Abstract
Hypoxia-inducible factor 1 (HIF-1) regulates many genes induced by low oxygen conditions. The expression of important hypoxic genes such as glucose transporter 1 and vascular endothelial growth factor are increased in macrophages during wound healing and in the presence of the endotoxin, lipopolysaccharide (LPS). Recent studies have demonstrated that nonhypoxic stimuli can also activate HIF-1 in a cell-specific manner. Here, we demonstrate that in macrophages, LPS can control the activation of hypoxia-regulated genes through the HIF-1 pathway. We show that in these cells, protein expression levels of HIF-1α are strongly increased to levels comparable to hypoxic induction. HIF-1α mRNA levels are markedly increased following LPS stimulation, suggesting a transcriptional induction. In functional studies, the LPS-induced HIF-1 complex could specifically bind to the HIF-1 DNA-binding motif. Additionally, when cells were transfected with an HIF-1-specific reporter construct, LPS could strongly activate the expression of the reporter to levels that surpassed those observed after hypoxic induction. This induction was blocked by the cotransfection of a dominant-negative form of HIF-1α. These results indicate that the HIF-1 complex is involved in macrophage gene activation following LPS exposure and identify a novel pathway that could play a determinant role during inflammation and wound healing. (Blood. 2004;103:1124-1130)
Introduction
Hypoxia activates a number of genes that are important in the cellular and tissue adaptation to low oxygen conditions.1 These genes include erythropoietin, glucose transporters, glycolytic enzymes, and vascular endothelial growth factor (VEGF). The hypoxic expression of these different genes is controlled at the transcriptional level by the ubiquitously expressed transcription factor, hypoxia-inducible factor 1 (HIF-1). HIF-1 is a member of the basic helix-loop-helix superfamily of transcription factors. Only active as a heterodimer, HIF-1 is composed of 2 subunits: HIF-1α and HIF-1β.2 Whereas the HIF-1β protein is readily found in all cells, HIF-1α is virtually undetectable in normal oxygen conditions. When cells are subjected to hypoxic conditions (1% oxygen), levels of the HIF-1α subunit are rapidly increased. Instead of acting on HIF-1α transcription or translation, hypoxia increases HIF-1α protein levels by inhibiting the rapid ubiquitination and degradation of HIF-1α by the proteasome. An elegant series of studies has shown that when oxygen is present, HIF-1α is modified by prolyl hydroxylation, which permits the binding of the von Hippel-Lindau protein (pVHL), a recognition component of the E3 ligase complex. This promotes the ubiquitin degradation of HIF-1α.3-8 In a very recent study, acetylation of HIF-1α in normoxic conditions also targets HIF-1α for proteasomal degradation.9 Under hypoxic conditions, prolyl hydroxylation of HIF-1α is blocked and acetylation is down-regulated, which permits HIF-1α protein stabilization. HIF-1α is then free to bind with HIF-1β to form the HIF-1 transcription complex. The heterodimer can then bind to hypoxic response elements (HREs) in the above-mentioned genes and increase their expression.
We and others have shown that a number of nonhypoxic stimuli can strongly increase the HIF-1 complex in normal oxygen conditions and modulate the transcription of hypoxic genes.10-19 Interestingly, treatment of macrophages with lipopolysaccharide (LPS) can increase the expression of a number of hypoxic genes.20-22 The mechanisms involved in the induction of these different genes by LPS have not all been clearly defined. In this study, we investigated whether the activation of LPS could increase HIF-1 and modulate hypoxic gene activation. We show here that LPS increases HIF-1α expression in a time- and dose-dependent manner. LPS induces an active form of HIF-1 because the complex can bind HIF-1-specific DNA sequences and strongly activate an HIF-1-specific reporter under these conditions. These results identify a novel pathway by which LPS can activate gene expression in macrophages.
Materials and methods
Chemicals and reagents
Actinomycin D, angiotensin II (Ang II), LPS, and 12, 13-phorbol myristate acetate (PMA) were from Sigma (St Louis, MO). GF109203X (bisindolylmaleimide I) and Ly294002 were from Calbiochem (San Diego, CA). All media for cell culture was from Invitrogen Life Technologies (Carlsbad, CA). Anti-HIF-1α antiserum 2087 was raised in rabbits immunized against the last 20 amino acids of the C termini of human HIF-1α.23 Monoclonal anti-HIF-1α antibody and polyclonal anti-HIF-1β antibody were from Novus Biologicals (Littleton, CO). Monoclonal antiphospho-p44/p42 mitogen-activated protein kinase (MAPK) antibody was from Sigma. The phospho-specific p70S6 kinase antibody was from Cell Signaling Technology (Beverly, MA). Horseradish peroxidase (HRP)-coupled antimouse and antirabbit antibodies were from Promega (Madison, WI). The PRE-tk-LUC reporter24 was a kind gift from Dr Steven McKnight (University of Texas).
Cell culture
Primary bone marrow-derived macrophages, kindly provided by Dr Kim Boulukos (INSERM, Université de Nice), were isolated from the femurs of 2- to 3-month-old male C57BL/6 mice and cultured as previously described.25 Murine macrophage-derived cell line RAW 264.7 gamma NO(-) and rat alveolar macrophage-derived cell line NR838326 were from American Type Culture Collection (Manassas, VA). The 2 cell lines were cultured in F-12K medium containing 15% inactivated fetal bovine serum (FBS), penicillin (50 U/mL), and streptomycin (50 μg/mL) in a humid atmosphere (5% CO2, 95% air) and passaged twice a week. Quiescent cells were obtained by total deprivation of FBS for 16 to 20 hours. Pretreatment of cells with drugs was performed 15 minutes prior to stimulation. Hypoxic conditions were obtained by placing the cells in a sealed hypoxic workstation (Ruskinn Technologies, Leeds, United Kingdom). The oxygen level in this workstation was maintained at 1% with the residual gas mixture containing 94% nitrogen and 5% carbon dioxide.
Western blot analysis
Cells were lysed in 2 × Laemmli sample buffer. Protein concentration was determined with the use of the Lowry assay. Whole cell extracts (25 μg) were resolved in sodium dodecyl sulfate-polyacrylamide gels (8%) and electrophoretically transferred onto a polyvinylidene difluoride membrane (Immobilon-P, Millipore, Billerica, MA). Proteins of interest were revealed with specific antibodies as indicated (1:1000 dilution). The bands were visualized with an enhanced chemiluminescence (ECL) system (Amersham Biosciences, Buckinghamshire, United Kingdom).
Northern blot analysis
Confluent NR8383 cells were lysed, and RNA was isolated with TRIzol reagent (Invitrogen Life Technologies). RNA resolved on 1% agarose/6% formaldehyde gels was transferred to Hybond N+ nylon membrane (Amersham Biosciences) and was hybridized with a radioactive cDNA probe comprising either the total coding sequence of the mouse VEGF gene, the rat plasminogen activator inhibitor 1 (PAI-1) and inducible nitric oxide synthase (iNOS) genes, the human glucose transporter (GLUT1) gene, or the first 900-base pair coding sequence of the human HIF-1α gene. An oligonucleotide probe against 18S rRNA was used as a loading control.
Transcription factor enzyme-linked immunoassay
High-bind NeutrAvidin-coated 96-well strip plates (Pierce Biotechnology, Rockford, IL) were incubated with 33 nM of a 5′-biotinylated 26-base pair dsDNA oligonucleotide sequence for 1 hour at room temperature. This sequence contains the previously described wild-type or mutant (bold underlined) HIF-1 binding motif.27 The sequences used here were: 5′-GATCGCCCTACGTGCTGTCTCAGATC-3′ for wild-type sequence W26 and 5′-GATCGCCCTAAAAGCTGTCTCAGATC-3′ for mutant sequence M26. Preparation of nuclear extracts was performed as described previously.27 DNA-binding reactions were carried out in a total volume of 50 μL containing 10 μg nuclear protein extract in a buffer containing 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.9), 50 mM NaCl, 5% glycerol, 1 mM EDTA (ethylenediaminetetraacetic acid), 1 mM dithiothreitol (DTT), and 3% nonfat milk for 1 hour at room temperature. For competition experiments, increasing concentrations of either the wild-type or mutant dsDNA oligonucleotide was added to binding buffer prior to the addition of protein extracts. Specific antibodies were then added at a 1:1000 concentration in phosphate-buffered saline (PBS) containing 3% nonfat milk for 1 hour at room temperature, followed by the addition of the corresponding HRP-coupled secondary antibody. Between each addition, wells were extensively washed in PBS containing 0.1% Tween-20. HRP activity was then detected by the addition of 100 μL TMB-One solution (Promega). After a 10-minute incubation period, the reaction was arrested by the addition of 0.5 M H2SO4. Color intensity was detected at 450 nM using a UVmax microplate reader (Molecular Devices, Sunnyvale, CA). HRP activity was normalized to control values (ie, nonstimulated cells). All experiments are an average ± SD of triplicate data representative of at least 3 independent experiments.
Transient transfection and luciferase assays
NR8383 cells were plated in 6-well plates at 5 × 105 cells/mL. The next day, transient transfections were performed with the use of 1 μg/well of PRE-tk-LUC reporter along with 500 ng/well cytomegalovirus Renilla reniformis luciferase expression vector (Promega) as a control for transfection efficiency. Transfection was performed using the Superfect transfection reagent (Qiagen, Valencia, CA) at a 1:5 DNA/reagent ratio. For dominant-negative studies, cells were transfected with 4 μg pcDNA3-HA-DN-HIF-1α.10 At 3 hours after transfection, cells were washed, and fresh medium was added. At 12 hours after transfection, cells were deprived of FBS for 16 hours. Stimulation with LPS and hypoxia was performed for 18 hours. Cells were then washed with cold PBS, and luciferase assays were performed with the Dual-Luciferase Reporter Assay System (Promega). Results were quantified with a Topcount NXT luminescence counter (Perkinelmer, Boston, MA). Results are expressed as a ratio of beetle luciferase activity over Renilla reniformis luciferase activity. All experiments are an average of at least 3 independent experiments performed in triplicate.
Results
Expression of hypoxic genes in response to LPS
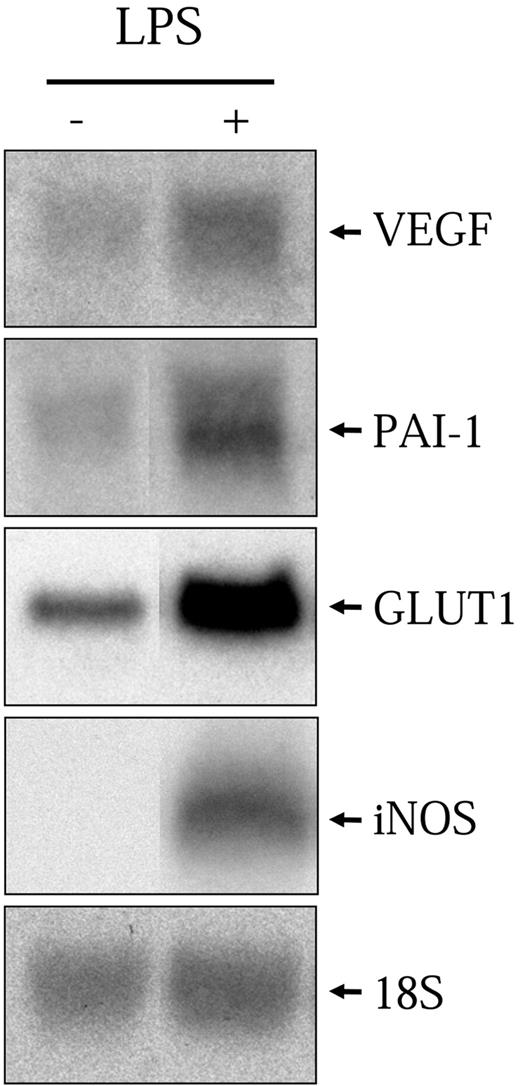
As mentioned previously, it has been shown that LPS can activate a number of hypoxic genes in various macrophage cell models. Here, we use a rat alveolar macrophage-derived cell line, NR8383, which was previously shown to recapitulate the properties of primary alveolar macrophages.26 NR8383 cells were stimulated with LPS (1 μg/mL) for 6 hours followed by total RNA extraction and Northern blot analysis with specific probes toward VEGF, PAI-1, iNOS, and GLUT1. As can be seen in Figure 1, LPS significantly increased the levels of all 4 transcripts. These results demonstrate that NR8383 cells can be stimulated by LPS to induce the transcription of known hypoxia-responsive genes.
LPS increases hypoxic gene expression. Quiescent NR8383 cells were maintained under control conditions or in the presence of LPS (1 μg/mL) for 6 hours. Total RNA was extracted from cells and resolved on formaldehyde/agarose gels. Northern blot was performed using a specific radiolabeled cDNA probe for VEGF, PAI-1, GLUT1, and iNOS. An 18S rRNA probe was used as a control for gel loading.
LPS increases hypoxic gene expression. Quiescent NR8383 cells were maintained under control conditions or in the presence of LPS (1 μg/mL) for 6 hours. Total RNA was extracted from cells and resolved on formaldehyde/agarose gels. Northern blot was performed using a specific radiolabeled cDNA probe for VEGF, PAI-1, GLUT1, and iNOS. An 18S rRNA probe was used as a control for gel loading.
Induction of HIF-1α protein in response to LPS
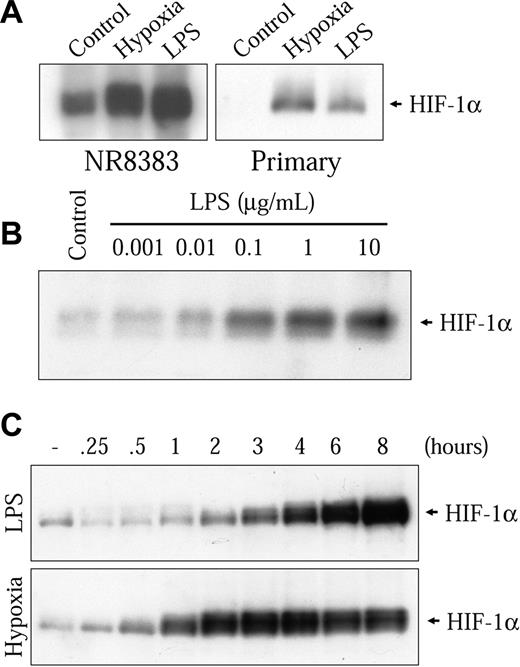
The genes mentioned all contain HREs that can be activated by HIF-1 in low oxygen conditions.28-31 Because different nonhypoxic cell stimulations have been shown to strongly increase HIF-1α, the hypoxia-sensitive subunit of the HIF-1 complex, we decided to examine the possibility that LPS could increase HIF-1α in macrophages. Therefore, we stimulated NR8383 cells in the presence of 1 μg/mL LPS for 6 hours followed by the evaluation of HIF-1α protein levels by Western blot with a specific antibody. In these conditions, we observed a strong increase in HIF-1α protein levels (Figure 2A left panel). This increase was comparable to the one seen in hypoxic conditions. This finding could also be seen in a mouse primary bone marrow-derived macrophage preparation (Figure 2A right panel) and the murine macrophage-derived cell model, RAW 264.7 gamma NO(-) cells (results not shown). The induction of HIF-1α by LPS was dose-dependent; an increase could be seen at 10 ng/mL, whereas maximal induction was attained at 1 μg/mL (Figure 2B). Time-course studies were then performed on NR8383 cells stimulated with 1 μg/mL LPS. As shown in the upper panel of Figure 2C, an increase can be seen after a 2-hour stimulation and the maximal induction was attained between 6 and 8 hours in the presence of LPS. After 8 hours in the presence of LPS, a decrease in HIF-1α levels was observed (results not shown). Taken together, these results demonstrate that the stimulation of macrophages with LPS increases the inducible subunit of the HIF-1 transcription complex.
LPS increases HIF-1α protein expression. (A) Primary mouse bone marrow-derived macrophages or NR8383 cells were rendered quiescent by FBS deprivation for 16 hours. Cells were then maintained either under control conditions (20.8% oxygen) or under hypoxic conditions (1% oxygen) or in the presence of LPS (1 μg/mL) for 6 hours. (B) Quiescent NR8383 cells were maintained under control conditions or in the presence of different concentrations of LPS for 6 hours. (C) Quiescent NR8383 cells were maintained under control conditions, in the presence of LPS (1 μg/mL) or under hypoxic conditions for different periods of time of up to 8 hours. Total cell extracts (25 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 8% gel) and immunoblotted using an anti-HIF-1α antiserum or an antiphospho-p44/p42 MAPK monoclonal antibody.
LPS increases HIF-1α protein expression. (A) Primary mouse bone marrow-derived macrophages or NR8383 cells were rendered quiescent by FBS deprivation for 16 hours. Cells were then maintained either under control conditions (20.8% oxygen) or under hypoxic conditions (1% oxygen) or in the presence of LPS (1 μg/mL) for 6 hours. (B) Quiescent NR8383 cells were maintained under control conditions or in the presence of different concentrations of LPS for 6 hours. (C) Quiescent NR8383 cells were maintained under control conditions, in the presence of LPS (1 μg/mL) or under hypoxic conditions for different periods of time of up to 8 hours. Total cell extracts (25 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 8% gel) and immunoblotted using an anti-HIF-1α antiserum or an antiphospho-p44/p42 MAPK monoclonal antibody.
LPS induces HIF-1α through mechanisms that are distinct from hypoxic induction
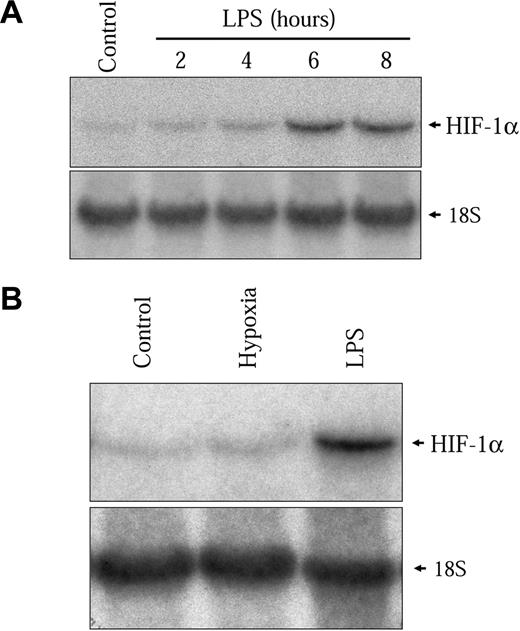
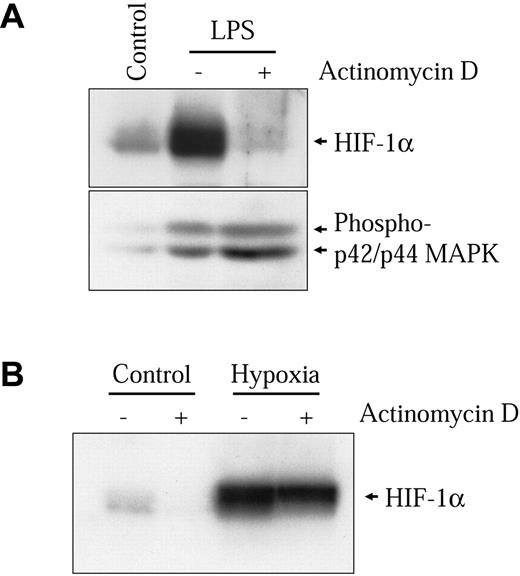
The kinetics of HIF-1α protein induction shown in Figure 2C closely follows the induction of the HIF-1 target genes VEGF, PAI-1, iNOS, and GLUT1 (results not shown). However, this induction is slow as compared to the hypoxic induction of HIF-1α in these same cells. In hypoxic conditions, HIF-1α induction in NR8383 cells commenced after 30 minutes and had already attained a maximum between 2 and 3 hours under hypoxia (Figure 2C lower panel). This indicates that different mechanisms are implicated in the activation of HIF-1α by hypoxia and by LPS in these cells. We have previously shown that increased transcription of the HIF-1α gene was important for HIF-1α protein induction by Ang II in vascular smooth muscle cells (VSMCs).32 We wanted to evaluate whether this mechanism could be implicated in the induction of HIF-1α by LPS in macrophages. Therefore, we evaluated whether LPS could modify the levels of HIF-1α mRNA in NR8383 cells. As seen in Figure 3A, LPS increased HIF-1α mRNA levels in these cells. The time frame of this induction closely resembles the induction of HIF-1α protein, with a maximum attained at 6 hours. Hypoxia did not increase the expression of HIF-1α mRNA in NR8383 cells (Figure 3B), as was the case in other cell models.32 To clearly demonstrate that increased mRNA levels were important for HIF-1α protein induction, NR8383 cells were pretreated with a potent transcriptional inhibitor, actinomycin D, prior to stimulation with LPS for 6 hours. Under these conditions, the induction of HIF-1α by LPS was completely blocked by actinomycin D (Figure 4A). The actinomycin D inhibition was not due to a toxic effect because the ability of LPS to activate p42/p44 MAPK, a pathway known to be activated by LPS in macrophages, was not decreased (Figure 4A). Also, in hypoxic cells, actinomycin D had only a very limited effect on HIF-1α protein induction (Figure 4B). Increases in HIF-1α mRNA stability do not seem to be responsible for increased mRNA levels because these levels remain high for extended periods of time (up to 2 hours) when actinomycin D is added following a 4-hour stimulation (results not shown). These experiments suggest that in an LPS-stimulated macrophage-derived cell line, enhanced transcription of the HIF-1α messenger is crucial to maintain elevated levels of HIF-1α. This pathway is distinct from hypoxic induction of HIF-1α in these same cells, in which HIF-1α protein stability plays a decisive role.4 It is interesting to note that under normoxic conditions actinomycin D also reduced basal levels of HIF-1α (Figure 4B). This indicates that in NR8383 transcriptional mechanisms are important to maintain normoxic levels.
LPS increases HIF-1α mRNA expression. (A) Quiescent NR8383 cells were maintained under control conditions or in the presence of LPS (1 μg/mL) for different periods of time of up to 8 hours. (B) Quiescent NR8383 cells were maintained either under control conditions, or under hypoxic conditions, or in the presence of LPS (1 μg/mL) for 6 hours. Total RNA was extracted from cells and resolved on formaldehyde/agarose gels. Northern blot was performed using a specific radiolabeled HIF-1α probe. An 18S rRNA probe was used as a control for gel loading.
LPS increases HIF-1α mRNA expression. (A) Quiescent NR8383 cells were maintained under control conditions or in the presence of LPS (1 μg/mL) for different periods of time of up to 8 hours. (B) Quiescent NR8383 cells were maintained either under control conditions, or under hypoxic conditions, or in the presence of LPS (1 μg/mL) for 6 hours. Total RNA was extracted from cells and resolved on formaldehyde/agarose gels. Northern blot was performed using a specific radiolabeled HIF-1α probe. An 18S rRNA probe was used as a control for gel loading.
Transcriptional activity is essential for HIF-1α induction by LPS. NR8383 cells were pretreated or not for 15 minutes with actinomycin D (100 ng/mL) and maintained under control conditions, in the presence of 1 μg/mL LPS (A) or under hypoxic conditions (B) for 6 hours. Total cell extracts (25 μg) were resolved by SDS-PAGE (8% gel) and immunoblotted using an anti-HIF-1α antiserum or an antiphospho-p44/p42 MAPK monoclonal antibody.
Transcriptional activity is essential for HIF-1α induction by LPS. NR8383 cells were pretreated or not for 15 minutes with actinomycin D (100 ng/mL) and maintained under control conditions, in the presence of 1 μg/mL LPS (A) or under hypoxic conditions (B) for 6 hours. Total cell extracts (25 μg) were resolved by SDS-PAGE (8% gel) and immunoblotted using an anti-HIF-1α antiserum or an antiphospho-p44/p42 MAPK monoclonal antibody.
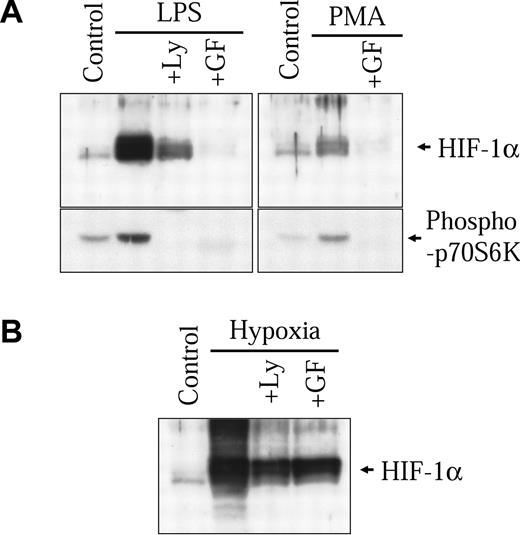
In our previous study, we demonstrated that classical diacylglycerol (DAG)-sensitive forms of protein kinase C (PKC) play an important part in modulating nonhypoxic HIF-1α induction through transcriptional mechanisms.32 In macrophages, LPS has been shown to signal through the activation of these same isoforms of PKC.33-36 Therefore, we wanted to evaluate the implication of DAG-sensitive forms of PKC in the activation of HIF-1α by LPS in our macrophage-derived cell model. As seen in Figure 5A (right panel), PMA, a potent activator of DAG-sensitive forms of PKC, can also increase HIF-1α protein levels in NR8383 cells. GF109203X, a known specific inhibitor of classical PKCs, completely inhibited HIF-1α protein induction by PMA. More interestingly, GF109203X could also strongly block the induction of HIF-1α protein by LPS (Figure 5A left panel). As was the case with actinomycin D, GF109203X caused only a modest inhibition on the hypoxic induction of the HIF-1α protein (Figure 5B). This last result demonstrates that (1) the effect of GF109203X is not due to a nonspecific effect and (2) different pathways are implicated in the hypoxic and LPS inductions of HIF-1α protein. Taken together, these results demonstrate that contrary to hypoxia, DAG-sensitive forms of PKC play an important role in the increase of HIF-1α protein levels in LPS-stimulated NR8383 cells.
Implication of the PKC and PI3K pathways in HIF-1α induction by LPS. NR8383 cells were pretreated or not for 15 minutes with Ly29004 (20 μM) or GF109203X (10 μM) and maintained under control conditions in the presence of 1 μg/mL LPS (A), 100 nM PMA (A), or under hypoxic conditions (B) for 6 hours. Total cell extracts (25 μg) were resolved by SDS-PAGE (8% gel) and immunoblotted using an anti-HIF-1α antiserum or an antiphospho-p70S6 kinase polyclonal antibody.
Implication of the PKC and PI3K pathways in HIF-1α induction by LPS. NR8383 cells were pretreated or not for 15 minutes with Ly29004 (20 μM) or GF109203X (10 μM) and maintained under control conditions in the presence of 1 μg/mL LPS (A), 100 nM PMA (A), or under hypoxic conditions (B) for 6 hours. Total cell extracts (25 μg) were resolved by SDS-PAGE (8% gel) and immunoblotted using an anti-HIF-1α antiserum or an antiphospho-p70S6 kinase polyclonal antibody.
Activation of the phosphoinositol 3-kinase (PI3K) pathway also plays a major role in the induction of HIF-1α protein levels by different nonhypoxic stimuli.12,13,15,17,19,32,37,38 Studies suggest that activation of the PI3K pathway preferentially increases HIF-1α protein translation.15,19,32 In the NR8383 cell line, LPS activated the PI3K pathway, as seen by the phosphorylation of a direct downstream target of PI3K, p70S6 kinase (Figure 5A left panel). The specific PI3K inhibitor, Ly294002, blocked the activation of p70S6 kinase and also strongly inhibited the increase in HIF-1α protein expression by LPS. Again, Ly294002 caused only a modest inhibition of the hypoxic induction of the HIF-1α protein (Figure 5B). Wortmannin (100 nM), another PI3K inhibitor, also inhibited HIF-1α induction by LPS in NR8383 cells (results not shown). These results demonstrate that, as was the case with other nonhypoxic stimuli, activation of the PI3K pathway is implicated in LPS-induced increases of HIF-1α in our macrophage-derived cell line. Additionally, Ly 294002 also blocked HIF-1α induction and p70S6 kinase activation following treatment of cells with PMA (results not shown). These results therefore demonstrate the essential role played by PI3K in nonhypoxic induction of HIF-1α and suggest that by activating this pathway in macrophages, LPS increases HIF-1α translation.
LPS activates the HIF-1 complex
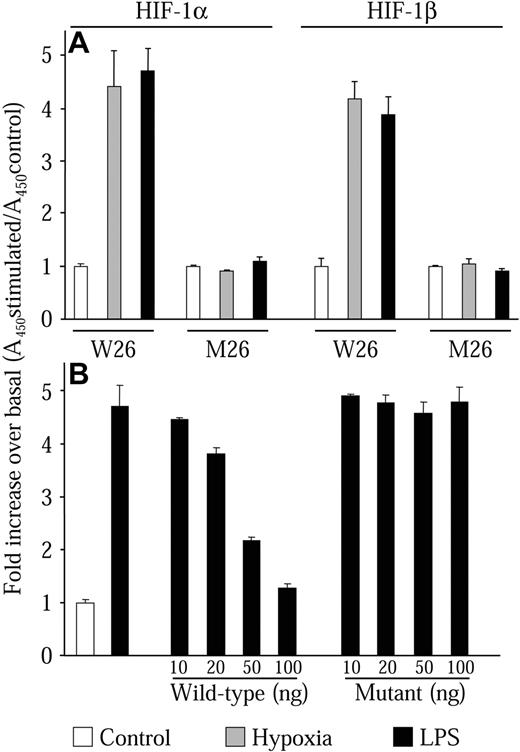
We next wanted to investigate whether HIF-1α protein levels induced by LPS could form an active HIF-1 transcription complex. To perform these studies, we developed an HIF-1 transcription factor enzyme-linked immunoassay (TF-EIA) similar to assays previously described.39-41 This assay has the advantage of being 10 times more sensitive than traditional electrophoretic mobility shift assays (EMSAs) and allows greater flexibility in the experimental setup. This assay uses a specific HIF-1-binding dsDNA oligonucleotide sequence (W26) immobilized on a 96-well microplate. Protein complexes are then bound to the dsDNA oligonucleotides. The identity of the bound complexes is then revealed by an enzyme-linked colorimetric reaction using specific antibodies coupled to HRP. Prior to use in this work, the assay was validated using other cell types such as HeLa, HEK-293, and VSMCs. These trial experiments always gave results that were consistent with EMSA experiments. Nuclear extracts from NR8383 cells maintained in hypoxic conditions or in the presence of 1 μg/mL LPS both demonstrated increased DNA-binding activity for HIF-1α and HIF-1β (Figure 6A, W26). The increase was similar for each stimulus, which concurred with results seen in Western blot experiments (Figure 2A). These results indicate that LPS induces the formation of the heterodimeric complex HIF-1, which binds to the HIF-1-binding sequence. To verify specificity, we used plates that were coated with a dsDNA oligonucleotide sequence that was mutated at crucial residues of the HIF-1-binding sequence.27 In this case, no increase in HIF-1α or HIF-1β binding could be observed (Figure 6A, M26). We also performed competition experiments with increasing concentrations of either wild-type or mutant nonimmobilized dsDNA oligonucleotide. Only the wild-type dsDNA oligonucleotide could effectively compete for the binding of the HIF-1 complex with the W26-coated plate (Figure 6B). These results demonstrate that HIF-1α protein induced by LPS can form the HIF-1 complex with HIF-1β and bind to the HIF-1 DNA-binding motif.
LPS induces the HIF-1 transcription factor complex. Quiescent NR8383 cells were maintained either under control conditions (20.8% oxygen, □), or under hypoxic conditions (1% oxygen, ▦), or in the presence of LPS (1 μg/mL, ▪) for 6 hours followed by preparation of nuclear extracts. (A) Nuclear protein (10 μg) was incubated in a 96-well plate coated with an oligonucleotide containing the wild-type (W26) or mutant (M26) HIF-1-binding site. Presence of HIF-1 transcription complex was evaluated with an antibody to either HIF-1α or HIF-1β. (B) Nuclear protein (10 μg) was incubated in a 96-well plate coated with an oligonucleotide containing the wild-type (W26) in the presence or absence of increasing concentrations of wild-type or mutant competitor oligonucleotide. Presence of HIF-1 transcription complex was evaluated with an antibody to HIF-1α. HIF-1 binding was then revealed by incubation with an HRP-conjugated secondary antibody and substrate. Results are expressed as the fold increase of the absorbance at 450 nM over control conditions. This experiment is an average ± SD of an experiment performed in triplicate and is representative of at least 3 independent experiments.
LPS induces the HIF-1 transcription factor complex. Quiescent NR8383 cells were maintained either under control conditions (20.8% oxygen, □), or under hypoxic conditions (1% oxygen, ▦), or in the presence of LPS (1 μg/mL, ▪) for 6 hours followed by preparation of nuclear extracts. (A) Nuclear protein (10 μg) was incubated in a 96-well plate coated with an oligonucleotide containing the wild-type (W26) or mutant (M26) HIF-1-binding site. Presence of HIF-1 transcription complex was evaluated with an antibody to either HIF-1α or HIF-1β. (B) Nuclear protein (10 μg) was incubated in a 96-well plate coated with an oligonucleotide containing the wild-type (W26) in the presence or absence of increasing concentrations of wild-type or mutant competitor oligonucleotide. Presence of HIF-1 transcription complex was evaluated with an antibody to HIF-1α. HIF-1 binding was then revealed by incubation with an HRP-conjugated secondary antibody and substrate. Results are expressed as the fold increase of the absorbance at 450 nM over control conditions. This experiment is an average ± SD of an experiment performed in triplicate and is representative of at least 3 independent experiments.
We then evaluated whether the HIF-1 complex induced by LPS is transcriptionally active. We measured HIF-1-dependent transcription in NR8383 cells transiently transfected with a luciferase reporter gene (PRE-tk-LUC) driven by 3 specific 50-bp HRE sequences.24,27 As shown in Figure 7A, an almost 7-fold increase in reporter activity was attained after an 18-hour incubation period in 1% oxygen. For the same time period, LPS increased reporter activity to a level that even surpassed that elicited by hypoxia (9.4-fold over basal levels). These results suggest that the HIF-1 complex induced by LPS is strongly active. To clearly demonstrate the involvement of HIF-1α and the HIF-1 complex in the stimulation of HRE reporter activity, we used a dominant-negative form of HIF-1α. The pcDNA3-HA-DN-HIF-1α construct encodes a form of HIF-1α lacking the C-terminal transactivation domains. DN-HIF-1α can dimerize with HIF-1β and block HRE-driven reporter genes.10,29 NR8383 cells were cotransfected with the PRE-tk-LUC reporter plasmid and the pcDNA3-HA-DN-HIF-1α construct followed by incubation of cells in the presence of LPS. When cells were transfected with the dominant-negative form of HIF-1α, a strong inhibition of reporter activity could be observed in cells that were stimulated with LPS (Figure 7B). This result could also be seen in cells maintained in hypoxic conditions (results not shown). These results demonstrate that LPS can induce hypoxic gene expression by increasing HIF-1α transcription and protein expression, resulting in the formation of a transcriptionally active HIF-1 complex.
LPS stimulates HIF-1 transcriptional activity. (A) NR8383 cells (5 × 105 cells/well, 6-well plate) were transfected with 500 ng PRE-tk-LUC reporter construct and 250 ng of an expression vector coding for Renilla reniformis luciferase was used to normalize for transfection efficiency. (B) NR8383 cells were cotransfected with luciferase vectors as in panel A and either 4 μg pcDNA3 (□)or 4 μg pcDNA3-HA-DN-HIF-1α (▦). At 12 hours after transfection, cells were deprived of FBS for 16 hours. Cells were then maintained under either control conditions, hypoxic conditions, or in the presence of LPS (1 μg/mL) for 18 hours. At this point, NR8383 cells were lysed and luciferase activity was measured using the Dual-Luciferase Reporter Assay System. Results are expressed as a ratio of beetle luciferase activity over Renilla reniformis luciferase activity. Data expressed are an average ± SD of at least 3 independent experiments performed in triplicate.
LPS stimulates HIF-1 transcriptional activity. (A) NR8383 cells (5 × 105 cells/well, 6-well plate) were transfected with 500 ng PRE-tk-LUC reporter construct and 250 ng of an expression vector coding for Renilla reniformis luciferase was used to normalize for transfection efficiency. (B) NR8383 cells were cotransfected with luciferase vectors as in panel A and either 4 μg pcDNA3 (□)or 4 μg pcDNA3-HA-DN-HIF-1α (▦). At 12 hours after transfection, cells were deprived of FBS for 16 hours. Cells were then maintained under either control conditions, hypoxic conditions, or in the presence of LPS (1 μg/mL) for 18 hours. At this point, NR8383 cells were lysed and luciferase activity was measured using the Dual-Luciferase Reporter Assay System. Results are expressed as a ratio of beetle luciferase activity over Renilla reniformis luciferase activity. Data expressed are an average ± SD of at least 3 independent experiments performed in triplicate.
Discussion
Recent evidence has identified a link between inflammation, wound healing, and the activation of HIF-1 expression. Hollander et al have demonstrated that HIF-1α protein was abundantly expressed by macrophages in inflamed rheumatoid synovia while being absent in healthy synovia.42 A second study demonstrated that HIF-1α expression is strongly increased in inflammatory cells from wounds.43 This induction was suggested to be caused by the release of tumor necrosis factor α (TNF-α), which can strongly increase HIF-1α protein levels in certain cells.18,43-45 In a very elegant study, Cramer et al demonstrated that a conditional knockout of HIF-1α in macrophages and other myeloid lineage cells leads to decreased myeloid cell infiltration and activation, to impaired chronic cutaneous inflammation, and to decreased joint inflammation in a rheumatoid arthritis.46 Taken together, these studies implicate HIF-1 as an important mediator of inflammatory responses in macrophages. However, very little information is available on the cellular aspects of these important effects.
The potent inflammatory factor LPS is a strong stimulator of gene expression in macrophages.33 Among the genes that are activated by LPS, a number are also genes known to be modified by hypoxia.20-22 In this study we set out to investigate whether HIF-1 could be involved in the LPS-mediated increase of hypoxia-regulated genes. Recent studies, including our own work, have shown that a number of nonhypoxic stimuli could strongly increase HIF-1 in a cell-specific manner.10-19 Interestingly, some of these increases have been shown to be equal or greater than the hypoxic induction of HIF-1α. In this study, we convincingly show that in macrophages, LPS is an excellent nonhypoxic stimulator and activator of the HIF-1 transcription factor.
We report here that LPS can increase HIF-1α mRNA levels in a macrophage-derived cell line. This was also the case in VSMCs stimulated with Ang II. However, in a number of other cell lines this is not the case. It seems that in growth-arrested VMSCs and macrophages, signals are shut down that are critical in the maintenance of elevated levels of HIF-1α mRNA. In these cells, stimulation with a number of growth factors, including the readdition of serum to growth media, will strikingly increase HIF-1α mRNA levels, which will permit increased protein expression.10,32 DAG-sensitive forms of PKC seem to play an important part in increasing HIF-1α mRNA transcription, possibly by stimulating the HIF-1α gene promoter on specific transcriptional regulatory elements such as Sp1.32 A second pathway, involving the activation of the PI3K pathway, would specifically increased the translation of the elevated mRNA, leading to increased HIF-1α protein levels in nonhypoxic cells.15,19,32 These mechanisms are in striking contrast to the hypoxic induction of HIF-1α, which relies heavily on the stabilization of HIF-1α protein. In hypoxia, mRNA levels and protein translation are similar to those found in normal oxygen conditions.19,32,47-50 In this study we did not investigate the possibility that LPS could increase HIF-1α stability. No evidence exists that links extracellular cell stimulation to changes in the specific HIF-1α modification and degradation pathways. We are currently investigating this area of research.
Our results suggest that HIF-1 should be involved in the activation of known hypoxic genes by LPS. However, we do not suggest that the HIF-1 pathway is the only mechanism involved in the activation of these genes. Multiple signaling pathways are activated by LPS in these cells and a number of these pathways have been shown to play an important part in the activation of certain hypoxia activated genes. We believe that along with the other pathways and transcription factors, HIF-1 permits the maximal activation of these genes.
In conclusion, our study provides convincing data that identifies an interesting mechanism by which LPS could induce hypoxic gene expression in macrophages. LPS binds specific cell surface receptors, which increases HIF-1α protein expression. An active HIF-1 complex is formed, which activates its target genes. This phenomenon likely has physiologic implication given the wide spectrum of genes activated and their implication in situations where macrophages are present such as inflammation and wound healing.
Prepublished online as Blood First Edition Paper, October 2, 2003; DOI 10.1182/blood-2003-07-2427.
Supported by grants from the Canadian Institutes of Health Research (MOP-49609), the Anemia Institute for Research and Education, les Fonds de la Recherche en Santé du Québec, and the Heart and Stroke Foundations of Québec. D.E.R. is a recipient of the McDonald Scholarship from the Heart and Stroke Foundation of Canada. E.L.P. is a recipient of a postgraduate scholarship from the Natural Sciences and Engineering Council of Canada.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Jacques Pouysségur and Marcel Lebel for support and reagents, Dr Steven McKnight for the PRE-tk-LUC plasmid, Dr Richard Lariviére for PAI-1 and iNOS cDNA, Dr Francesc Viñals for GLUT1 cDNA, Dr Gilles Pagés for VEGF cDNA, Drs Kim Boulukos and Martine Torres for help with cell lines, Geneviéve Robitaille for excellent technical assistance, and Dr Fergus R. McKenzie for helpful comments.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal