cells to purified OPN led to a 5-fold decrease in Notch1 gene expression (Figure 6C), while the expression of other molecules such as integrin alpha 6 (ITGA6) was not affected.
Decreased expression of Notch1 gene in CD34+ cells cocultured with HS-27a and monocytes. (A) CD34+ cells were isolated from apheresis products obtained from G-CSF-mobilized healthy donors. The CD34+ cells were seeded in a Transwell insert and cultured in media containing G-CSF, IL-3, IL-6, SCF, and 10% FCS. The bottom chamber contained either no cells (control), HS-27a cells (+HS-27a), CD14+ cells (+CD14), or HS-27a plus CD14+ cells (+HS-27a and CD14). The CD34+ cells were harvested after 4 days, and total RNA was isolated for determination of Notch1 gene expression by a Syber Green real-time PCR assay, normalized to G3PDH gene expression. (B) Conditioned media from the above cultures were collected, and OPN secretion was measured by ELISA. (C-D) The CD34+ cells were seeded in a Transwell insert and cultured in the presence or absence of 1 μg/mL OPN in addition to the growth factor cocktail for 4 days (+OPN or control, respectively). (C) Notch1 and (D) integrin alpha 6 (ITGA6) gene expression were determined by a Syber Green assay as described for panel A. The data represent the average ± SD for at least 2 samples.
Decreased expression of Notch1 gene in CD34+ cells cocultured with HS-27a and monocytes. (A) CD34+ cells were isolated from apheresis products obtained from G-CSF-mobilized healthy donors. The CD34+ cells were seeded in a Transwell insert and cultured in media containing G-CSF, IL-3, IL-6, SCF, and 10% FCS. The bottom chamber contained either no cells (control), HS-27a cells (+HS-27a), CD14+ cells (+CD14), or HS-27a plus CD14+ cells (+HS-27a and CD14). The CD34+ cells were harvested after 4 days, and total RNA was isolated for determination of Notch1 gene expression by a Syber Green real-time PCR assay, normalized to G3PDH gene expression. (B) Conditioned media from the above cultures were collected, and OPN secretion was measured by ELISA. (C-D) The CD34+ cells were seeded in a Transwell insert and cultured in the presence or absence of 1 μg/mL OPN in addition to the growth factor cocktail for 4 days (+OPN or control, respectively). (C) Notch1 and (D) integrin alpha 6 (ITGA6) gene expression were determined by a Syber Green assay as described for panel A. The data represent the average ± SD for at least 2 samples.
Discussion
The in vivo hematopoietic microenvironment (ME) consists of a complex mixture of cells that work in concert to regulate hematopoiesis. Primary LTCs, which provide an in vitro model of the ME, are also complex, containing stromal elements as well as hematopoietic-derived monocyte/macrophages. Understanding the interactions that occur among cells in this model has been a challenge because of the complexity of the system. Many investigators have simplified the system by using cloned stromal cell lines that retain relevant ME functions.
In this report we have investigated the interactions between monocytes and stroma in an in vitro coculture system using HS-27a cells, a cloned human marrow stromal cell line. HS-27a expresses a Notch ligand, Jagged-1, and supports cobblestone area formation by early hematopoietic progenitors, thus representing at least one important ME function. We used DNAmicroarray technology to investigate changes in gene expression that occur as a consequence of monocyte-HS-27a interactions. We identified OPN as the most strongly up-regulated gene in coculture and found that its source was the CD14+ monocytes. The increase in OPN gene expression in monocytes was induced by an activity secreted by HS-27a cells. Given that HS-27a is an immortalized stromal line, it is possible that this OPN-inducing activity is unique to these cells. We did, however, detect OPN mRNA in primary LTCs that contained 10% to 40% macrophages. Additional studies are needed to determine if the source and induction of OPN gene expression in primary cultures is similar to that of our experimental system.
OPN is a 60-kDa secreted phosphoglycoprotein expressed in many different cell types including activated macrophages, natural killer cells, myeloid dendritic cells, osteoblasts, tumor cells, and epithelial cells.28,36-39 OPN was originally isolated as an extracellular matrix protein and then found to be an important component of T-cell activation.28,40,41 OPN is a multifunctional molecule reported to function as a chemoattractant, as both an anti-inflammatory and proinflammatory factor, as well as an antiapoptotic signal depending on the system studied. Its precise role in hematopoiesis is unclear.
Our studies found that OPN produced in the coculture could down-regulate Notch1 gene expression in CD34+ cells. These data suggest that OPN produced in the ME may function to reduce Jagged 1-Notch 1 signaling. The inability of Jagged 1-expressing stroma to signal through Notch could change the consequences of CD34+ cell-stromal cell interactions, depending on what additional signals are available to the CD34 cells. For example, Notch signaling is reported to prevent G-CSF-stimulated myeloid differentiation.17 Consequently, monocytes releasing OPN in the microenvironment and thereby reducing Notch 1 expression by progenitor cells could result in a greater proportion of progenitor cells responding to myeloid differentiation signals. It is easy to speculate here that macrophage-derived OPN may be responsible for the gradual decline in progenitor production seen in primary LTCs as the macrophage numbers increase. This, too, requires formal testing.
Notch receptor signaling is important at several stages of hematopoiesis—that is, hematopoietic stem cell generation and self-renewal, T-cell commitment, B-cell development, and myeloid differentiation.20,42-44 Recently, Notch1 was shown to be critical for the genesis of stem cells but was not required once the stem cell pool was established.45 Separate studies, using overexpression of Notch, indicated that the Notch pathway is important to stem cell self-renewal and the establishment of immortalized cytokine-dependent cell lines with both lymphoid and myeloid characteristics.19,46,47 The transcription factor RBP-Jk is known to mediate most of the well-characterized effects of Notch.48
While the Notch pathway clearly plays an important role in hematopoietic cell fate decisions, very little is known about the transcriptional regulation of the Notch receptor genes themselves. Our studies presented here suggest that cellular interactions within the hematopoietic microenvironment may participate in controlling mRNA levels of Notch in CD34+ cells. Further studies using CD34+ cells, monocytes, and stroma should provide new insights into the regulation of Notch gene expression and its role in the ME.
Supported by grants HL62923, DK56465, and CA15704 from the National Institutes of Health, Bethesda, MD; a fellowship from Deutsche Krebshilfe, Dr Mildred-Scheel-Stiftung für Krebsforschung (C.K.); and a fellowship from Naito Foundation, Japan (N.A.).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Prepublished online as Blood First Edition Paper, March 2, 2004; DOI 10.1182/blood-2004-01-0256.
The authors thank Ludmilla Golubev and Gretchen Johnson for maintaining the cell lines and Bonnie Larson and Helen Crawford for preparing the manuscript. The anti-OPN antibody, MPIIIB10, developed by M. Solursh and A. Franzen, was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development (NICHD) and maintained by the University of Iowa, Department of Biological Sciences, Iowa City.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal