Abstract
X-linked ectodermal dysplasia and immunodeficiency (XL-EDA-ID) is an X-linked recessive disease caused by a mutation in the nuclear factor-κB (NF-κB) essential modulator (NEMO). Here we report an XL-EDA-ID patient with atypical features of very few naive-phenotype T cells and defective mitogen-induced proliferation of peripheral blood mononuclear cells (PBMCs). The patient's NEMO defect was diagnosed by flow cytometric analysis of intracellular NEMO staining. Specific cell lineages (monocytes and neutrophils) expressed reduced levels of NEMO, but 2 populations of T, B, and NK cells were detected with normal and reduced expression of NEMO. Genomic analysis revealed that duplication of a 4.4-kb sequence ranging from intron 3 to exon 6 caused the reduced expression of NEMO. Polymorphism analysis showed that the patient's B- and T-cell lines with reduced and normal expression of NEMO had the same X chromosome, indicating that the somatic mosaicism was not due to fetomaternal transfusion but was most likely due to postzygotic reversion. This XLEDA-ID case adds to our understanding of NEMO biology, indicating that NEMO is critical for T-cell development and/or survival in humans as well as in mice. (Blood. 2004;103:4565-4572)
Introduction
Ectodermal dysplasia and immunodeficiency (EDA-ID) is a disease whose clinical features include hypohidrosis, delay of eruption of teeth, coarse hair, and immunodeficiency associated with frequent bacterial infections.1-5 Two genes responsible for EDA-ID have been identified: nuclear factor-κB (NF-κB) essential modulator (NEMO; in X-linked-EDA-ID [XL-EDAID])6-8 and IκB (in autosomal-dominant EDA-ID).9 NEMO is necessary for the function of IκB kinase, which phosphorylates and degrades IκB to activate NF-κB.10 Thus, the defect in NEMO causes various kinds of abnormalities in signal transduction involving NF-κB, the interleukin 1 (IL-1) family protein receptors, the Toll-like receptors, vascular endothelial growth factor receptor-3 (VEGFR-3), receptor activator of nuclear factor κB (RANK), the ectodysplasin-A receptor, CD40, and the tumor necrosis factor (TNF) receptor.11
NEMO is also responsible for X-linked-dominant incontinentia pigmenti (IP).12 Males with IP usually die before birth. The XL-EDA-ID cases reported so far have also been male (with one exception13 ) but have only one mutated NEMO allele. Residual NEMO activity in XL-EDA-ID and a total lack of NEMO activity in IP can explain the phenotype differences in these 2 populations of males with NEMO defects.7,8,12 Furthermore, the majority of NEMO mutations in IP patients are large deletions due to recombination, while XL-EDA-ID patients have small mutations such as missense mutations, early stop codons, and stop codon mutations.12,14
The immunologic features of XL-EDA-ID reported so far consist of dysregulated immunoglobulin synthesis or hyper-immunoglobulin M (hyper-IgM) syndrome, defective antipolysaccharide antibody synthesis (antipneumococcal antibody and isohemagglutinin), reduced lipopolysaccharide (LPS) and IL-1 family protein responses, and defective natural killer (NK) cell activity.3,4,6-8,15-19 Complete loss of NEMO function is lethal in mice due to liver failure,20 but studies using conditional knockout mice or recombination-activating gene (RAG) chimera reconstitution have suggested that T and B cells do not develop in the complete absence of NEMO in the mouse.21-24 Although XL-EDA-ID is phenotypically different in individuals with different NEMO mutations, there have been no XL-EDA-ID cases reported so far that show a role for NEMO in T-cell development and survival. Now we report a patient with a novel type of XL-EDA-ID whose NEMO expression varied among cell lineages due to reversion mosaicism of a 4.4-kb duplication of a portion of the NEMO gene. The patient provided us with a unique opportunity to elucidate NEMO biology in humans because in this patient we could correlate the NEMO level with cell function in various cell types. In particular, we showed that T cells with normal levels of NEMO (due to reversion) predominated over T cells with reduced levels of NEMO, indicating that NEMO is critical for human T-cell development and/or survival.
Patients, materials, and methods
Informed consent
Informed consent was obtained for the patient and his family according to the protocol of the internal review board of Kyoto University Hospital.
Proliferation assay
Peripheral blood mononuclear cells (PBMCs) were stimulated with a plate-bound antihuman CD3 monoclonal antibody (mAb; clone OKT3, 1 μg/mL; Ortho Diagnostics Systems, Raritan, NJ), a plate-bound antihuman CD3 mAb (1 μg/mL) plus a soluble antihuman CD28 mAb (clone 9.3, 2 μg/mL; kindly provided by Dr Martin Paul, Fred Hutchinson Cancer Institute, Seattle, WA), phytohemagglutinin (PHA; 1:100 dilution; Invitrogen, Carlsbad, CA), and phorbol myristate acetate (PMA; 20 ng/mL)/ionomycin (1 μM; Sigma, St Louis, MO) with or without additional IL-2 (100 IU/mL; kindly provided by Shionogi & Co, Osaka, Japan). Forty-eight hours later, 3 H-thymidine uptake was measured in triplicate using a 6-hour pulse of 0.5 μCi (0.0185 MBq) 3 H-thymidine.
Cytokine production study
PBMCs were stimulated with varying doses of IL-18 with IL-12 (20 ng/mL; R&D Systems Inc, Minneapolis, MN) or varying doses of LPS (Sigma) with interferon γ (IFN-γ; 5000 U/mL; R&D Systems Inc). Forty-eight hours later, the supernatant was harvested and the cytokines were measured using human BD OptEIA enzyme-linked immunosorbent assay (ELISA) kits (BD Biosciences Pharmingen, San Diego, CA).
Western blotting
Human T-cell lymphotropic virus I (HTLV-I)-transformed cell lines were made as previously described by Ueno et al.25 Cell lysates (50 μg) were electrophoresed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting was performed as previously described using a mouse anti-NEMO mAb (C73-764; BD Biosciences Pharmingen) and a horseradish peroxidase (HRP)-conjugated rabbit antimouse IgG polyclonal antibody.26 The same blots were reprobed with a mouse anti-β-actin mAb (AC-74; Sigma) and an HRP-conjugated goat antimouse IgG polyclonal antibody.
Flow cytometry analysis
For NEMO intracellular staining, the cells were fixed, permeabilized, washed with Permfix and Permwash (BD Biosciences Pharmingen), and stained sequentially with a mouse anti-NEMO mAb (1 μg/mL; C73-764 or clone 54; BD Biosciences Pharmingen) and a phycoerythrin (PE)-labeled goat antimouse IgG Ab (DAKO Japan Co, Kyoto, Japan). The cells were stained for the following lineage markers after staining for NEMO: CD4, CD8, CD14, CD15, CD19, CD56 (BD Biosciences Pharmingen), and IgD (DAKO Japan Co). For intracellular staining of human IFN-γ, PBMCs were stimulated with PMA/ionomycin/monensin (2 μM; Calbiochem-Novabiochem, San Diego, CA) for 6 hours, fixed, permeabilized, and stained for NEMO, human IFN-γ (BD Biosciences Pharmingen), and lineage markers, sequentially. For CD40L stimulation, PBMCs were cultured with recombinant human soluble CD40L (2.5 μg/mL; PeproTech Inc, Rocky Hill, NJ) for 48 hours; stained for CD23, CD54, CD86, CD95, and CD19 (BD Biosciences Pharmingen); fixed; permeabilized; and stained with an fluorescein isothiocyanate (FITC)-labeled rabbit anti-NEMO polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). For analysis of naive T cells, PBMCs were stained with CCR7, CD45RA, and lineage makers and stained for NEMO with an FITC-labeled rabbit anti-NEMO polyclonal antibody. The stained cells were collected by FACSCalibur (BD Biosciences, San Jose, CA) and analyzed with FlowJo (Tree Star, San Carlos, CA).
Southern blotting and Northern blotting
Genomic DNA was isolated with a Puregene isolation kit according to the manufacturer's protocol (Gentra, Minneapolis, MN). DNA (5 μg) was digested with HindIII, electrophoresed, transferred to a Hybond-N nylon membrane (Amersham Biosciences, Piscataway, NJ), and cross-linked by UV irradiation. Southern hybridization was performed as previously described with a DNA probe containing exon 2 of the NEMO gene.26 Total RNA was isolated with a Trizol RNA isolation kit according to the manufacturer's protocol (Invitrogen). Total RNA (10 μg) was electrophoresed in a formaldehyde gel, transferred to a Hybond-N nylon membrane, and cross-linked by UV irradiation. Northern hybridization was performed as previously described with a NEMO cDNA-comprising NEMO exon 3.26 The blot was stripped and reprobed with a glyceraldehyde phosphate dehydrogenase (GAPDH) cDNA as a loading control.
PCR
Genomic polymerase chain reaction (PCR) was performed using the long-range amplification system Takara LA Taq (TaKaRa Shuzo, Otsu, Shiga, Japan) with GC high buffer I according to the manufacturer's protocol. The PCR primers (Figure 3C) were as follows: NEMO1, 5′-CAATACCGAGCATCTGAGGGCAGGCACAC-3′; NEMO2, 5′-AGAGACGAAGGAGCACAAAGCTGCCTTGAG-3′; NEMO3, 5′-ACTGCAGGGACAATGGTGGGTGCATCTGTC-3′; NEMO4, 5′-TGTGGACACGCAGTGAAACGTGGTCTGGAG-3′; NEMO5, 5′-TGGCCTGCTGACACTCCTGAGAGCAACATC-3′; NEMO6, 5′-GTGGCTCAGAAGCTTACGAAAGGCCACTAC-3′; NEMO7, 5′-TGGAAGCTCAGGTGAGAGGAGAGG-3′; NEMO8, 5′-TCTTTGATACAGTCATGCCATC-TGCTCTCC-3′; and NEMO9, 5′-AGTCCCTGAGATGGAGAGAAAGGAGTATCC-3′.
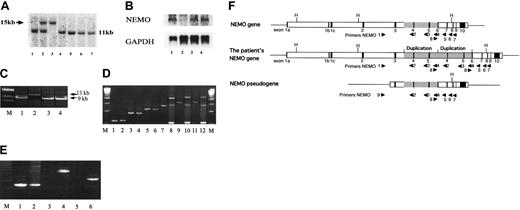
Reduced NEMO expression was caused by duplication of the NEMO gene. (A) Southern blotting analysis of the NEMO gene. Genomic DNA was digested with HindIII and electrophoresed, and the blot was hybridized with exon 2 DNA as a probe. DNA from the patient's EBV-transformed cells (lane 3) showed a 15-kb band, which was shared by his mother (lane 2), instead of the 11-kb bands, which were detected in the DNA from the patient's HTLV-I-transformed cells (lane 4), his father's EBV-transformed cells (lane 1), his mother's EBV-transformed cells (lane 2), and EBV-transformed cells from healthy controls (lanes 5-7). (B) Northern blotting analysis of the NEMO gene. The blot was hybridized with exon 3 DNA as a probe. RNA from the patient's EBV-transformed cells (lane 2) expressed very little NEMO mRNA compared with his HTLV-I-transformed cells (lane 1) and the EBV-transformed cells from healthy controls (lanes 3-4). (C) PCR reaction using primers 1 and 7. Only the NEMO gene was amplified. EBV-transformed cells (lane 2) had a 13-kb band instead of the 9-kb band seen in HTLV-I-transformed cells (lane 1) and healthy controls (lanes 3-4). M indicates lambda/HindIII digestion marker. (D) PCR reaction using primers 1 and 2 (lanes 1-2), primers 1 and 3 (lanes 3-4), primers 1 and 4 (lanes 5-6), primers 1 and 5 (lanes 7-8), primers 1 and 6 (lanes 9-10), and primers 1 and 7 (lanes 11-12). The patient's EBV-transformed cells (lanes 2, 4, 6, 8, 10, and 12) and those of healthy controls (lanes 1, 3, 5, 7, 9, 11) were used as templates. The larger bands detected in lanes 8, 10, and 12 indicate that the insertion is between primers 4 and 5 (between introns 5 and 6). M indicates 2.5-kb ladder. (E) PCR reaction using primers 8 and 5 (lanes 1-2), primers 8 and 3 (lanes 3-4), and primers 8 and 2 (lanes 5-6). The patient's EBV-transformed cells (lanes 2, 4, and 6) and those of healthy controls (lanes 1, 3, and 5) were used as templates. M indicates 1-kb marker. (F) Schematic representation of the normal NEMO gene, the patient's NEMO gene, and the NEMO pseudogene. Primers are shown as triangles. The duplicated region found in the patient is shaded. H indicates HindIII digestion sites.
Reduced NEMO expression was caused by duplication of the NEMO gene. (A) Southern blotting analysis of the NEMO gene. Genomic DNA was digested with HindIII and electrophoresed, and the blot was hybridized with exon 2 DNA as a probe. DNA from the patient's EBV-transformed cells (lane 3) showed a 15-kb band, which was shared by his mother (lane 2), instead of the 11-kb bands, which were detected in the DNA from the patient's HTLV-I-transformed cells (lane 4), his father's EBV-transformed cells (lane 1), his mother's EBV-transformed cells (lane 2), and EBV-transformed cells from healthy controls (lanes 5-7). (B) Northern blotting analysis of the NEMO gene. The blot was hybridized with exon 3 DNA as a probe. RNA from the patient's EBV-transformed cells (lane 2) expressed very little NEMO mRNA compared with his HTLV-I-transformed cells (lane 1) and the EBV-transformed cells from healthy controls (lanes 3-4). (C) PCR reaction using primers 1 and 7. Only the NEMO gene was amplified. EBV-transformed cells (lane 2) had a 13-kb band instead of the 9-kb band seen in HTLV-I-transformed cells (lane 1) and healthy controls (lanes 3-4). M indicates lambda/HindIII digestion marker. (D) PCR reaction using primers 1 and 2 (lanes 1-2), primers 1 and 3 (lanes 3-4), primers 1 and 4 (lanes 5-6), primers 1 and 5 (lanes 7-8), primers 1 and 6 (lanes 9-10), and primers 1 and 7 (lanes 11-12). The patient's EBV-transformed cells (lanes 2, 4, 6, 8, 10, and 12) and those of healthy controls (lanes 1, 3, 5, 7, 9, 11) were used as templates. The larger bands detected in lanes 8, 10, and 12 indicate that the insertion is between primers 4 and 5 (between introns 5 and 6). M indicates 2.5-kb ladder. (E) PCR reaction using primers 8 and 5 (lanes 1-2), primers 8 and 3 (lanes 3-4), and primers 8 and 2 (lanes 5-6). The patient's EBV-transformed cells (lanes 2, 4, and 6) and those of healthy controls (lanes 1, 3, and 5) were used as templates. M indicates 1-kb marker. (F) Schematic representation of the normal NEMO gene, the patient's NEMO gene, and the NEMO pseudogene. Primers are shown as triangles. The duplicated region found in the patient is shaded. H indicates HindIII digestion sites.
The PCR cycle consisted of 94°C for 25 seconds and 68°C for 1 minute/kb with a 3-minute initial denaturation at 94°C and a 7-minute final extension at 72°C. Thirty-five cycles were used for all PCR reactions. To clone the duplicated NEMO gene, we subcloned 3 overlapping fragments using the TOPO XL PCR cloning kit (Invitrogen) and sequenced all 3 fragments with an ABI BigDye Terminator kit on an ABI3100 (Applied Biosystems, Foster City, CA). The mutation-specific PCR reaction was performed using primers NEMO8 and NEMO2 (or NEMO3), and no compatible bands were detected from any of the 20 healthy controls tested.
For the assay of the human androgen receptor trinucleotide repeat polymorphism with X inactivation, PCR was performed as previously described by Allen et al.27 For pseudogene delta NEMO-specific PCR amplification, primer pairs NEMO9 and NEMO7 were used.
Results
Clinical and immunologic features of the patient with EDA-ID
The patient was a 2-year-old boy, born at term to healthy, nonconsanguineous parents. He had hypohidrosis, coarse hair, delayed eruption of teeth, and transient lower extremity lymphedema but did not develop the osteopetrosis or skin lesions typical of IP. At the age of 3 months, he was admitted to a local hospital because of interstitial pneumonitis. Since then he has had frequent bacterial infections and febrile episodes due to a Staphylococcus aureus skin abscess (7 and 9 months of age), pneumonia (13 months of age), pneumococcal sepsis (19 and 26 months of age), Pseudomonas aeruginosa otitis media (26 months of age), and Pseudomonas aeruginosa sepsis (28 months of age). In addition, he suffered from intractable diarrhea (22 months of age) that resulted in failure to thrive. Tests for Cryptosporidium parvum and Giardia lamblia were negative in his stool. The patient received one oral live polio vaccination, which resulted in no detectable anti-polio virus titers.
The clinical symptoms described above led us to speculate that the patient had XL-EDA-ID, which is known to be caused by a mutation in NEMO. First we evaluated the patient's immunologic profile. A laboratory examination when the patient was 9 months old showed increased levels of IgG and IgA (IgG, 10.63 g/L [1063 mg/dL] [healthy controls 4.55-10.15 g/L (455-1015 mg/dL)]; IgA, 1.36 g/L [136 mg/dL] [healthy controls 0.14-0.84 g/L (14-84 mg/dL)]; IgM, 0.4 g/L [40 mg/dL] [healthy controls 0.35-1.83 g/L (35-183 mg/dL)]) but no isohemagglutinin at the age of 2.5 years and no antipneumococcal IgG2 antibody after 2 episodes of pneumococcal sepsis. C3, C4, and CH50 were within normal limits, and phagocytosis and hydrogen superoxide production by neutrophils were intact.
Notably, the patient had few CD4+ cells (8%-12%), especially CD4+CD45RA+ cells (20.4% of CD4+ cells), although an HIV RNA analysis was negative (Table 1). In addition, a proliferation assay showed that the patient's cells did not proliferate in response to PHA and concanavalin A (ConA; PHA, 2426 counts per minute [cpm] [healthy control 27 799-79 995 cpm]; ConA, 1230 cpm [healthy control 26 782-73 506 cpm]). These features have not previously been reported in XL-EDA-ID.
Surface marker analysis of the XL-EDA-ID patient's PBMCs
. | % . | Healthy control, % . |
|---|---|---|
| CD3 | 41.2 | 66.1 ± 5.9 |
| CD4 | 8.0 | 37.7 ± 9.6 |
| CD8 | 31.8 | 23.7 ± 4.2 |
| TCRαβ | 38.0 | 56.5 ± 10.5 |
| TCRγδ | 3.0 | 9.5 ± 5.5 |
| CD4+CD45RA | 1.6 | 32.5 ± 10.8 |
| CD4+CD45RO | 7.2 | 10.2 ± 3.5 |
| CD8+CD45RA | 32.3 | 20.7 ± 5.1 |
| CD8+CD45RO | 9.9 | 7.6 ± 3.6 |
| CD4+CD25+ | 2.2 | 5.5 ± 1.2 |
| CD19 | 32.9 | 23.1 ± 4.1 |
| CD20 | 32.6 | 23.3 ± 4.0 |
| CD19/Sm-IgG | 0.2 | 1.2 ± 1.0 |
| CD19/Sm-IgA | 0.4 | 0.6 ± 0.5 |
| CD19/Sm-IgM | 32.1 | 21.0 ± 3.8 |
| CD19/Sm-IgD | 32.1 | 20.8 ± 3.7 |
| CD16 | 25.0 | 10.5 ± 5.2 |
| CD56 | 24.9 | 9.2 ± 4.8 |
. | % . | Healthy control, % . |
|---|---|---|
| CD3 | 41.2 | 66.1 ± 5.9 |
| CD4 | 8.0 | 37.7 ± 9.6 |
| CD8 | 31.8 | 23.7 ± 4.2 |
| TCRαβ | 38.0 | 56.5 ± 10.5 |
| TCRγδ | 3.0 | 9.5 ± 5.5 |
| CD4+CD45RA | 1.6 | 32.5 ± 10.8 |
| CD4+CD45RO | 7.2 | 10.2 ± 3.5 |
| CD8+CD45RA | 32.3 | 20.7 ± 5.1 |
| CD8+CD45RO | 9.9 | 7.6 ± 3.6 |
| CD4+CD25+ | 2.2 | 5.5 ± 1.2 |
| CD19 | 32.9 | 23.1 ± 4.1 |
| CD20 | 32.6 | 23.3 ± 4.0 |
| CD19/Sm-IgG | 0.2 | 1.2 ± 1.0 |
| CD19/Sm-IgA | 0.4 | 0.6 ± 0.5 |
| CD19/Sm-IgM | 32.1 | 21.0 ± 3.8 |
| CD19/Sm-IgD | 32.1 | 20.8 ± 3.7 |
| CD16 | 25.0 | 10.5 ± 5.2 |
| CD56 | 24.9 | 9.2 ± 4.8 |
Surface markers of the XL-EDA-ID patient PBMCs are shown as a percentage of gated mononuclear cells. Health control values are based on 1- to 6-year-old children for whom informed consent was obtained. Health control values are shown as mean ± SD. Sm indicates surface membrane.
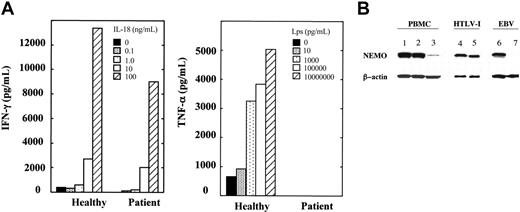
A study of the patient's cytokine response to LPS and IL-18, which activate the NF-κB signaling pathway through NEMO, showed a mixed defect (Figure 1A). LPS stimulation of the patient's PBMCs in the presence of IFN-γ caused no TNF-α secretion, while treatment with IL-18 and IL-12 induced almost as much IFN-γ as produced by healthy control cells.
The differential responses of the patient's PBMCs are due to differential expression of NEMO in each lineage. (A) Defective response of the patient's PBMCs to LPS but not to IL-18. PBMCs from the patient and healthy controls were stimulated with varying doses of IL-18 plus IL-12 (20 ng/mL) or with varying doses of LPS plus IFN-γ (5000 U/mL). A representative of 2 consistent results is shown. (B) Western blotting analysis of NEMO. Fifty micrograms of PBMC cell lysates (lanes 1-3), HTLV-I-transformed cells (lanes 4-5), and EBV-transformed cells (lanes 6-7) from the patient (lanes 3, 5, and 7) and healthy controls (lanes 1, 2, 4, and 6) were used. β-actin was used as a control for equal loading (lanes 1-7).
The differential responses of the patient's PBMCs are due to differential expression of NEMO in each lineage. (A) Defective response of the patient's PBMCs to LPS but not to IL-18. PBMCs from the patient and healthy controls were stimulated with varying doses of IL-18 plus IL-12 (20 ng/mL) or with varying doses of LPS plus IFN-γ (5000 U/mL). A representative of 2 consistent results is shown. (B) Western blotting analysis of NEMO. Fifty micrograms of PBMC cell lysates (lanes 1-3), HTLV-I-transformed cells (lanes 4-5), and EBV-transformed cells (lanes 6-7) from the patient (lanes 3, 5, and 7) and healthy controls (lanes 1, 2, 4, and 6) were used. β-actin was used as a control for equal loading (lanes 1-7).
Thus, the patient had both the typical immunologic features of XL-EDA-ID (recurrent streptococcal pneumoniae infections, no anti-streptococcal pneumoniae antibodies, no isohemagglutinin, and no LPS response) and atypical features (low CD4 lymphocyte concentration, differential response to LPS and IL-18, and reduced mitogen-induced proliferation).
Reduced protein expression of NEMO in EBV-transformed but not HTLV-I-transformed cells obtained from the patient
The NEMO gene is located on the X chromosome and the patient had a 46XY karyotype as confirmed by a G-banding chromosome analysis and an XY fluorescence in situ hybridization (FISH) study of his PBMCs. We sequenced a NEMO cDNA and PCR-amplified genomic DNA from the patient's PBMCs that corresponded to the cDNA. The DNA at first seemed to be intact and did not contain the NEMO mutations in the coding region and the stop codon that have been reported previously in XL-EDA-ID.6-8,11 In a complementary approach to define the defect, we performed Western blotting for NEMO expression (Figure 1B). We found reduced expression but not an absolute loss of NEMO in the patient's PBMCs. We then checked NEMO expression in an Epstein-Barr virus (EBV)-transformed cell line and an HTLV-I-transformed cell line derived from the patient's PBMCs. Surprisingly, the patient's HTLV-I-transformed cell lines expressed as much NEMO protein as healthy control HTLV-I-transformed cell lines, while the patient's EBV-transformed cell line expressed no or very little NEMO protein. These data suggested that the differential response to LPS and IL-18 could be derived from the differential expression of NEMO protein among these lineages.
Analysis of NEMO expression by flow cytometry
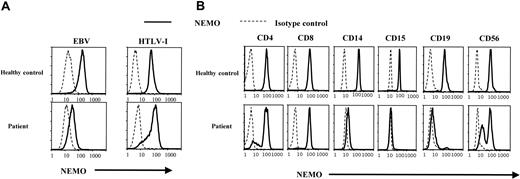
To further analyze cell lineage-specific NEMO expression, we performed intracellular staining for NEMO and flow cytometry analysis at the single-cell level. As shown in Figure 2A, flow cytometry showed a lower expression of NEMO in the patient's EBV-transformed cells than in the EBV-transformed cells of healthy controls. This is consistent with the reduced expression of NEMO seen with Western blotting. In contrast, the NEMO expression level in HTLV-I-transformed cells was similar between the patient and healthy controls.
Analysis of NEMO protein expression using intracellular staining and flow cytometry. (A) Intracellular staining of NEMO in EBV-transformed and HTLV-I-transformed cell lines. Solid lines indicate the anti-NEMO mAb, and dotted lines indicate the isotype controls. We confirmed the NEMO specificity with 2 different anti-NEMO mAbs (C73-764, BD; or clone 54, BD) and obtained identical results. (B) Intracellular staining for NEMO in PBMCs from the patient and healthy controls. The results shown were gated on lineage markers. We performed intracellular staining on PBMCs from 10 different healthy controls, all of which showed specific staining for NEMO on every lineage tested.
Analysis of NEMO protein expression using intracellular staining and flow cytometry. (A) Intracellular staining of NEMO in EBV-transformed and HTLV-I-transformed cell lines. Solid lines indicate the anti-NEMO mAb, and dotted lines indicate the isotype controls. We confirmed the NEMO specificity with 2 different anti-NEMO mAbs (C73-764, BD; or clone 54, BD) and obtained identical results. (B) Intracellular staining for NEMO in PBMCs from the patient and healthy controls. The results shown were gated on lineage markers. We performed intracellular staining on PBMCs from 10 different healthy controls, all of which showed specific staining for NEMO on every lineage tested.
In further studies along these lines, we measured NEMO expression in each PBMC lineage by flow cytometry. As shown in Figure 2B, healthy controls displayed a single peak of NEMO expression in all cells. In contrast, there were 2 peaks of NEMO expression in the patient's CD4+, CD8+, CD19+, and CD56+ cells and virtually no NEMO expression in the patient's CD14+ and CD15+ cells. Furthermore, the main population of CD4+ and CD8+ T cells had normal levels of NEMO (NEMOnormal), while the CD19+ cells had a reduced level of NEMO (NEMOlow), and CD56+ cells were almost equally divided between normal and low NEMO expression. Thus, the patient's cells had levels of NEMO expression that varied between cell lineages and even in the same cell lineages there were sometimes 2 peaks of NEMO expression.
Molecular mechanism underlying the reduced expression of NEMO
To elucidate the mechanism underlying this chimeric NEMO expression, we performed Southern blotting of the NEMO gene using exon 2 DNA as a probe. We chose exon 2 because the NEMO pseudogene, which is almost identical to the NEMO gene, does not contain exon 1 to exon 2 (Figure 3F). As shown in Figure 3A, analysis of the patient's EBV-transformed cells yielded a 15-kb band instead of the 11-kb band that was detected in healthy controls; both bands were present in his mother's cells. The patient's HTLV-I-transformed cells had the 11-kb band, which correlated with the normal protein expression of NEMO. To evaluate the NEMO mRNA level in these cells, we performed Northern blotting on the patient's EBV-transformed cells and HTLV-I-transformed cells. As shown in Figure 3B, the patient's EBV-transformed cells expressed a much-reduced amount of NEMO mRNA, while the HTLV-I-transformed cells expressed a normal amount of NEMO mRNA. To identify the aberrant sequence, we performed genomic PCR of the patient's EBV-transformed cells using primers in intron 2 (NEMO1) and intron 8 (NEMO7), which do not amplify the NEMO pseudogene (Figure 3C,F). We obtained a 13-kb band from the EBV-transformed cells and a 9-kb band from healthy control cells and the patient's HTLV-I-transformed cells (Figure 3C). To identify the region in which the extra sequence was located, we performed PCR using the intron 2 primer and various 3′ primers (at intron 3 [NEMO2], intron 4 [NEMO3], intron 5 [NEMO4], intron 6 [NEMO5], and intron 7 [NEMO6]; Figure 3D). We obtained bands that were approximately 4-kb larger when using the 3′ primers at intron 6, intron 7, and intron 8, indicating that the extra sequence was inserted between intron 5 and intron 6. In contrast, when we performed PCR amplifying the sequence between the 3′ primers at intron 5 (NEMO8) and intron 6 (NEMO5) (Figure 3E), no size differences in the major bands were detected between EBV-transformed cells from healthy controls and the patient's cells. We also performed a restriction enzyme digestion analysis (PstI, SacI, BglII) to compare the 9-kb and 13-kb bands obtained from the EBV-transformed cells (Figure 3C) after isolation of both bands, which showed that the digested bands were almost identical (data not shown). These data led us to speculate that the insertion sequence could be a duplication of the 5′ adjacent sequence. To test this hypothesis, we performed PCR at the 5′ region adjacent to intron 5 using 5′ reverse (NEMO2 and NEMO3) and 3′ forward (NEMO8) primers (Figure 3E-F). As in Figure 3E, we obtained bands from the patient's EBV-transformed cells and his mother's EBV-transformed cells (data not shown) but not from healthy controls (20 healthy controls checked). We sequenced the patient-specific band and found that the third base of exon 6 (nucleotide 19 426, GenBank accession number AJ271718) was connected to intron 3 (nucleotide 14 961). We sequenced the whole 13 kb using 3 overlapping fragments, which revealed the duplication of a 4.4-kb sequence including the region from intron 3 to exon 6. Sequencing analysis demonstrated that the NEMO gene of the patient's HTLV-I-transformed cells was intact. Thus, the reduced expression of NEMO in the EBV-transformed cells was due to insertion of a duplicated sequence spanning intron 3 to exon 6 (Figure 3F).
A mutated X-chromosome allele was shared by cell lines with normal and reduced NEMO expression: reversion mosaicism
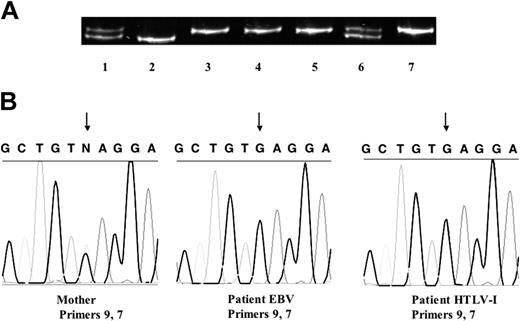
To identify the sources of the cells expressing normal and reduced amounts of NEMO, we used a genetic polymorphism on the X chromosome (Figure 4A). Analysis of the human androgen receptor trinucleotide repeat polymorphism showed that the patient carried the same allele from his mother in both HTLV-I-transformed cells with normal levels of NEMO and EBV-transformed cells with reduced levels of NEMO and did not carry the other allele from his mother nor the allele from his father. Thus, expansion of maternal cells after fetomaternal transfusion was unlikely (also confirmed by FISH with a Y probe). An X-inactivation assay showed that the allele shared by the patient and his mother was methylated and inactivated in the mother's cells (Figure 4A). Furthermore, both the patient's EBV-transformed cells and HTLV-I-transformed cells have the same single-nucleotide polymorphism found in his mother's NEMO pseudogene, corresponding to exon 8 of the NEMO coding region (Figure 4B). Taken together, these data strongly imply that the patient's normal and duplicated NEMO genes were derived from the same X chromosome, indicating that postzygotic reversion was responsible for the somatic cell NEMO mosaicism.
Reversion mosaicism of NEMO. (A) Human androgen receptor trinucleotide repeat polymorphism. Cells from the patient's mother (lanes 1, 6, and 7), the patient's father (lane 2), and the patient (PBMCs, lane 3; EBV-transformed cells, lane 4; and HTLV-I-transformed cells, lane 5) were used as templates. The patient's cells all had the same allele, which matched one of his mother's. This allele was methylated and inactivated in the mother, which was shown by an X-inactivation assay using HpaII (with HpaII, lane 7; without HpaII, lane 6). (B) A single-nucleotide polymorphism in the pseudogene delta NEMO in the patient and his mother. The mother is heterozygous G/C, whereas both EBV- and HTLV-I-transformed cells from the patient had G. The arrows indicate the single-nucleotide polymorphism.
Reversion mosaicism of NEMO. (A) Human androgen receptor trinucleotide repeat polymorphism. Cells from the patient's mother (lanes 1, 6, and 7), the patient's father (lane 2), and the patient (PBMCs, lane 3; EBV-transformed cells, lane 4; and HTLV-I-transformed cells, lane 5) were used as templates. The patient's cells all had the same allele, which matched one of his mother's. This allele was methylated and inactivated in the mother, which was shown by an X-inactivation assay using HpaII (with HpaII, lane 7; without HpaII, lane 6). (B) A single-nucleotide polymorphism in the pseudogene delta NEMO in the patient and his mother. The mother is heterozygous G/C, whereas both EBV- and HTLV-I-transformed cells from the patient had G. The arrows indicate the single-nucleotide polymorphism.
Immunologic re-evaluation of the XL-EDA-ID with NEMO gene duplication
Given the conclusion of postzygotic reversion, we were in a unique position to correlate NEMO expression with cellular function using NEMOnormal and NEMOlow cells (T, B, and NK cells) developing in the same environment. Thus, we re-evaluated the patient's immunologic phenotype, focusing on the expression level of NEMO, to explore the unique characteristics of this XL-EDA-ID patient (reduced mitogen-induced proliferation, low CD4 lymphocyte number in PBMCs, and relatively higher serum IgG concentration).
Since the CD40-CD40L system, and especially the NF-κB pathway, is important for immunoglobulin class switching,28 XL-EDA-ID patients usually have normal-to-reduced IgG levels.6-8,15,17 We examined the CD40 signaling pathway of the patient's B cells by stimulating the patient's PBMCs with recombinant human soluble CD40L (Figure 5A). Because the patient's B cells contained NEMOlow and NEMOnormal cells, we compared control CD19+ cells and the patient's NEMOlow CD19+ cells. While recombinant human soluble CD40L induced less up-regulation of CD23, CD54, CD86, and CD95 in the patient's NEMOlow CD19+ cells than in control CD19+ cells, CD40L signaling was conserved to some extent in the patient's NEMOlow B cells.7,8 Next we examined NEMO expression on CD19+IgD+ mature B cells and CD19+IgD- class-switched B cells in the patient. As shown in Figure 5B, the majority of CD19+IgD- cells were NEMOnormal, while CD19+IgD+ cells were NEMOlow. These data suggest that the increased level of IgG arises mainly from CD19+IgD-NEMOnormal reverted cells, even though these cells were a minority of the B-cell population.
Analysis of the XL-EDA-ID patient's B cells. (A) Recombinant human soluble CD40L stimulation of the patient's B cells induced CD23, CD54, CD86, and CD95 up-regulation to a lesser extent. PBMCs from the patient and healthy controls were treated with (open histograms) or without (shaded histograms) recombinant human soluble CD40L (2.5 μg/mL) for 48 hours. Histograms are shown gated on CD19+ cells of a control, CD19+NEMOlow, and CD19+NEMOnormal cells of the patient. (B) IgD-CD19+ class-switched B cells were NEMOnormal. PBMCs from the patient and healthy controls were stained with CD19, NEMO, and IgD, and CD19+-gated cells are shown.
Analysis of the XL-EDA-ID patient's B cells. (A) Recombinant human soluble CD40L stimulation of the patient's B cells induced CD23, CD54, CD86, and CD95 up-regulation to a lesser extent. PBMCs from the patient and healthy controls were treated with (open histograms) or without (shaded histograms) recombinant human soluble CD40L (2.5 μg/mL) for 48 hours. Histograms are shown gated on CD19+ cells of a control, CD19+NEMOlow, and CD19+NEMOnormal cells of the patient. (B) IgD-CD19+ class-switched B cells were NEMOnormal. PBMCs from the patient and healthy controls were stained with CD19, NEMO, and IgD, and CD19+-gated cells are shown.
The patient's CD4+ and CD8+ T cells were composed mainly of NEMOnormal cells, which indicated that low levels of NEMO are disadvantageous for the development and/or survival of T cells. On the other hand, even though the majority of T cells were NEMOnormal, the patient's cells displayed reduced mitogen-induced proliferation and the patient had low numbers of CD4+ lymphocytes. To elucidate the mechanism underlying these findings, we performed a T-cell receptor Vβ (TCR Vβ) repertoire analysis to see whether normal T-cell development occurred after the reversion events (Figure 6A). While the CD4+ T cells consisted of relatively varied types of Vβ-expressing T cells, 58% of the CD8+ T cells were Vβ16+, which indicated that T-cell development was impaired. The marker analysis of this repertoire showed they were CD28-, CD27-, CD57+, CD62Llow, CCR7-, CD45RO-, and CD45RA+, which is a CD8+ memory/effector phenotype (data not shown).30 In addition, naive-phenotype CD4+ T cells (CD45RA+, CCR7+) and naive-phenotype CD8+ T cells (CD45RA+, CCR7+) with either reduced or normal NEMO expression were markedly reduced in number (1.7% of NEMOnormal CD4+ cells, 3.9% of NEMOlow CD4+ cells, 0.4% of NEMOnormal CD8+ cells, and 14.3% of NEMOlow CD8+ cells; Figure 6B). To explore the functional aspects of these T cells, we performed intracellular cytokine staining for IFN-γ. As shown in Figure 6C, reverted NEMOnormal cells (both CD4 and CD8) produced more IFN-γ than control cells, while NEMOlow cells produced reduced amounts of IFN-γ. Thus, NEMO is critical both for the production and/or survival of T cells and for the development of IFN-γ-producing T cells. In addition, the abnormal T-cell repertoire, especially of CD8+ cells, and the reduced number of naive T cells showed that T-cell development was impaired, although the majority of the T cells were NEMOnormal.
Analysis of the XL-EDA-ID patient's T cells. (A) TCR Vβ repertoire analysis of the patient's CD4+ and CD8+ cells. The patient's PBMCs were stained for the TCR Vβ panel and CD4 or CD8 as described by Kobayashi et al.29 The healthy controls consisted of children 1 to 10 years old (n = 10). The values are shown as mean ± SD. (B) Naive and memory phenotypes of the patient's T cells. PBMCs from the patient and control were stained with NEMO, CCR7, CD45RA, and CD4 or CD8. Cells gated on NEMOlow or NEMOnormal of CD4 or CD8 are shown. (C) Reduced IFN-γ production of NEMOlow T cells. The patient and control PBMCs were stimulated with PMA/ionomycin/monensin for 6 hours and stained for intracellular IFN-γ along with NEMO and CD4 or CD8. Cells gated on CD4 or CD8 are shown. (D) Proliferation assay of PBMCs stimulated by an anti-CD3 mAb plus an anti-CD28 mAb. PBMCs from the patient or healthy controls were stimulated for 48 hours with an anti-CD3 mAb, an anti-CD3 mAb plus the anti-CD28 mAb ± IL-2 (100 IU/mL), PHA, or PMA/ionomycin. Data are shown as mean ± SD. (E) Reduced production of IL-2 and IFN-γ by anti-CD3 mAb plus anti-CD28 mAb. PBMCs from the patient or healthy controls were stimulated for 48 hours by the anti-CD3 mAb, anti-CD3 mAb plus anti-CD28 mAb, or PMA/ionomycin, and the supernatants were harvested for ELISA assays.
Analysis of the XL-EDA-ID patient's T cells. (A) TCR Vβ repertoire analysis of the patient's CD4+ and CD8+ cells. The patient's PBMCs were stained for the TCR Vβ panel and CD4 or CD8 as described by Kobayashi et al.29 The healthy controls consisted of children 1 to 10 years old (n = 10). The values are shown as mean ± SD. (B) Naive and memory phenotypes of the patient's T cells. PBMCs from the patient and control were stained with NEMO, CCR7, CD45RA, and CD4 or CD8. Cells gated on NEMOlow or NEMOnormal of CD4 or CD8 are shown. (C) Reduced IFN-γ production of NEMOlow T cells. The patient and control PBMCs were stimulated with PMA/ionomycin/monensin for 6 hours and stained for intracellular IFN-γ along with NEMO and CD4 or CD8. Cells gated on CD4 or CD8 are shown. (D) Proliferation assay of PBMCs stimulated by an anti-CD3 mAb plus an anti-CD28 mAb. PBMCs from the patient or healthy controls were stimulated for 48 hours with an anti-CD3 mAb, an anti-CD3 mAb plus the anti-CD28 mAb ± IL-2 (100 IU/mL), PHA, or PMA/ionomycin. Data are shown as mean ± SD. (E) Reduced production of IL-2 and IFN-γ by anti-CD3 mAb plus anti-CD28 mAb. PBMCs from the patient or healthy controls were stimulated for 48 hours by the anti-CD3 mAb, anti-CD3 mAb plus anti-CD28 mAb, or PMA/ionomycin, and the supernatants were harvested for ELISA assays.
Finally, we re-evaluated the proliferation potential of the T cells using an anti-CD3 mAb and an anti-CD28 mAb as stimulants, since reduced costimulatory molecule expression on B cells has been reported in XL-EDA-ID patients7,8 and was shown in the patient under study (Figure 5B). The patient's cells did not proliferate as well as the healthy control's cells in response to the anti-CD3 mAb, while the addition of the anti-CD28 mAb or IL-2 increased the proliferation capacity (Figure 6D). It is known that memory phenotype CD4+ and CD8+ T cells are poor IL-2 producers.30,31 Therefore we performed a cytokine secretion study, which showed that IL-2 production was reduced even after treatment with the anti-CD28 mAb, although the reduction in IFN-γ production was less prominent (Figure 6E). Thus, both defects in costimulation and IL-2 production contributed to the diminished mitogen-induced proliferation.
Discussion
Reversion mosaicism has been reported in primary immunodeficiency syndromes such as adenosine deaminase deficiency, X-linked recessive severe combined immunodeficiency, and Wiskott-Aldrich syndrome.32-34 However, these mutations are small, such as missense mutations, and reversion mosaicism of a large gene duplication has not been reported in primary immunodeficiency diseases. There have been some reports that large gene duplications have been followed by reversion mosaicism in mice (pink-eyed unstable),35,36 Drosophila (white-ivory mutation),37 and humans (Lesch-Nyhan syndrome).38 Characteristically, the reversion rate was very high and the duplicated gene deletions were very precise. Four possible mechanisms underlying the reversion of duplicated genes may be considered: intrachromosomal exchange, unequal sister chromatid exchange, single-strand annealing, and sister chromatid conversion.39,40 In our case, HTLV-I-transformed cells had a normal NEMO gene as confirmed by sequence analysis, and cells with the gene reversion expressed the same amount of NEMO protein as healthy control cells (Figure 2B). These features suggest that the exact deletion of the gene duplication had also occurred in the patient.
The duplication of the NEMO gene extending from intron 3 to exon 6 greatly reduced the expression of NEMO protein. Using the gene structure prediction program GENSCAN,41 we speculated that the duplication would insert exon 4 and exon 5 between exons 5 and 6. This would cause the NEMO protein to be truncated to 223 amino acids with the addition of 9 new amino acids as a result of a frameshift (K224fsX9, new peptide NRWLRTRPL), which is similar to the dominant-negative form of NEMO.42 It should be pointed out that the patient's mother had completely skewed X-inactivation (also confirmed by intracellular NEMO staining, no NEMOlow cells in the mother's PBMCs), which is not typical of XL-EDA-ID cases.15,16 Furthermore, the appearance of transient lymphedema in the patient implied that the NEMO defect of the patient was profound.8 Although alternative explanations exist for the reduced expression of NEMO, such as enhancer/silencer effects of the inserted sequence or changes in the stability of NEMO mRNA, we suspect that the NEMO mutation described here is the most detrimental among XL-EDA-ID cases in terms of NEMO residual activity.
It has been reported that genetically mutated mice with reduced NF-κB activity show abnormalities in T-cell development, T-cell proliferation, IL-2 production, and survival of activated T cells.43-47 Recently, Schmidt-Supprian et al24 reported that the number of mature T cells in T-cell-specific conditional NEMO knockout mice was markedly reduced due to apoptotic death of single-positive NEMO-deficient thymocytes. This is reminiscent of our results in that the number of mature peripheral NEMOlow (or NEMOnegaitve)T cells was low and the phenotype was more severe in CD8+ NEMOlow T cells than in CD4+ NEMOlow T cells (Figure 2B). Thus, the phenotype of NEMOlow T cells in our study, which is uncharacteristic of XL-EDA-ID, can be explained by much-reduced NEMO activity as reported in previous mouse studies.24
Although the majority of PBMC T cells had reverted to the normal form of NEMO, CD4+ and CD8+ T cells (both NEMOnormal and NEMOlow) were mainly of the memory phenotype. In addition, the TCR repertoire was skewed, especially among CD8+ T cells. The frequent infections suffered by the patient could be part of the reason for the TCR repertoire skewing and the dominance of memory T cells. However, it is doubtful that frequent infection was the only reason because the TCR repertoire abnormality (also checked by CDR3 analysis; data not shown) and the lack of naive T cells were too profound. It seems more likely that the reversion in the patient was not sufficient for complete T-cell restoration and that the few reverted NEMOnormal T cells expanded homeostatically, resulting in T cells with memory phenotype markers.48,49 Similar CD4+ lymphocytopenia and reduced mitogen-induced proliferation with a skewed CD8 Vβ repertoire has been reported in one case of reversion mosaicism of X-linked recessive severe combined immunodeficiency with reverted T cells and nonreverted B cells, monocytes, and neutrophils.33 The similarity would indicate a requirement for NEMO in T-cell development and/or survival as observed previously for the IL-2Rγ chain.
Prepublished online as Blood First Edition Paper, January 15, 2004; DOI 10.1182/blood-2003-10-3655.
Supported by the Program for Promotion of Fundamental Studies in Health Science of the Organization for Pharmaceutical Safety and Research of Japan; a Grant-in-Aid for Creative Scientific Research (13GS0009) from the Ministry of Education, Science, Technology, Sports, and Culture of Japan; and The Shimizu Foundation Research Grant for 2003.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to T. Yorifuji, M. Mamada, and K. Kurokawa for helpful suggestions about the genetic analysis; to M. Iseki for parasite examination/identification; and to W. Strober and A. Jain for critical reading of the manuscript.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal