Abstract
Somatic mitochondrial DNA (mtDNA) mutations accumulate with age in postmitotic tissues but have been postulated to be diluted and lost in continually proliferating tissues such as bone marrow (BM). Having observed marked sequence variation among healthy adult individuals' total BM cell mtDNA, we undertook analysis of the mtDNA control region in a total of 611 individual CD34+ clones from 6 adult BM donors and comparison of these results with the sequences from 580 CD34+ clones from 5 umbilical cord blood (CB) samples. On average, 25% (range, 11% to 50%) of individual CD34+ clones from adult BM showed mtDNA heterogeneity, or sequence differences from the aggregate mtDNA sequence of total BM cells of the same individual. In contrast, only 1.6% of single CD34+ clones from CB showed mtDNA sequence variation from the aggregate pattern. Thus, age-dependent accumulation of mtDNA mutations appears relatively common in a mitotically active human tissue and may provide a method to approximate the mutation rate in mammalian cells, to assess the contribution of reactive oxygen species to genomic instability, and for natural “marking” of hematopoietic stem cells; our data also have important implications for the aging process, forensic identifications, and anthropologic conclusions dependent on the mtDNA sequence.
Introduction
Mitochondrial DNA (mtDNA) comprises 0.1% to 1.0% of the total DNA in most mammalian cells, and 2 to 10 copies come packaged in each nucleated cell in each of up to 1000 mitochondria. Human mtDNA is a double-stranded, maternally inherited circular molecule of 16 569 base pairs; its 37 genes encode for polypeptides of the mitochondrial electron transport chain, transfer RNAs, and ribosomal RNAs necessary for their synthesis.1,2 In comparison with the nuclear genome, mtDNA has a modified genetic code,3 a paucity of introns, and lack of histone protection.4 Past evidence had indicated that mtDNA repair capacity was limited and that the proximity of mtDNA to sites of reactive oxygen species generation suggested that mtDNA may be more susceptible to mutation than nuclear DNA. Although the limited repair capacity hypothesis has been validated experimentally in some experimental systems, recent data have shown that base excision repair mechanisms do occur in mammalian mtDNA.5,6 Another major difference between mtDNA and nuclear DNA is that multiple species of mtDNA can coexist in a single cell, a condition called heteroplasmy. During development and with aging, at least in nonmitotic tissues of the neuromuscular system, mtDNA mutations not only accumulate but achieve homoplasmy within tissues, perhaps as a result of random genetic drift or of a selective replicative advantage conferred on 1 of the 2 heteroplasmic species.7
Because of its abundance and inherent variability, mtDNA has been widely used for forensic identification and in anthropologic studies. Furthermore, several hundred human diseases have been associated with maternally inherited specific deletions and mutations.8 Somatically acquired mtDNA mutations also have been linked to aging and degenerative diseases, cancer, and autoimmunity.9 A large deletion of mtDNA is a hallmark of Pearson syndrome, a constitutional disorder that includes sideroblastic anemia.10 mtDNA mutations recently were reported also in apparently acquired sideroblastic anemia and in myelodysplastic syndromes in general.11 While we were unable to confirm these results by amplification and direct sequencing of the entire mtDNA genome in patients and healthy controls, we coincidentally observed numerous sequence changes in bulk samples of bone marrow cells from our healthy controls as well as in patients in these experiments.12 While mtDNA changes have been postulated to underlie the aging process,13 mutant mtDNA genomes also have been assumed to be lost by dilution from rapidly dividing tissues such as bone marrow.8 We therefore undertook investigation of the possibility that mtDNA mutations might accumulate with aging in individual human CD34+ cells by examining a portion of mtDNA thought to have a high rate of somatic mutation.14
Materials and methods
Normal bone marrow and cord blood
Bone marrow (BM) specimens from 6 healthy adult donors were collected after informed consent was obtained following protocols approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute. Of 5 cord blood (CB) samples, 2 came from the Blood Bank at the National Institutes of Health (Bethesda, MD) and 3 from the New York Blood Center (New York, NY).
Single CD34+ cell sorting
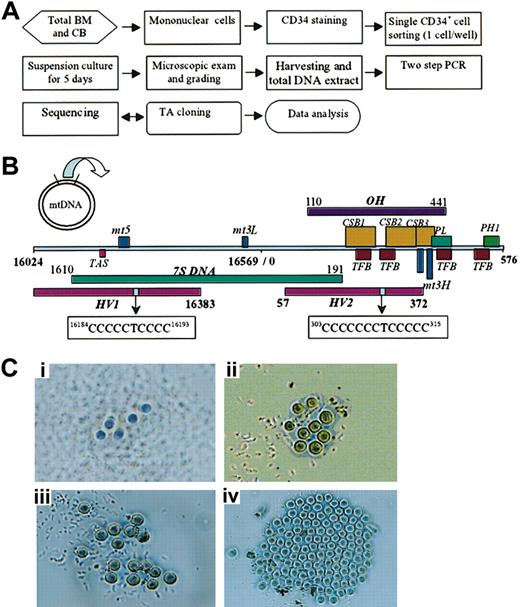
Mononuclear cells from BM and CB were separated by density gradient centrifugation and washed twice in phosphate-buffered saline (PS). The number of cells suspended in PBS was adjusted to 2 × 107 cells per milliliter. Ten microliters of anti-CD34 phycoerythrin (PE)–conjugated antibody (BD Bioscience, Franklin Lakes, NJ) were added to each 12 × 75 mm tube containing 100 μL cell suspension. After incubation for 30 minutes at 4°C, cells were washed using cold PBS and resuspended in 0.5 mL buffer. Cell sorting was performed on a MoFlo Cytometer (Dako-Cytomation, Ft Collins, CO) using 100 mW of the 488 nm line of an argon laser (I-90, Coherent, Palo Alto, CA) for excitation. Forward scatter was the triggering parameter. Fluorescence of PE was detected using a 580/30 bandpass filter. Single cell deposition was accomplished using the CyClone automated cloner (Dako-Cytomation); in the 0.5 single drop mode and with gating based on forward scatter and PE fluorescence, individual CD34+ cells were placed into each well of a 96-well plate containing 100 μL culture media (Figure 1A).
Flow chart, linearized map of mtDNA control region, and single CD34+ clones. (A) Mononuclear cells from BM and CB were separated by density gradient centrifugation and washed twice in phosphate-buffered saline. CD34+ cells were sorted by single cell deposition into 96-well microtiter plates using a phycoerythrin anti-CD34 monoclonal antibody (BD Biosciences), a MoFlo cytometer, and a CyClone automated cloner (Dako-Cytomation) in the 0.5 single drop mode. After 5 days of culture in media containing stem cell factor, Flt-3, thrombopoietin, and G-CSF, each well of the microplate was carefully examined and scored for cell number. To directly sequence the control region of mtDNA, DNA in these colonies, derived from single CD34+ cells, was subjected to nested gene amplification (see “Materials and methods”). Sequencing was performed on an ABI Prism 3100 Genetic Analyzer in both orientations. Evidence of mtDNA heterogeneity was further confirmed by reamplification of the original lysate. (B) Linearized map and function location of mtDNA control region between nucleotides 16 024 to 16 569 and 1 to 576 (D-loop); HV1 (hypervariable segment 1, nucleotides 16 024 to 16 383), HV2 (hypervariable segment 2, nucleotides 57 to 372), OH (H-stand origin, nucleotides 110 to 441), CSB (conserved sequence block, nucleotides 213 to 235, nucleotides 299 to 315, nucleotides 346 to 363), mt5 (control element, nucleotides 16 194 to 16 208), mt3L (L-strand control element, nucleotides 16 499 to 16 506), TAS (termination-associated sequence, nucleotides 16 157 to 16 172), PL (L-strand promoter, nucleotides 392 to 345), PH1 (major H-strand promoter, nucleotides 545 to 567), TFB (mitochondrial transcription factor binding site, nucleotides 233 to 260, 276 to 303, 418 to 445, 523 to 550), mt4H (H-strand control element, nucleotides 371 to 379), mt3H (H-strand control element, nucleotides 384 to 391), and 7S DNA (nucleotides 16 106 to 16 191). The homopolymeric C tracts located on HV1 (nucleotides 16 184 to 16 193; 5CT4C) and HV2 (nucleotides 303 to 315; 7CT5C). (C) The number and morphology of CD34+ clones after 5-day suspension culture. Each picture (original magnification, × 200) shows a different grade: (i) 5 or fewer cells per well, grade 1; (ii) 6 to 10 cells per well, grade 2; (iii) 11 to 20 cells per well, grade 3; and (iv) 21 or more cells per well, grade 4.
Flow chart, linearized map of mtDNA control region, and single CD34+ clones. (A) Mononuclear cells from BM and CB were separated by density gradient centrifugation and washed twice in phosphate-buffered saline. CD34+ cells were sorted by single cell deposition into 96-well microtiter plates using a phycoerythrin anti-CD34 monoclonal antibody (BD Biosciences), a MoFlo cytometer, and a CyClone automated cloner (Dako-Cytomation) in the 0.5 single drop mode. After 5 days of culture in media containing stem cell factor, Flt-3, thrombopoietin, and G-CSF, each well of the microplate was carefully examined and scored for cell number. To directly sequence the control region of mtDNA, DNA in these colonies, derived from single CD34+ cells, was subjected to nested gene amplification (see “Materials and methods”). Sequencing was performed on an ABI Prism 3100 Genetic Analyzer in both orientations. Evidence of mtDNA heterogeneity was further confirmed by reamplification of the original lysate. (B) Linearized map and function location of mtDNA control region between nucleotides 16 024 to 16 569 and 1 to 576 (D-loop); HV1 (hypervariable segment 1, nucleotides 16 024 to 16 383), HV2 (hypervariable segment 2, nucleotides 57 to 372), OH (H-stand origin, nucleotides 110 to 441), CSB (conserved sequence block, nucleotides 213 to 235, nucleotides 299 to 315, nucleotides 346 to 363), mt5 (control element, nucleotides 16 194 to 16 208), mt3L (L-strand control element, nucleotides 16 499 to 16 506), TAS (termination-associated sequence, nucleotides 16 157 to 16 172), PL (L-strand promoter, nucleotides 392 to 345), PH1 (major H-strand promoter, nucleotides 545 to 567), TFB (mitochondrial transcription factor binding site, nucleotides 233 to 260, 276 to 303, 418 to 445, 523 to 550), mt4H (H-strand control element, nucleotides 371 to 379), mt3H (H-strand control element, nucleotides 384 to 391), and 7S DNA (nucleotides 16 106 to 16 191). The homopolymeric C tracts located on HV1 (nucleotides 16 184 to 16 193; 5CT4C) and HV2 (nucleotides 303 to 315; 7CT5C). (C) The number and morphology of CD34+ clones after 5-day suspension culture. Each picture (original magnification, × 200) shows a different grade: (i) 5 or fewer cells per well, grade 1; (ii) 6 to 10 cells per well, grade 2; (iii) 11 to 20 cells per well, grade 3; and (iv) 21 or more cells per well, grade 4.
Culture and harvest of CD34+ cells
Individual CD34+ cells placed into separate wells of 96-well plates were cultured in serum-free medium containing 100 ng/mL stem cell factor (SCF), 100 ng/mL Flt-3, 100 ng/mL thrombopoietin (TPO), with or without 50 ng/mL granulocyte colony-stimulating factor (G-CSF) (all from Stem Cell Technologies, Vancouver, BC, Canada). After culture for 5 days, each well of the microtiter plate was carefully observed using an inverted microscope (Olympus IX50, Olympus, Melville, NY) to determine growth and plating efficiency of single CD34+ cells and to grade growth with the following scoring system based on cell number in each CD34+ clone: grade 1, 5 or fewer cells per well; grade 2, 6 to 10 cells per well; grade 3, 11 to 20 cells per well; grade 4, 21 or more cells per well (Figure 1C). Plating efficiency was defined as the number of positive (cells were present) wells divided by total wells × 100. Each CD34+ clone was harvested from the well by vigorous pipetting and dispensed into a 1.5 mL microcentrifuge tube and rinsed with 200 μL PBS. Cells were collected after centrifugation at 300g for 5 minutes and then washed with PBS. Cell pellets were stored at –80°C.
DNA extraction from individual CD34+ clones
A total of 30 μLof 1 × Tris-EDTA (TE) buffer was placed in each 1.5 mL tube containing a cell pellet. The cells were lysed by incubation at 95°C for 10 minutes with occasional shaking to liberate the total DNA. The lysate was briefly microcentrifuged and stored at –20°C.
PCR amplification of the mtDNA control region
Cell lysates of individual CD34+ clones were subjected to amplification of mtDNA using the long-and-accurate polymerase chain reaction (LA PCR) kit (TaKaRa LA Taq, Panvera, Madison, WI). Two-step PCR amplification was performed with outer and inner pairs of primers to generate sufficient template from CD34+ clones for sequencing of the mtDNA control region. The outer pair of primers (5′-CGCCTACACAATTCTCCGATC-3′ and 5′-ACTTGGGTTAATCG TGTGACC-3′) was used for amplification of the fragment spanning nucleotides 15 574 to 16 569 and 1 to 921 of the revised human mtDNA Revised Cambridge Reference Sequence. The inner nested pair of primers (5′-TTAACTCCACCATTAGCACC-3′ and 5′-GAAAGGCTA GGACCAAACCTA-3′) amplified the fragment spanning nucleotides 15 971 to 16 569 and 1 to 670 (Figure 1B). The primary PCR mixture contained 400 μM of each deoxyribonucleoside triphosphate (dNTP), 2 units of LA Taq (TaKaRa LA Taq), 0.8 μM outer primers, and 3 μL cell lysate in a total volume of 30 μL. PCR amplification was carried out in a thin-wall 0.5 mL PCR tube using the DNA thermal cycler 9700 (Perkin-Elmer, Foster City, CA): 1 cycle of 96°C for 1 minute; then 35 cycles of 94°C for 30 seconds, 52°C for 50 seconds, and 72°C for 1 minute with a 10-second increase per cycle; ending with 1 cycle of 72°C for 5 minutes. The secondary PCR was performed in 50 μL reaction mixture containing 400 μM of each dNTP, 2 units of LA Taq, 0.8 μM inner nested primers, and 2 μL primary PCR product under the same amplification conditions as described above. Secondary PCR samples were electrophoresed on 1% agarose gels and stained with ethidium bromide to assess the purity and size of the DNA fragments and subsequently purified using the QIA quick PCR purification kit (Qiagen, Valencia, CA). The negative controls, reaction mixtures without DNA templates, were subjected to the same PCR amplification conditions and in all cases confirmed to be negative. To prevent DNA cross-contamination, special precautions were taken for each procedure of cell harvest, DNA extraction, PCR amplification, and DNA sequencing.
Sequence analysis of the mtDNA control region
We amplified and then directly sequenced the 1121–base pair control region (nucleotides 16 024 to 16 569 and 1 to 576) (Figure 1B) using the BigDye Terminator v3.0 Ready Reaction kit (Applied Biosystems, Foster City, CA) and the ABI Prism 3100 Genetic Analyzer (Applied Biosystems). The following oligonucleotide primers were used for sequencing: 5′-CAGTGTATTGCTTTGAGGAGG-3′, 5′-CATCTGGTTCCTACTTCAGGGTC-3′, 5′-TTAACTCCACCATTAGCACC-3′, 5′-GCATGGAGAGCTCCCGTGAGTGG3-3′, 5′-CACCCTATTAACCACTCACG-3′, and 5′-TACATTACTGCCAGCCACCATG-3′. mtDNA sequences experimentally obtained were compared with the Revised Cambridge Reference Sequence (http://www.mitomap.org)2 using the BLAST2 program (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html) and the database search tool MitoAnalyzer (http://www.cstl.nist.gov/biotech/strbase/mitoanalyzer.html)15 to determine polymorphisms and mutations. All automated results were manually confirmed. To exclude potential artifacts, PCR amplifications from original cell lysates were additionally replicated 1 or 2 more times, and when nucleotide changes were reproduced in all independent PCR amplifications, they were considered to be confirmed. Several CD34+ clones with potential mtDNA-nucleotide changes were eliminated because no mtDNA was amplified when the PCR was replicated. The PCR replication probably failed due to the low concentration of mtDNA.
TA cloning
In preliminary experiments, 285-bp amplicons were generated by gene amplification of wild-type mtDNA and mtDNA altered in a single base and then mixed in varying proportions. On sequencing of the mixtures, the lower limit of detection of a minor species of mtDNA was approximately 20%. Mixed nucleotide signals on sequencing chromatograms, when observed in the current study, were assumed to represent at least 20% heteroplasmy but could also result from gene amplification artifacts. To confirm heteroplasmy and mixed nucleotide signals in the sequences of the mtDNA control region, PCR products were directly inserted into the pCR 2.1-TOPO vector and transformed into competent Escherichia coli (TOP10 cells) using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Recombinant plasmids isolated from 8 to 12 white colonies were sequenced.
Statistical analysis
A χ2 test was used to determine statistical differences in the frequency of heteroplasmy in adult bone marrow and cord blood. The 1-way analysis of variance (ANOVA) test was performed to examine whether the number of cells in each clone or the culture media produced significant statistical differences in mtDNA heterogeneity; P < .05 was considered significant.
Results
Culture of single CD34+ cells
After sorting, single CD34+ cells were cultured in individual wells of 96-well plates in serum-free medium containing selected hematopoietic growth factors, with or without G-CSF. Plating efficiency was microscopically determined by the presence of clusters of viable cells. CD34+ cell-derived colonies were classified according to the cell number per well (Figure 1C). Although there was some variation of plating efficiency of CD34+ cells among 6 healthy BM and 5 CB donors, overall average plating efficiency in BM and CB was 30% (30.0% ± 11.7%, mean ± SD) and 79% (78.6% ± 11.7%), respectively (Table 1) and was not affected by G-CSF. As expected, the colony size was increased in the presence of G-CSF.
Plating efficiency and grade of CD34+ clones from normal adult bone marrow and cord blood samples after 5-day culture
. | . | Grade . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | 1 . | . | 2 . | . | 3 . | . | 4 . | . | Subtotal no. . | . | . | Microplate* . | . | Plating Efficiency, % . | . | |||||||
| Donor . | Age, y/sex . | + . | - . | + . | - . | + . | - . | + . | - . | + . | - . | Total no. . | + . | - . | + . | - . | |||||||
| BM | |||||||||||||||||||||||
| 1 | 47/F | 14 | 19 | 9 | 12 | 16 | 14 | 36 | 2 | 75 | 47 | 122 | 2 | 2 | 39.1 | 24.5 | |||||||
| 2 | 38/F | 27 | 22 | 1 | 2 | 1 | 3 | 6 | 0 | 35 | 27 | 62 | 5 | 5 | 7.3 | 5.6 | |||||||
| 3 | 43/M | 103 | 176 | 65 | 84 | 49 | 42 | 122 | 5 | 339 | 307 | 646 | 10 | 10 | 35.3 | 32.0 | |||||||
| 4 | 34/M | 45 | 71 | 33 | 74 | 50 | 53 | 112 | 21 | 240 | 219 | 459 | 10 | 10 | 25.0 | 22.8 | |||||||
| 5 | 54/M | 98 | 208 | 67 | 110 | 60 | 59 | 135 | 5 | 360 | 382 | 742 | 10 | 10 | 37.5 | 39.8 | |||||||
| 6 | 34/F | 82 | 131 | 43 | 103 | 79 | 70 | 150 | 16 | 354 | 320 | 674 | 10 | 10 | 36.9 | 33.3 | |||||||
| BM subtotal | 369 | 627 | 218 | 385 | 255 | 241 | 561 | 49 | 1403 | 1302 | 2705 | 47 | 47 | 31.1 | 28.9 | ||||||||
| BM total | 996 | 603 | 496 | 610 | 2705 | 94 | 30.0 | ||||||||||||||||
| CB | |||||||||||||||||||||||
| 1 | — | 46 | 85 | 79 | 155 | 167 | 309 | 398 | 124 | 690 | 673 | 1363 | 10 | 10 | 71.9 | 70.1 | |||||||
| 2 | — | 8 | 8 | 6 | 21 | 23 | 78 | 369 | 309 | 406 | 416 | 822 | 5 | 5 | 84.6 | 86.7 | |||||||
| 3 | — | 28 | 55 | 33 | 45 | 57 | 44 | 280 | 245 | 398 | 389 | 787 | 5 | 5 | 82.9 | 81.0 | |||||||
| 4 | — | 23 | 30 | 21 | 26 | 30 | 59 | 346 | 299 | 420 | 414 | 834 | 5 | 5 | 87.5 | 86.3 | |||||||
| 5 | — | 22 | 33 | 23 | 36 | 29 | 52 | 286 | 243 | 360 | 364 | 724 | 5 | 5 | 75.0 | 75.8 | |||||||
| CB subtotal | 127 | 211 | 162 | 283 | 306 | 542 | 1679 | 1220 | 2274 | 2256 | 4530 | 30 | 30 | 79.0 | 78.3 | ||||||||
| CB total | 338 | 445 | 848 | 2899 | 4530 | 60 | 78.6 | ||||||||||||||||
. | . | Grade . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | 1 . | . | 2 . | . | 3 . | . | 4 . | . | Subtotal no. . | . | . | Microplate* . | . | Plating Efficiency, % . | . | |||||||
| Donor . | Age, y/sex . | + . | - . | + . | - . | + . | - . | + . | - . | + . | - . | Total no. . | + . | - . | + . | - . | |||||||
| BM | |||||||||||||||||||||||
| 1 | 47/F | 14 | 19 | 9 | 12 | 16 | 14 | 36 | 2 | 75 | 47 | 122 | 2 | 2 | 39.1 | 24.5 | |||||||
| 2 | 38/F | 27 | 22 | 1 | 2 | 1 | 3 | 6 | 0 | 35 | 27 | 62 | 5 | 5 | 7.3 | 5.6 | |||||||
| 3 | 43/M | 103 | 176 | 65 | 84 | 49 | 42 | 122 | 5 | 339 | 307 | 646 | 10 | 10 | 35.3 | 32.0 | |||||||
| 4 | 34/M | 45 | 71 | 33 | 74 | 50 | 53 | 112 | 21 | 240 | 219 | 459 | 10 | 10 | 25.0 | 22.8 | |||||||
| 5 | 54/M | 98 | 208 | 67 | 110 | 60 | 59 | 135 | 5 | 360 | 382 | 742 | 10 | 10 | 37.5 | 39.8 | |||||||
| 6 | 34/F | 82 | 131 | 43 | 103 | 79 | 70 | 150 | 16 | 354 | 320 | 674 | 10 | 10 | 36.9 | 33.3 | |||||||
| BM subtotal | 369 | 627 | 218 | 385 | 255 | 241 | 561 | 49 | 1403 | 1302 | 2705 | 47 | 47 | 31.1 | 28.9 | ||||||||
| BM total | 996 | 603 | 496 | 610 | 2705 | 94 | 30.0 | ||||||||||||||||
| CB | |||||||||||||||||||||||
| 1 | — | 46 | 85 | 79 | 155 | 167 | 309 | 398 | 124 | 690 | 673 | 1363 | 10 | 10 | 71.9 | 70.1 | |||||||
| 2 | — | 8 | 8 | 6 | 21 | 23 | 78 | 369 | 309 | 406 | 416 | 822 | 5 | 5 | 84.6 | 86.7 | |||||||
| 3 | — | 28 | 55 | 33 | 45 | 57 | 44 | 280 | 245 | 398 | 389 | 787 | 5 | 5 | 82.9 | 81.0 | |||||||
| 4 | — | 23 | 30 | 21 | 26 | 30 | 59 | 346 | 299 | 420 | 414 | 834 | 5 | 5 | 87.5 | 86.3 | |||||||
| 5 | — | 22 | 33 | 23 | 36 | 29 | 52 | 286 | 243 | 360 | 364 | 724 | 5 | 5 | 75.0 | 75.8 | |||||||
| CB subtotal | 127 | 211 | 162 | 283 | 306 | 542 | 1679 | 1220 | 2274 | 2256 | 4530 | 30 | 30 | 79.0 | 78.3 | ||||||||
| CB total | 338 | 445 | 848 | 2899 | 4530 | 60 | 78.6 | ||||||||||||||||
PhE indicates plating efficiency (see “Materials and methods”); grade 1, 5 or fewer cells per well; grade 2, 6 to 10 cells per well; grade 3, 11 to 20 cells per well; grade 4, 21 or more cells per well; +, culture media containing 100 ng/m of each stem cell factor, Flt-3, thrombopoietin, serum-free media, and 50 ng/mL G-CSF; -, same culture media without G-CSF; BM, bone marrow; CB, cord blood; and —, not applicable.
No. of 96-well microplates.
mtDNA control region
To assess heterogeneity of the mtDNA sequences among CD34+ cells from healthy BM and CB donors, we targeted the 1121 bp of mtDNA control region known to contain multiple mutational hotspots.14 More than 100 CD34+ clones per each donor except for donor no. 2 were subjected to sequencing analysis, resulting in a total number of 611 and 580 CD34+ clones from adult BM and CB, respectively (Table 2).
Assayed number of individual CD34+ clones from normal adult bone marrow and cord blood samples after 5-day culture
. | Grade . | . | . | . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | 1 . | . | 2 . | . | 3 . | . | 4 . | . | Subtotal no. . | . | Total assayed no. . | |||||||
| Donor . | + . | - . | + . | - . | + . | - . | + . | - . | + . | - . | . | |||||||
| BM | ||||||||||||||||||
| 1 | 14 | 14 | 8 | 12 | 16 | 14 | 35 | 2 | 73 | 42 | 115 | |||||||
| 2 | 13 | 10 | 1 | 2 | 1 | 3 | 6 | 0 | 21 | 15 | 36 | |||||||
| 3 | 15 | 15 | 15 | 15 | 15 | 15 | 25 | 5 | 70 | 50 | 120 | |||||||
| 4 | 13 | 13 | 14 | 14 | 15 | 13 | 15 | 14 | 57 | 54 | 111 | |||||||
| 5 | 12 | 14 | 15 | 13 | 15 | 15 | 25 | 5 | 67 | 47 | 114 | |||||||
| 6 | 14 | 11 | 15 | 15 | 15 | 15 | 15 | 15 | 59 | 56 | 115 | |||||||
| BM subtotal | 81 | 77 | 68 | 71 | 77 | 75 | 121 | 41 | 347 | 264 | 611 | |||||||
| BM total | 158 | 139 | 152 | 162 | 611 | |||||||||||||
| CB | ||||||||||||||||||
| 1 | 15 | 15 | 14 | 15 | 15 | 15 | 15 | 15 | 59 | 60 | 119 | |||||||
| 2 | 8 | 8 | 6 | 15 | 15 | 16 | 26 | 26 | 55 | 65 | 120 | |||||||
| 3 | 14 | 13 | 14 | 15 | 15 | 15 | 5 | 13 | 48 | 56 | 104 | |||||||
| 4 | 15 | 14 | 15 | 15 | 15 | 15 | 15 | 15 | 60 | 59 | 119 | |||||||
| 5 | 14 | 15 | 15 | 14 | 15 | 15 | 15 | 15 | 59 | 59 | 118 | |||||||
| CB subtotal | 66 | 65 | 64 | 74 | 75 | 76 | 76 | 84 | 281 | 299 | 580 | |||||||
| CB total | 131 | 138 | 151 | 160 | 580 | |||||||||||||
. | Grade . | . | . | . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | 1 . | . | 2 . | . | 3 . | . | 4 . | . | Subtotal no. . | . | Total assayed no. . | |||||||
| Donor . | + . | - . | + . | - . | + . | - . | + . | - . | + . | - . | . | |||||||
| BM | ||||||||||||||||||
| 1 | 14 | 14 | 8 | 12 | 16 | 14 | 35 | 2 | 73 | 42 | 115 | |||||||
| 2 | 13 | 10 | 1 | 2 | 1 | 3 | 6 | 0 | 21 | 15 | 36 | |||||||
| 3 | 15 | 15 | 15 | 15 | 15 | 15 | 25 | 5 | 70 | 50 | 120 | |||||||
| 4 | 13 | 13 | 14 | 14 | 15 | 13 | 15 | 14 | 57 | 54 | 111 | |||||||
| 5 | 12 | 14 | 15 | 13 | 15 | 15 | 25 | 5 | 67 | 47 | 114 | |||||||
| 6 | 14 | 11 | 15 | 15 | 15 | 15 | 15 | 15 | 59 | 56 | 115 | |||||||
| BM subtotal | 81 | 77 | 68 | 71 | 77 | 75 | 121 | 41 | 347 | 264 | 611 | |||||||
| BM total | 158 | 139 | 152 | 162 | 611 | |||||||||||||
| CB | ||||||||||||||||||
| 1 | 15 | 15 | 14 | 15 | 15 | 15 | 15 | 15 | 59 | 60 | 119 | |||||||
| 2 | 8 | 8 | 6 | 15 | 15 | 16 | 26 | 26 | 55 | 65 | 120 | |||||||
| 3 | 14 | 13 | 14 | 15 | 15 | 15 | 5 | 13 | 48 | 56 | 104 | |||||||
| 4 | 15 | 14 | 15 | 15 | 15 | 15 | 15 | 15 | 60 | 59 | 119 | |||||||
| 5 | 14 | 15 | 15 | 14 | 15 | 15 | 15 | 15 | 59 | 59 | 118 | |||||||
| CB subtotal | 66 | 65 | 64 | 74 | 75 | 76 | 76 | 84 | 281 | 299 | 580 | |||||||
| CB total | 131 | 138 | 151 | 160 | 580 | |||||||||||||
Abbreviations are explained in Table 1.
Aggregate genotype of the mtDNA control region from total BM and CB cells
To identify mtDNA heterogeneity in individual CD34+ clones, we first determined the aggregate genotype of total BM cells from each donor. There was marked variation in the number of nucleotide changes among individual healthy BM donors, with ranges of 6 (donor no. 1) to 23 (donor no. 4) (11.3 ± 6.1, mean ± SD) (Table 3). A total of 68 mtDNA sequence variants were found in aggregate BM cells from 6 healthy donors; among these 66 variants were already listed in the polymorphism database (http://www.mitomap.org). Two new nucleotide variants were classified mutations (A478G and A517G in donor nos. 2 and 6, respectively). The number of mtDNA sequence changes from aggregate CB cells also showed individual variations (Table 3) but, interestingly, 3 (CB nos. 2, 3, and 5) of 5 CB donors had length variations of poly C tract at nucleotide position 303 to 315 and 16 183 to 16 193.
Nucleotide sequence changes of mtDNA control region from aggregate cells
Donor no. and polymorphism . | Affected mtDNA gene . |
|---|---|
| BM donor 1 | |
| 73A>G | HV2, 7S |
| 150C>T | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 16192C>T | HV1, 7S |
| 16270C>T | HV1, 7S |
| BM donor 2 | |
| 73A>G | HV2, 7S |
| 185G>A | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| A478G† | — |
| 16093T>C | HV1 |
| 16158A>T | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16183A>C | HV1, 7S |
| 16189T>C (12C)* | HV1, 7S |
| 16219A>G | HV1, 7S |
| 16278C>T | HV1, 7S |
| BM donor 3 | |
| 73A>G | HV2, 7S |
| 146T>C | HV2, 7S, OH |
| 152T>C | HV2, 7S, OH |
| 195T>C | HV2, OH |
| 263A>G | HV2, OH |
| 8CT6C*, 9CT6C* | HV2, OH, CSB2 |
| 514-515delCA | — |
| 16223C>T | HV1, 7S |
| 16278C>T | HV1, 7S |
| 16294C>T | HV1, 7S |
| 16390G>G | 7S |
| BM donor 4 | |
| 93A>G | HV2, 7S |
| 95A>C | HV2, 7S |
| 185G>A | HV2, 7S, OH |
| 189A>G | HV2, 7S, OH |
| 236T>C | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 247G>A | HV2, OH, TFB1 |
| 263A>G | HV2, OH |
| 514-515delCA | — |
| 16093T>C | HV1 |
| 16129G>A | HV1, 7S |
| 16148C>T | HV1, 7S |
| 16168C>T | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16187C>T* | HV1, 7S |
| 16188C>G* | HV1, 7S |
| 16189T>C* | HV1, 7S |
| 16223C>T | HV1, 7S |
| 16230A>G | HV1, 7S |
| 16278C>T | HV1, 7S |
| 16293A>G | HV1, 7S |
| 16311T>C | HV1, 7S |
| 16320C>T | HV1, 7S |
| BM donor 5 | |
| 73A>G | HV2, 7S |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| 514-515delCA | — |
| 16126T>C | HV1, 7S |
| 16294C>T | HV1, 7S |
| 16296C>T | HV1, 7S |
| 16519T>C | 7S |
| BM donor 6 | |
| 73A>G | HV2, 7S |
| 150C>T | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 517A>G† | — |
| 16270C>T | HV1, 7S |
| 16292C>T | HV1, 7S |
| 16362T>C | HV1, 7S |
| CB donor 1 | |
| 73A>G | HV2, 7S |
| 146T>C | HV2, 7S, OH |
| 195T>A | HV2, OH |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| 489T>C | — |
| 514-515delCA | — |
| 16166A>T | — |
| 16169delC† | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16223C>T | HV1, 7S |
| 16354C>T | HV1, CSB3 |
| 16519T>C | 7S |
| CB donor 2 | |
| 72T>C | HV2, 7S |
| 253C>T | HV2, OH, TFB1 |
| 263A>G | HV2, OH |
| 9CT6C* | HV2, OH, CSB2 |
| 10CT6C* | HV2, OH, CSB2 |
| 8CT6C* | HV2, OH, CSB2 |
| 16256C>T | HV1, 7S |
| 16298T>C | HV1, 7S |
| 16519T>C | 7S |
| CB donor 3 | |
| 73A>G | HV2, 7S |
| 146T>C | HV2, 7S, OH |
| 189A>G | HV2, 7S, OH |
| 194C>T | HV2, OH |
| 195T>C | HV2, OH |
| 204T>C | HV2, OH |
| 207G>A | HV2, OH |
| 263A>G | HV2, OH |
| 279T>C | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 9CT6C* | HV2, OH, CSB2 |
| 16223C>T | HV1, 7S |
| 16292C>T | HV1, 7S |
| 16519T>C | 7S |
| CB donor 4 | |
| 73A>G | HV2, 7S |
| 249delA | HV2, OH, TFB1 |
| 290-291delAA | HV2, OH, TFB2 |
| 7CT6C* | HV2, OH, CSB2 |
| 489T>C | — |
| 493A>G | — |
| 514-515delCA | — |
| 16223C>T | HV1, 7S |
| 16298T>C | HV1, 7S |
| 16325T>C | HV1, 7S |
| 16327C>T | HV1, 7S |
| 16519T>C | 7S |
| CB donor 5 | |
| 73A>G | HV2, 7S |
| 150C>T | HV2, 7S, OH |
| 195T>C | HV2, OH |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| 16171A>G | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16189T>C* | HV1, 7S |
| 16193C>C/CC* | HV1, 7S |
| 16223C>T | HV1, 7S |
| 16320C>T | HV1, 7S |
| 16519T>C | 7S |
Donor no. and polymorphism . | Affected mtDNA gene . |
|---|---|
| BM donor 1 | |
| 73A>G | HV2, 7S |
| 150C>T | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 16192C>T | HV1, 7S |
| 16270C>T | HV1, 7S |
| BM donor 2 | |
| 73A>G | HV2, 7S |
| 185G>A | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| A478G† | — |
| 16093T>C | HV1 |
| 16158A>T | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16183A>C | HV1, 7S |
| 16189T>C (12C)* | HV1, 7S |
| 16219A>G | HV1, 7S |
| 16278C>T | HV1, 7S |
| BM donor 3 | |
| 73A>G | HV2, 7S |
| 146T>C | HV2, 7S, OH |
| 152T>C | HV2, 7S, OH |
| 195T>C | HV2, OH |
| 263A>G | HV2, OH |
| 8CT6C*, 9CT6C* | HV2, OH, CSB2 |
| 514-515delCA | — |
| 16223C>T | HV1, 7S |
| 16278C>T | HV1, 7S |
| 16294C>T | HV1, 7S |
| 16390G>G | 7S |
| BM donor 4 | |
| 93A>G | HV2, 7S |
| 95A>C | HV2, 7S |
| 185G>A | HV2, 7S, OH |
| 189A>G | HV2, 7S, OH |
| 236T>C | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 247G>A | HV2, OH, TFB1 |
| 263A>G | HV2, OH |
| 514-515delCA | — |
| 16093T>C | HV1 |
| 16129G>A | HV1, 7S |
| 16148C>T | HV1, 7S |
| 16168C>T | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16187C>T* | HV1, 7S |
| 16188C>G* | HV1, 7S |
| 16189T>C* | HV1, 7S |
| 16223C>T | HV1, 7S |
| 16230A>G | HV1, 7S |
| 16278C>T | HV1, 7S |
| 16293A>G | HV1, 7S |
| 16311T>C | HV1, 7S |
| 16320C>T | HV1, 7S |
| BM donor 5 | |
| 73A>G | HV2, 7S |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| 514-515delCA | — |
| 16126T>C | HV1, 7S |
| 16294C>T | HV1, 7S |
| 16296C>T | HV1, 7S |
| 16519T>C | 7S |
| BM donor 6 | |
| 73A>G | HV2, 7S |
| 150C>T | HV2, 7S, OH |
| 263A>G | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 517A>G† | — |
| 16270C>T | HV1, 7S |
| 16292C>T | HV1, 7S |
| 16362T>C | HV1, 7S |
| CB donor 1 | |
| 73A>G | HV2, 7S |
| 146T>C | HV2, 7S, OH |
| 195T>A | HV2, OH |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| 489T>C | — |
| 514-515delCA | — |
| 16166A>T | — |
| 16169delC† | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16223C>T | HV1, 7S |
| 16354C>T | HV1, CSB3 |
| 16519T>C | 7S |
| CB donor 2 | |
| 72T>C | HV2, 7S |
| 253C>T | HV2, OH, TFB1 |
| 263A>G | HV2, OH |
| 9CT6C* | HV2, OH, CSB2 |
| 10CT6C* | HV2, OH, CSB2 |
| 8CT6C* | HV2, OH, CSB2 |
| 16256C>T | HV1, 7S |
| 16298T>C | HV1, 7S |
| 16519T>C | 7S |
| CB donor 3 | |
| 73A>G | HV2, 7S |
| 146T>C | HV2, 7S, OH |
| 189A>G | HV2, 7S, OH |
| 194C>T | HV2, OH |
| 195T>C | HV2, OH |
| 204T>C | HV2, OH |
| 207G>A | HV2, OH |
| 263A>G | HV2, OH |
| 279T>C | HV2, OH |
| 8CT6C* | HV2, OH, CSB2 |
| 9CT6C* | HV2, OH, CSB2 |
| 16223C>T | HV1, 7S |
| 16292C>T | HV1, 7S |
| 16519T>C | 7S |
| CB donor 4 | |
| 73A>G | HV2, 7S |
| 249delA | HV2, OH, TFB1 |
| 290-291delAA | HV2, OH, TFB2 |
| 7CT6C* | HV2, OH, CSB2 |
| 489T>C | — |
| 493A>G | — |
| 514-515delCA | — |
| 16223C>T | HV1, 7S |
| 16298T>C | HV1, 7S |
| 16325T>C | HV1, 7S |
| 16327C>T | HV1, 7S |
| 16519T>C | 7S |
| CB donor 5 | |
| 73A>G | HV2, 7S |
| 150C>T | HV2, 7S, OH |
| 195T>C | HV2, OH |
| 263A>G | HV2, OH |
| 7CT6C* | HV2, OH, CSB2 |
| 16171A>G | HV1, 7S, TAS |
| 16172T>C | HV1, 7S, TAS |
| 16189T>C* | HV1, 7S |
| 16193C>C/CC* | HV1, 7S |
| 16223C>T | HV1, 7S |
| 16320C>T | HV1, 7S |
| 16519T>C | 7S |
HV1 indicates hypervariable segment 1; HV2, hypervariable segment 2; 7S, 7S DNA; OH, H-strand origin; CSB2, conserved sequence block II; TAS, termination-associated sequence; TFB1, mitochondrial transcription factor 1 binding site; and —, not applicable.
Homopolymeric C tracts at nucleotides position at 303 to 315 (for example, 8CT6C defined CCCCCCCCTCCCCCC) and 16184 to 16193 in HV2 and HV1, respectively.
New mtDNA polymorphisms (not listed in accepted database).
mtDNA heterogeneity among individual CD34+ clones
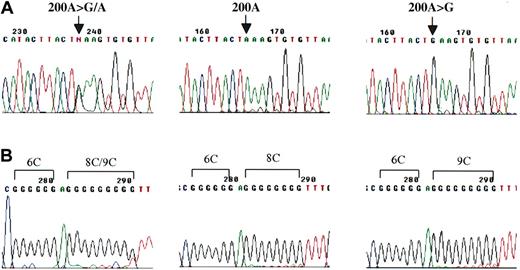
Analysis of 611 CD34+ clones from the 6 healthy BM donors revealed that a total of 152 clones (24.9% ± 17.2%, mean ± SD) displayed mtDNA heterogeneity distinct from the donor's corresponding aggregate mtDNA sequences (Table 4). Common patterns of mtDNA heterogeneity in CD34+ clones among 6 BM donors were 1 or 2 nucleotide changes (substitution, insertion, or deletion) in addition to the polymorphisms detected in the respective aggregate mtDNA. Among them, most differences were due to single nucleotide substitutions at various positions and length alterations in the poly C tract localized between nucleotides 303 to 315 (Figure 2B). The heterogeneous mtDNA of CD34+ clones in 6 BM donors was classified into several unique patterns according to nucleotide changes; 8, 5, 7, 6, 14, and 6 patterns in donor nos. 1 to 6, respectively (Table 4). The mean proportion of unique heterogeneous pattern of mtDNA among single CD34+ clones was 7.5% (7.5% ± 3.3%) (Table 5). Figure 2A disclosed one of the typical heteroplasmic mutations in CD34+ clone derived from BM donor no. 6: 200A>G/A heteroplasmy in one CD34+ clone of adult BM donor no. 6 was clearly identified using TA cloning. Neither the presence of G-CSF nor the colony size was statistically correlated with the proportion of CD34+ clones with variant mtDNA (Table 6).
Summary of mtDNA heterogeneity among individual single CD34+ clones from normal adult bone marrow and cord blood
. | . | Heterogeneity . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| . | . | Different . | . | Unique . | . | |||
| Donor and mtDNA sequence . | Clone no. . | No. . | % . | No. . | % . | |||
| BM donor 1 | ||||||||
| “Aggregate” sequence of total BM cells | 85 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 30 | 26.1 | 8 | 7.0 | ||||
| +8CT6C*, 9CT6C* | 22 | |||||||
| +9CT6C* | 2 | |||||||
| +7CT6C* | 1 | |||||||
| +189A>G/A | 1 | |||||||
| +204T>C | 1 | |||||||
| +277C>T | 1 | |||||||
| +514insCA | 1 | |||||||
| +16114C>T | 1 | |||||||
| BM donor 2 | ||||||||
| “Aggregate” sequence of total BM cells | 27 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 9 | 25.0 | 5 | 13.9 | ||||
| +16184C>CC (11C)* | 5 | |||||||
| +16131T>C/T | 1 | |||||||
| +16145G>A | 1 | |||||||
| +16184C>CCCC (13C)* | 1 | |||||||
| 73A>G, 263A>G, 191A>AA, 194C>T, 199T>C, 207G>A, 8CT6C*, 489T>C, 16147C>T, 16173C>T, 16245C>T, 16362T>C | 1 | |||||||
| BM donor 3 | ||||||||
| “Aggregate” sequence of total BM cells | 96 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 24 | 20.0 | 7 | 5.8 | ||||
| +9CT6C*, 10CT6C* | 11 | |||||||
| +9CT6C* | 6 | |||||||
| +8CT6C* | 2 | |||||||
| +182C>T/C, 8CT6C*, 9CT6C* | 2 | |||||||
| +71delG, 9CT6C*, 10CT6C* | 1 | |||||||
| +279T>C/T | 1 | |||||||
| +16153G>A | 1 | |||||||
| BM donor 4 | ||||||||
| “Aggregate” sequence of total BM cells | 92 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 19 | 17.1 | 6 | 5.4 | ||||
| +8CT6C*, 9CT6C* | 11 | |||||||
| +514-515delCA, 514insCA† | 3 | |||||||
| +7CT6C*, 8CT6C* | 2 | |||||||
| +9CT6C*, 10CT6C* | 1 | |||||||
| +89T>C | 1 | |||||||
| +8CT6C*, 9CT6C*, 16093T‡ | 1 | |||||||
| BM donor 5 | ||||||||
| “Aggregate” sequence of total BM cells | 57 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 57 | 50.0 | 14 | 12.3 | ||||
| +514insCA‡ | 19 | |||||||
| +264C>T | 18 | |||||||
| +514-515delCA, 514insCA† | 5 | |||||||
| +264C>T/C | 3 | |||||||
| +7CT6C*, 8CT6C* | 2 | |||||||
| +146T>C, 514insCA‡ | 2 | |||||||
| +146T>C, 264C>T/C | 1 | |||||||
| +146T>C/T, 514insCA‡ | 1 | |||||||
| +146T>C | 1 | |||||||
| +189A>G | 1 | |||||||
| +264C>T/C, 514insCA‡ | 1 | |||||||
| +161T>C/T, 264C>T, 514insCA‡ | 1 | |||||||
| +16189T>C* | 1 | |||||||
| +16296C>C/T | 1 | |||||||
| BM donor 6 | ||||||||
| “Aggregate” sequence of total BM cells | 102 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 13 | 11.3 | 6 | 5.2 | ||||
| +8CT6C*, 9CT6C* | 5 | |||||||
| +200A>G/A | 3 | |||||||
| +200A>G | 2 | |||||||
| +200A>G/A, 7CT6C*, 8CT6C* | 1 | |||||||
| +200A>G/A, 8CT6C*, 9CT6C* | 1 | |||||||
| +7CT6C*, 8CT6C* | 1 | |||||||
| CB donor 1 | ||||||||
| “Aggregate” sequence of total CB cells | 118 | 0 | 0 | 0 | 0 | |||
| 73A>G, 153A>G, 183A>G, 263A>G, 7CT6C*, 8CT6C*, 325C>T, 463C>T, 485T>C, 489T>C, 514-515delCA, 16198T>C, 16223C>T, 16268C>T, 16354C>T, 16381T>A, 16519T>C | 1 | 1 | 0.8 | 1 | 0.8 | |||
| CB donor 2 | ||||||||
| “Aggregate” sequence of total CB cells | 120 | 0 | 0 | 0 | 0 | |||
| CB donor 3 | ||||||||
| “Aggregate” sequence of total CB cells | 100 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 4 | 3.8 | 2 | 1.9 | ||||
| +316T>G/C | 3 | |||||||
| +305C>A, 306C>A, 307C>A | 1 | |||||||
| CB donor 4 | ||||||||
| “Aggregate” sequence of total CB cells | 118 | 0 | 0 | 0 | 0 | |||
| +6CT6C*, 16022T>C | 1 | 1 | 0.8 | 1 | 0.8 | |||
| CB donor 5 | ||||||||
| “Aggregate” sequence of total CB cells | 115 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 3 | 2.5 | 3 | 2.5 | ||||
| +16184C>CCC (12C)* | 1 | |||||||
| +16189T>C* | 1 | |||||||
| +16184C>CCCC (13C)* | 1 | |||||||
. | . | Heterogeneity . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|
| . | . | Different . | . | Unique . | . | |||
| Donor and mtDNA sequence . | Clone no. . | No. . | % . | No. . | % . | |||
| BM donor 1 | ||||||||
| “Aggregate” sequence of total BM cells | 85 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 30 | 26.1 | 8 | 7.0 | ||||
| +8CT6C*, 9CT6C* | 22 | |||||||
| +9CT6C* | 2 | |||||||
| +7CT6C* | 1 | |||||||
| +189A>G/A | 1 | |||||||
| +204T>C | 1 | |||||||
| +277C>T | 1 | |||||||
| +514insCA | 1 | |||||||
| +16114C>T | 1 | |||||||
| BM donor 2 | ||||||||
| “Aggregate” sequence of total BM cells | 27 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 9 | 25.0 | 5 | 13.9 | ||||
| +16184C>CC (11C)* | 5 | |||||||
| +16131T>C/T | 1 | |||||||
| +16145G>A | 1 | |||||||
| +16184C>CCCC (13C)* | 1 | |||||||
| 73A>G, 263A>G, 191A>AA, 194C>T, 199T>C, 207G>A, 8CT6C*, 489T>C, 16147C>T, 16173C>T, 16245C>T, 16362T>C | 1 | |||||||
| BM donor 3 | ||||||||
| “Aggregate” sequence of total BM cells | 96 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 24 | 20.0 | 7 | 5.8 | ||||
| +9CT6C*, 10CT6C* | 11 | |||||||
| +9CT6C* | 6 | |||||||
| +8CT6C* | 2 | |||||||
| +182C>T/C, 8CT6C*, 9CT6C* | 2 | |||||||
| +71delG, 9CT6C*, 10CT6C* | 1 | |||||||
| +279T>C/T | 1 | |||||||
| +16153G>A | 1 | |||||||
| BM donor 4 | ||||||||
| “Aggregate” sequence of total BM cells | 92 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 19 | 17.1 | 6 | 5.4 | ||||
| +8CT6C*, 9CT6C* | 11 | |||||||
| +514-515delCA, 514insCA† | 3 | |||||||
| +7CT6C*, 8CT6C* | 2 | |||||||
| +9CT6C*, 10CT6C* | 1 | |||||||
| +89T>C | 1 | |||||||
| +8CT6C*, 9CT6C*, 16093T‡ | 1 | |||||||
| BM donor 5 | ||||||||
| “Aggregate” sequence of total BM cells | 57 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 57 | 50.0 | 14 | 12.3 | ||||
| +514insCA‡ | 19 | |||||||
| +264C>T | 18 | |||||||
| +514-515delCA, 514insCA† | 5 | |||||||
| +264C>T/C | 3 | |||||||
| +7CT6C*, 8CT6C* | 2 | |||||||
| +146T>C, 514insCA‡ | 2 | |||||||
| +146T>C, 264C>T/C | 1 | |||||||
| +146T>C/T, 514insCA‡ | 1 | |||||||
| +146T>C | 1 | |||||||
| +189A>G | 1 | |||||||
| +264C>T/C, 514insCA‡ | 1 | |||||||
| +161T>C/T, 264C>T, 514insCA‡ | 1 | |||||||
| +16189T>C* | 1 | |||||||
| +16296C>C/T | 1 | |||||||
| BM donor 6 | ||||||||
| “Aggregate” sequence of total BM cells | 102 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 13 | 11.3 | 6 | 5.2 | ||||
| +8CT6C*, 9CT6C* | 5 | |||||||
| +200A>G/A | 3 | |||||||
| +200A>G | 2 | |||||||
| +200A>G/A, 7CT6C*, 8CT6C* | 1 | |||||||
| +200A>G/A, 8CT6C*, 9CT6C* | 1 | |||||||
| +7CT6C*, 8CT6C* | 1 | |||||||
| CB donor 1 | ||||||||
| “Aggregate” sequence of total CB cells | 118 | 0 | 0 | 0 | 0 | |||
| 73A>G, 153A>G, 183A>G, 263A>G, 7CT6C*, 8CT6C*, 325C>T, 463C>T, 485T>C, 489T>C, 514-515delCA, 16198T>C, 16223C>T, 16268C>T, 16354C>T, 16381T>A, 16519T>C | 1 | 1 | 0.8 | 1 | 0.8 | |||
| CB donor 2 | ||||||||
| “Aggregate” sequence of total CB cells | 120 | 0 | 0 | 0 | 0 | |||
| CB donor 3 | ||||||||
| “Aggregate” sequence of total CB cells | 100 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 4 | 3.8 | 2 | 1.9 | ||||
| +316T>G/C | 3 | |||||||
| +305C>A, 306C>A, 307C>A | 1 | |||||||
| CB donor 4 | ||||||||
| “Aggregate” sequence of total CB cells | 118 | 0 | 0 | 0 | 0 | |||
| +6CT6C*, 16022T>C | 1 | 1 | 0.8 | 1 | 0.8 | |||
| CB donor 5 | ||||||||
| “Aggregate” sequence of total CB cells | 115 | 0 | 0 | 0 | 0 | |||
| Nonaggregate sequences | 3 | 2.5 | 3 | 2.5 | ||||
| +16184C>CCC (12C)* | 1 | |||||||
| +16189T>C* | 1 | |||||||
| +16184C>CCCC (13C)* | 1 | |||||||
Different indicates different from aggregate cell sequence; unique, uniquely different heterogeneity; +, mtDNA nucleotide changes in comparison with aggregate cell mtDNA sequence.
Homopolymeric C tracts at nucleotides position at 303 to 315 (for example, 8CT6C defined CCCCCCCCTCCCCCC) and 16184 to 16193 in HV2 and HV1, respectively.
Mixed pattern of 514-515delCA and 514insCA.
The same as the Cambridge Reference Sequence but different from the aggregate sequence.
mtDNA heterogeneity in individual CD34+ clones. (A) Sequence chromatogram with mixed nucleotide signal (G/A) at nucleotide position 200 in HV2 from 1 CD34+ clone of BM donor no. 6 (left), and then was clearly divided into wild type (center) and mutation (200A>G/A) (right) after TA cloning. (B) Poly C length heteroplasmy between nucleotide position 303 and 315 from one of the CD34+ clones of BM donor no. 3 (left). Reverse-sequence analysis after TA cloning revealed poly C length heteroplasmy of 8CT6C (center) and 9CT6C (right).
mtDNA heterogeneity in individual CD34+ clones. (A) Sequence chromatogram with mixed nucleotide signal (G/A) at nucleotide position 200 in HV2 from 1 CD34+ clone of BM donor no. 6 (left), and then was clearly divided into wild type (center) and mutation (200A>G/A) (right) after TA cloning. (B) Poly C length heteroplasmy between nucleotide position 303 and 315 from one of the CD34+ clones of BM donor no. 3 (left). Reverse-sequence analysis after TA cloning revealed poly C length heteroplasmy of 8CT6C (center) and 9CT6C (right).
mtDNA heterogeneity characteristics of CD34+ clones from adult BM and CB
. | Adult BM CD34+ . | CB CD34+ . |
|---|---|---|
| Plating efficiency, % | 30.0 | 78.6* |
| Aggregate cell genotype | Uniform pattern | Frequent mixed nucleotide signal† |
| Heterogeneity of CD34+ clones, % | ||
| Total rate | 24.9* | 1.6 |
| Unique pattern | 7.5* | 1.2 |
| Substitution (no.) | 10.5 (64 of 611)* | 0.0 (0 of 580) |
| Poly C tract (no.)† | 11.9 (73 of 611)* | 1.2 (7 of 580) |
| Length heteroplasmy at 303 to 315 | 10.8 (66 of 611)* | 0.0 (0 of 580) |
| Length heteroplasmy at 16184 to 16193 | 1.1 (7 of 611) | 0.5 (3 of 580) |
| Nucleotide change | 0.0 (0 of 611) | 0.7 (4 of 580) |
| Substitution plus poly C tract and others (no.) | 2.5 (15 of 611) | 0.4 (2 of 580) |
. | Adult BM CD34+ . | CB CD34+ . |
|---|---|---|
| Plating efficiency, % | 30.0 | 78.6* |
| Aggregate cell genotype | Uniform pattern | Frequent mixed nucleotide signal† |
| Heterogeneity of CD34+ clones, % | ||
| Total rate | 24.9* | 1.6 |
| Unique pattern | 7.5* | 1.2 |
| Substitution (no.) | 10.5 (64 of 611)* | 0.0 (0 of 580) |
| Poly C tract (no.)† | 11.9 (73 of 611)* | 1.2 (7 of 580) |
| Length heteroplasmy at 303 to 315 | 10.8 (66 of 611)* | 0.0 (0 of 580) |
| Length heteroplasmy at 16184 to 16193 | 1.1 (7 of 611) | 0.5 (3 of 580) |
| Nucleotide change | 0.0 (0 of 611) | 0.7 (4 of 580) |
| Substitution plus poly C tract and others (no.) | 2.5 (15 of 611) | 0.4 (2 of 580) |
Data are expressed as mutations per total assayed CD34+ clones.
Statistically significant difference (P < .05) between 2 groups.
Homopolymeric C tracts at nucleotides position at 303 to 315 and 16184 to 16193 in HV2 and HV1, respectively.
Distribution of mtDNA heterogeneity from adult BM CD34+ clones according to each grade and culture media
. | Grade . | . | . | . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | 1 . | . | 2 . | . | 3 . | . | 4 . | . | Subtotal no. . | . | . | |||||||
| Donor . | + . | - . | + . | - . | + . | - . | + . | - . | + . | - . | Total no. . | |||||||
| 1 | 3 | 3 | 1 | 1 | 5 | 7 | 10 | 0 | 19 | 11 | 30 | |||||||
| 2 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | |||||||
| 3 | 2 | 3 | 3 | 3 | 3 | 4 | 5 | 1 | 13 | 11 | 24 | |||||||
| 4 | 1 | 1 | 1 | 3 | 4 | 2 | 0 | 4 | 6 | 10 | 16 | |||||||
| 5 | 6 | 6 | 6 | 6 | 10 | 7 | 10 | 1 | 32 | 20 | 52 | |||||||
| 6 | 0 | 1 | 3 | 5 | 2 | 0 | 1 | 1 | 6 | 7 | 13 | |||||||
| Subtotal no. | 14 | 14 | 15 | 18 | 24 | 20 | 26 | 7 | 79 | 59 | 138 | |||||||
| Assayed clone no. | 81 | 77 | 68 | 71 | 77 | 75 | 121 | 41 | 347 | 264 | 611 | |||||||
| Proportion, %* | 17.3 | 18.2 | 22.1 | 25.4 | 31.2 | 26.7 | 21.5 | 17.1 | 22.8 | 22.3 | 22.6 | |||||||
. | Grade . | . | . | . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | 1 . | . | 2 . | . | 3 . | . | 4 . | . | Subtotal no. . | . | . | |||||||
| Donor . | + . | - . | + . | - . | + . | - . | + . | - . | + . | - . | Total no. . | |||||||
| 1 | 3 | 3 | 1 | 1 | 5 | 7 | 10 | 0 | 19 | 11 | 30 | |||||||
| 2 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | |||||||
| 3 | 2 | 3 | 3 | 3 | 3 | 4 | 5 | 1 | 13 | 11 | 24 | |||||||
| 4 | 1 | 1 | 1 | 3 | 4 | 2 | 0 | 4 | 6 | 10 | 16 | |||||||
| 5 | 6 | 6 | 6 | 6 | 10 | 7 | 10 | 1 | 32 | 20 | 52 | |||||||
| 6 | 0 | 1 | 3 | 5 | 2 | 0 | 1 | 1 | 6 | 7 | 13 | |||||||
| Subtotal no. | 14 | 14 | 15 | 18 | 24 | 20 | 26 | 7 | 79 | 59 | 138 | |||||||
| Assayed clone no. | 81 | 77 | 68 | 71 | 77 | 75 | 121 | 41 | 347 | 264 | 611 | |||||||
| Proportion, %* | 17.3 | 18.2 | 22.1 | 25.4 | 31.2 | 26.7 | 21.5 | 17.1 | 22.8 | 22.3 | 22.6 | |||||||
No significant statistical differences were found between culture media with and without G-CSF and each grade. Abbreviations (+, -) are explained in the footnote to Table 1.
Only 9 clones of 580 CD34+ clones (1.6%, 1.6% ± 1.5%) from the 5 CB donors showed mtDNA heterogeneity distinct from the sequences of the donor's corresponding aggregate mtDNA as well as from other CD34+ clones (Table 5). The mean proportion of unique mtDNA heterogeneity pattern among single CD34+ clones from CB was 1.2% (1.2% ± 1.0%). As noted in a single CD34+ clone from BM donor no. 2, the mtDNA sequence of a single CD34+ clone from CB donor no. 1 showed an extremely distinct pattern as compared with the aggregate CB mtDNA and other CD34+ clones (Table 4).
Characteristics of CD34+ clones derived from adult BM and CB
Genetic changes in normal marrow and cord blood samples are summarized in Table 5. Most striking, about a quarter of the more than 600 CD34+ cell clones of adult bone marrow differed from the aggregate mtDNA sequence of each specific donor while, in contrast, under 2% of the almost 600 cord blood CD34+ cell clones were different from their respective individual aggregate sequence. The differences for unique clones (different from aggregate sequence and other CD34+ cell clones) were also marked. No solitary nucleotide substitutions were observed in CB mtDNA CD34+ clones.
Hotspots of the mtDNA control region in CD34+ clones from adult BM
As anticipated, a high incidence of nucleotide variations was observed in both HV2 (110 of 152 total genetic changes observed, or 72%) and HV1 (12 of 152 total genetic changes, or 7.9%) segments; most mutations were localized in the HV2 homopolymeric C tracts between nucleotides 303 and 315 (43%, or 66 of 152) (Figure 2B).
Discussion
As we previously observed, a wide range of polymorphisms and mutations not previously described in accepted databases were noted among healthy individuals in the aggregate genotype (the sequence obtained from total BM cells), even in this relatively short region of mtDNA. When more than 600 CD34+ cell clones from 6 adult BMs were analyzed, we found marked sequence heterogeneity in all donors. In comparison with each person's aggregate BM mtDNA sequence, individual CD34+ cells showed sequence differences in 11% to 50% of the CD34+ cell clones; sequence that was uniquely different from both the aggregate sequence and the other cell clones was present in 5% to 14% of the CD34+ cells. Approximately half the observed changes in individual clones' mtDNA sequence occurred in a homopolymeric C tract (nucleotides 303 to 315), and the remaining were point substitutions outside the poly C region. Low levels of heteroplasmy (where the minor species is less than 20% that of the major heteroplasmic species) may be technically difficult to detect; nevertheless, we saw evidence of heteroplasmy in sequence variants among individual adult CD34+ cells. Neither the combination of growth factors utilized during the brief period of in vitro culture nor the colony size correlated with the proportion of CD34+ clones with variant mtDNA, suggesting that our observed results were not secondary to the proliferation of cells during the few days of tissue culture.
The marked degree of sequence heterogeneity among individual CD34+ cell clones was unexpected, because sequence variations in mtDNA have not been thought to be sustainable in dividing tissues. Because somatic mutations in general have been correlated with aging of the organism, we next undertook similar experiments utilizing CD34+ cells derived from CB. CB hematopoietic progenitor cells have a higher proliferative capacity compared with adult CD34+ cells, as reflected in both better plating efficiency and large colony size in tissue culture. Sequencing 580 clones from 5 samples of CB from different donors (Table 2) indicated that only an average of 1.6% of the CD34+ clones had an mtDNA sequence divergent from that of the aggregate CB (Tables 4 and 5).
The mtDNA control region contains many regulatory elements, some of which are important in the initiation of transcription and replication of mtDNA; control region mutants might alter the ratio of proteins or transcripts derived from mtDNA to relevant nuclear DNA gene products, potentially affecting the efficiency of mitochondrial oxidative phosphorylation.16 Control region mtDNA mutations and variants therefore have been hypothesized to impact longevity, climatic adaptation, and some diseases such as type II diabetes16,17 and human tumors.18 One possible speculation based on our results is that age-related control region mutations in CD34+ clones could lead to derangement of marrow function in older individuals. Empirically, coding regions might be experimentally correlated with respiratory function in marrow cell populations. However, cells appear to have remarkable tolerance for decreased mitochondrial activity,19 and the likely presence of mutations elsewhere in the mtDNA genome might also affect function. Indeed, functional measurements of oxidative phosphorylation in myelodysplastic bone marrow samples have suggested abnormal function.20 Nevertheless, a likely alternative possibility is that accumulated mtDNA mutations, while advantageous to the DNA species by, for example, favoring replicative efficiency, are neutral as related to overall cell or organ function.
Our data suggest age-related accumulation of somatic mtDNA mutations in normal human hematopoietic tissue. As in other organs, it appears probable that reactive oxygen species, generated in the mitochondria, lead to oxidative damage of the mtDNA.9 Mutated mtDNA copies almost always would be underrepresented in aggregate samples due to their small number and only detectable by analysis of individual cells undergoing clonal expansion—in our experiments after in vitro cell culture but in vivo after malignant transformation. Our data are consistent with some recent publications; for example, acquisition of a specific, easily detected substitution that leads to a well-described polymorphism (C150T) was far more common in the leukocytes of very aged individuals and showed an apparent tendency to increase with age.21 Single cell analysis of human cardiomyocytes and buccal epithelial cells also revealed a remarkably high frequency of homoplasmic expansion of mtDNA point mutations and deletions in persons over 50 years of age.22 Multiple mtDNA genotypes were present in closely proximate hair follicles; while individual cells were not analyzed, the interpretation of the investigators was that mtDNAmutations occur with aging and resolve to homoplasmy within cells.23,24 Primary tissue and cell lines of colon tumor origin also show a high frequency of mtDNA mutations, presumably due to fixation of established homoplasmic nucleotide changes.25 More generally, our results are concordant with observations of surprisingly rapid divergence and resolution to homoplasmy of mtDNA sequence among inbred Holstein cows26 and in carefully studied human kindreds.27,28
The molecular mechanisms responsible for expansion of mutant mtDNA molecules to homoplasmy within a cell are obscure. mtDNA properties of polyploidy and relaxed replication and the need to regulate intracellular mtDNA copy number are believed to contribute to random genetic drift13 and ultimately also to the clonal expansion of mtDNA forms over extended periods of time.29 This physiology has been more amenable to computer simulation than to empirical experimentation.7 Also unclear is the regulation of replicative fidelity in the homopolymeric C tract, in which we localized much cell-to-cell heterogeneity and which has been described by others as a mutational “hotspot”.30,31 The relative proportion of variable length poly C tracts appears to be actively maintained during cell division despite evidence of random mtDNA segregation, suggesting de novo regeneration of specific pattern following cell division by an as yet unknown molecular mechanism.32 Poly C and other hypervariable region variants have been associated with several common human diseases such as diabetes mellitus,17 low birth weight,33 and dilated cardiomyopathy.34 The pathophysiology of mtDNA disease is confusing, and alterations in mtDNA may have tissue-specific consequences. For BM, accumulation of mtDNA mutations may relate to normal aging of the hematopoietic compartment, and specific mtDNA lesions may affect the phenotype of an expanded population of abnormal cells in clonal hematologic diseases such as leukemia or myelodysplasia.
Heterogeneity of mtDNA sequence in normal adult CD34+ cells may have several implications. The possible relationship of mtDNA sequence changes to hematopoietic aging has been discussed above. For blood samples and, if our results apply to other tissues, in general caution may be needed in forensic identifications and anthropologic studies that depend primarily on the considerable sequence variation found in the 2 hypervariable segments (HV1 and HV2). For the study of hematopoiesis, mtDNAvariant sequences may provide a natural genetic “marker” for the determination of the contribution of individual stem cells to blood cell production. Fixed mtDNA changes in leukemic cells might provide a simple and nearly universally applicable method of monitoring minimal residual disease. Finally, mtDNAsequence variability might be adapted to the measurement of the mammalian mtDNA mutation rate. CD34+ cells in blood and marrow have virtually identical properties, so blood sampling rather than marrow aspiration could be utilized to determine the mutational history of an individual, following exposure to endogenous reactive oxygen species or environmental mutagens. Because mitochondria are believed to have derived from bacteria, mtDNA mutations in individual cells may allow development of an “intracellular” in vivo Ames test for mutations.
Prepublished online as Blood First Edition Paper, September 22, 2003; DOI 10.1182/blood-2003-05-1724.
M.G.S. and S.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This paper is a contribution of the US National Institutes of Health (NIH) and the National Institute of Standards and Technology (NIST) and is not subject to copyright. Certain commercial equipment, instruments, materials, or companies are identified in this paper to specify the experimental procedure. Such identification does not imply recommendation or endorsement by NIH and NIST, nor does it imply that the materials or equipment identified are the best available for this purpose.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal