Abstract
Glycoprotein (GP) VI is the major receptor responsible for platelet activation by collagen, but the collagen-binding surface of GPVI is unknown. To address this issue we expressed, from insect cells, the immunoglobulin (Ig)–like ectodomains (residues 1-185) of human and murine GPVI, called hD1D2 and mD1D2, respectively. Both proteins bound specifically to collagen-related peptide (CRP), a GPVI-specific ligand, but hD1D2 bound CRP more strongly than did mD1D2. Molecular modeling and sequence comparison identified key differences between hD1D2 and mD1D2. Ten mutant hD1D2s were expressed, of which 4 had human residues replaced by their murine counterpart, and 6 had replacements by alanine. CRP binding studies with these mutants demonstrated that the exchange of lysine at position 59 for the corresponding murine glutamate substantially reduced binding to CRP. The position of lysine59 on the apical surface of GPVI suggests a mode of CRP binding analogous to that used by the related killer cell Ig-like receptors to bind HLA. This surface was confirmed as critical for collagen binding by epitope mapping of an inhibitory phage antibody against GPVI. This anti-GPVI, clone 10B12, gave dose-dependent inhibition of the hD1D2-collagen interaction. Clone 10B12 inhibited activation of platelets by CRP and collagen in aggregometry and thrombus formation by the latter in whole blood perfusion. Antibody 10B12 showed significantly reduced binding to the hD1D2-E59, and, on that basis, the GPVI:10B12 interface was modeled.
Introduction
Damage to blood vessels exposes circulating platelets to the extracellular matrix. Here, collagen supports adhesion and stimulates platelet activation by acting as a ligand for a number of platelet receptors.1 Platelets are first tethered by the interaction of glycoprotein (GP) Ibα with the A1 domain of von Willebrand factor (VWF),2 a plasma protein that binds to exposed collagen.3 Firm platelet adhesion results from the concerted action of the collagen receptor GPIaIIa (integrin α2β1) and the fibrinogen receptor GPIIbIIIa (αIIbβ3) which also binds immobilized VWF.4 Collagen-mediated activation of platelets is dependent on the engagement and clustering of GPVI,1,5 an immunoglobulin (Ig) superfamily member with homology to killer cell Ig-like receptors (KIRs) and the FcαRI.6 We and others have described the ligand-binding sites of α2 integrin7 and GPIbα2 at the structural level.
Recent work has demonstrated the key role played by GPVI in arterial thrombus formation in mice and provides a clear basis for the development of potentially therapeutic GPVI inhibitors.8 Indeed, blockade of GPVI is attractive for several reasons. First, the expression of GPVI appears to be restricted to platelets and megakaryocytes.9 Second, collagen is required for prothrombotic “COAT” platelet formation,10 for which blockade of GPVI may provide a specific control point. Third, patients with congenital or acquired autoantibody-mediated GPVI deficiency have only a mild bleeding disorder, despite having a significantly reduced platelet response to collagen.11,12 These observations have been recently confirmed in mice.13,14 However, the development of successful GPVI antagonists requires a better understanding of the GPVI-collagen interaction.
The GPVI gene is part of the human leukocyte receptor cluster (LRC) on chromosome 19q13.4.15 GPVI has 2 extracellular C2-type Ig-like domains (D1 and D2). These Ig-like domains are connected by a glycosylated stem of approximately 60 amino acids to a transmembrane sequence within which an arginine residue (R252) forms a salt-bridge with a transmembrane aspartate (D11) in the FcR γ chain. GPVI signaling is dependent on its interaction with the γ chain.16,17 The 51 residue cytoplasmic domain of human GPVI has binding sites for various signaling proteins.18,19 Two modes of ligand binding for LRC members have been described. The KIRs bind to HLA via a surface that contains the interdomain linker,20,21 whereas FcαRI binds IgA via its N-terminal Ig-like domain.22 Previous studies have shown that GPVI recognizes glycine-proline-hydroxyproline (GPO) repeat motifs in the triple helical structure of collagen.23 It was also shown that collagen-related peptide (CRP), which contains 10 GPO motifs and spontaneously forms helices under physiologic conditions,24 is a powerful and specific agonist for GPVI. GPVI specifically recognizes CRP but not a peptide of similar structure (GPP10) lacking hydroxyproline.25 Studies with GPVI-deficient murine platelets and the inhibitory monoclonal antibody JAQ-1 are suggestive of the existence of 2 collagen-binding sites on murine GPVI, the primary one being the binding site for CRP.26,27
We postulate that the CRP binding site on human GPVI maps to the same surface as in murine GPVI. Here, we seek to map the primary collagen-binding site of human GPVI by using the aforementioned peptides. By homology modeling of the Ig-like domains of human and murine GPVI on the basis of the KIR coordinates, residues of possible relevance to CRP binding were identified. On that basis, 12 D1D2 molecules (wild-type human and mouse and 10 mutants of human) were recombinantly expressed, and their relative binding affinities for CRP were determined. The wild-type hD1D2 was used as a bait to select inhibitory and noninhibitory phage antibodies specific for GPVI. Results of epitope mapping of the former and the modeling of interface of the antibodies and GPVI combined with the results obtained with the mutant D1D2 molecules allowed the identification of the primary collagen-binding surface on GPVI.
Materials and methods
Cloning of hGPVI and mGPVI and expression of D1D2 proteins
Full-length human and mouse GPVI cDNA were isolated by reverse transcriptase polymerase chain reaction (RT-PCR) from platelet-derived mRNA by using primers designed with reference to published sequences (Clemetson et al6 and Jandrot-Perrus et al,9 respectively). Amplified DNA was cloned into the vector pCR2.1 (Invitrogen, Paisley, United Kingdom) and sequenced to ensure that no PCR mutations had been introduced. Fragments encoding hD1D2 and mD1D2 (residues 1-185 in each case) and hD1 (residues Q1-G89) and hD2 (residues A87-T185) were then amplified for subsequent cloning into the plasmids pMT/BiP/CaM or pMT/BiP/V5/His, used to produce recombinant protein in the Drosophila expression system (DES; Invitrogen) with hexahistidine (His) or calmodulin (CaM) tags, respectively (N.S.J. et al, manuscript submitted, 2003).
All His-tagged proteins were purified with the use of HiTrap Chelating columns (Amersham Biosciences, Little Chalfont, United Kingdom) and were desalted into 150 mM NaCl, 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), pH 7.4 (HBS) prior to use. CaM-tagged proteins were purified on a W-7 resin affinity column.28 The purity of all proteins was verified using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the concentration was measured by using a bicinchoninic acid (BCA) assay kit (Perbio, Chester, United Kingdom). For affinity measurements, the molarity of purified proteins was calculated by using molecular masses of 38 kDa for each CaM-tagged D1D2 and 28.5 kDa for each single-chain variable domain antibody fragment (scFv).
Site-directed mutagenesis
Nine point mutations were introduced into the hD1D2 gene fragment in pCR2.1 with the use of Quickchange (Stratagene, La Jolla, CA). Primers are presented in Table 1.
PCR primers used in site-directed mutagenesis of human D1D2
K59Efor | CCAGCCAGACTTCTCTCCATGGCCGGGATG |
| K59Eback | CATCCCGGCCATGGAGAGAAGTCTGGCTGG |
| R60Aback | CCCGGCCATGAAGGCTAGCCTGGCTGGACGCTACC |
| R60Afor | GGTAGCGTCCAGCCAGGCTAGCCTTCATGGCCGGG |
| F91Aback | GGAGCTCGTTGCCACCGGTGTTGCTGCCAAACCCTCG |
| F91Afor | CGAGGGTTTGGCAGCAACACCGGTGGCAACGAGCTCC |
| R117Pback | GGTCAAAGCCATACGGAGTCTGACACTGTAGGG |
| R117Pfor | CCCTACAGTGTCAGACTCCGTATGGCTTTGACC |
| Y118Aback | CCTACAGTGTCAGACTCGGGCCGGCTTTGACCAATTTGC |
| Y118Afor | GCAAATTGGTCAAAGCCGGCCCGAGTCTGACACTGTAGG |
| F120Aback | GTGTCAGACTCGGTATGGCGCCGACCAATTTGCTCTG |
| F120Afor | CAGAGCAAATTGGTCGGCGCCATACCGAGTCTGACAC |
| R139Aback | GCCCTACAAGAATCCCGAAGCTTGGTACCGGGCTAGTTTCCC |
| R139Afor | GGGAAACTAGCCCGGTACCAAGCTTCGGGATTCTTGTAGGGC |
| S164Aback | CGATGCTACAGCTTCGCTAGCAGGGACCCATACC |
| S164Afor | GGTATGGGTCCCTGCTAGCGAAGCTGTAGCATCG |
| R166Sfor | CGACCACAGGTATGGGTCGCTGCTCGAGAAGC |
| R166Sback | GCTTCTCGAGCAGCGACCCATACCTGTGGTCG |
K59Efor | CCAGCCAGACTTCTCTCCATGGCCGGGATG |
| K59Eback | CATCCCGGCCATGGAGAGAAGTCTGGCTGG |
| R60Aback | CCCGGCCATGAAGGCTAGCCTGGCTGGACGCTACC |
| R60Afor | GGTAGCGTCCAGCCAGGCTAGCCTTCATGGCCGGG |
| F91Aback | GGAGCTCGTTGCCACCGGTGTTGCTGCCAAACCCTCG |
| F91Afor | CGAGGGTTTGGCAGCAACACCGGTGGCAACGAGCTCC |
| R117Pback | GGTCAAAGCCATACGGAGTCTGACACTGTAGGG |
| R117Pfor | CCCTACAGTGTCAGACTCCGTATGGCTTTGACC |
| Y118Aback | CCTACAGTGTCAGACTCGGGCCGGCTTTGACCAATTTGC |
| Y118Afor | GCAAATTGGTCAAAGCCGGCCCGAGTCTGACACTGTAGG |
| F120Aback | GTGTCAGACTCGGTATGGCGCCGACCAATTTGCTCTG |
| F120Afor | CAGAGCAAATTGGTCGGCGCCATACCGAGTCTGACAC |
| R139Aback | GCCCTACAAGAATCCCGAAGCTTGGTACCGGGCTAGTTTCCC |
| R139Afor | GGGAAACTAGCCCGGTACCAAGCTTCGGGATTCTTGTAGGGC |
| S164Aback | CGATGCTACAGCTTCGCTAGCAGGGACCCATACC |
| S164Afor | GGTATGGGTCCCTGCTAGCGAAGCTGTAGCATCG |
| R166Sfor | CGACCACAGGTATGGGTCGCTGCTCGAGAAGC |
| R166Sback | GCTTCTCGAGCAGCGACCCATACCTGTGGTCG |
Back primers amplify the antisense strand and for(ward) primers amplify the sense strand of pCR 2.1-hD1D2. Underlined bases correspond to restriction sites introduced by the primers.
Sequential mutation steps were used to create the triple mutant encoding 59E, 117P, and 166S. All mutant hD1D2 fragments were expressed and purified as described earlier.
Synthesis of peptides and conjugates
CRP and GPP10 were synthesized as previously described.25 N9A peptide (CAAARWKKAFIAVSAANRFKKIS)29 was synthesized as described previously.28 N9A-BSA was produced using standard techniques.30 Polymerized, maleimide-activated peroxidase (Sigma, St Louis, MO) was used for the production of horseradish peroxidase (HRP)–N9A, following manufacturer's instructions, using a molar ratio of 4 peptides per polymer.
Ligand-binding assay for D1D2 proteins
A ligand-binding assay was developed to study the interactions of recombinant proteins with CRP and GPP10. Ninety-six well Maxisorp microplates (Life Technologies, Paisley, United Kingdom) were coated overnight at 4°C with 100 μL per well of CRP, GPP10, or collagen fibers (Equine Type 1; Horm, Nycomed, Munich, Germany) at 10 μg/mL in 0.01 M acetic acid. The wells were then incubated with 200 μL blocking buffer (0.15 M NaCl, 0.05 M Tris (tris(hydroxymethyl)aminomethane), pH 7.4 [TBS] + 5% bovine serum albumin [BSA]) for 2 hours at 37°C. Three washes were then made with 200 μL assay buffer (TBS supplemented with 1 mM CaCl2 and 0.1% BSA). Then 100 μL recombinant protein (either alone or after preincubation with scFvs for 1 hour at room temperature) was added and incubated for 2 hours. After a further 4 washes with assay buffer, 100 μL HRP-N9A (0.16 μg/mL) was added for 1 hour. Finally, after an additional 6 washes, 100 μL HRP substrate (TMB substrate kit; Perbio) was added. The reaction was stopped after 20 minutes by the addition of 50 μL 2.5 M H2SO4, and the absorbance was measured at 450 nm on an enzyme-linked immunosorbent assay (ELISA) plate reader (Dynex Technologies, Middlesex, United Kingdom). Values for nonspecific binding to BSA-blocked wells were subtracted prior to analysis by using PRISM (GraphPad, San Diego, CA).
Selection of phage antibodies binding to hD1D2
Two human V gene phage display libraries, Marks31 and CAT,32 were used for the selection of scFvs specific for GPVI. For both libraries, an identical protocol was used for the first round of selection. Briefly, phage was mixed with hD1D2 in solution, and complexes were then captured via the CaM tag onto the surface of BSA-N9A–coated immunotubes. After extensive washing, the phage antibody-hD1D2 complexes were eluted by chelating calcium with 10 mM EDTA (ethylenediaminetetraacetic acid) and then propagated in Escherichia coli as previously described.33 For the Marks library, this protocol was repeated twice, and clones from the third round of selection were screened for binding to hD1D2 by ELISA. For the CAT library, the second round of selection was designed to enrich for scFvs that specifically blocked the CRP-D1D2 interaction. In detail, preformed, immobilized hD1D2-CRP complexes were obtained by incubating CaM-tagged hD1D2 in wells coated with CRP. Regrown phage particles from the first round were added to these wells and allowed to compete with CRP for binding to hD1D2, thus releasing phage-hD1D2 complexes into solution. These complexes were captured by way of the CaM tag, washed, and eluted with EDTA as before. Clones from this second round of selection were extensively screened for specificity.
Sequencing, recloning, and expression of scFvs
Purified double-stranded phagemid DNA was sequenced as previously described,33 and nucleotide sequences were compared with germ line V-gene sequences in the V-base directory.34 The scFv gene cassettes of selected clones were subcloned into the pUC119-Sfi/Not-His6 vector, and scFv was expressed as previously described.33
ELISA
The specificity of selected scFvs was investigated by ELISA with proteins either captured by way of the CaM tag to BSA-N9A–coated plates in the presence of Ca2+, or coated directly onto the microplate wells. For the capture method the wells were coated overnight at 4°C with 50 μL BSA-N9A at 5 μg/mL in 50 mM Na-Borate, pH 8.3. The wells were then washed once with 200 μL TBS and blocked with TBS containing 1 mM CaCl2 and 0.1% Tween 20 (TBS-TCa) for 30 minutes at 37°C. The wells were then washed twice with TBS-TCa, and 50 μL CaM-tagged protein (5 μL/mL) in TBS-TCa was added for 30 minutes at room temperature. After a further 4 washes with TBS-TCa, 100 μL scFv was added for 2 hours at room temperature. Following 4 washes, 100 μL HRP-labeled mouse monoclonal anti–c-myc (clone 9E10, 0.2 μg/mL; Roche, Lewes, United Kingdom) was added for 30 minutes at room temperature. Finally, after 6 additional washes, 100 μL TMB substrate was added, and the reaction was stopped and read as described earlier. For the directly coated method, the microplate wells were incubated overnight at 4°C with 50 μL protein (5 μg/mL) in 50 mM Na-Borate, pH 8.3. The wells were then washed once with TBS and blocked by incubation with TBS containing 3% BSA for 1 hour at 37°C. After a further 4 washes with TBS, scFv was added for 2 hours at room temperature and detected as described earlier.
Biosensor
Real-time interactions between recombinant proteins and selected scFvs were analyzed by using a resonant mirror biosensor (IAsys Auto+; ThermoLabsystems, Cambridge, United Kingdom) with a stirrer speed of 100 at 25°C in a volume of 50 to 70 μL per cell. For CaM-tagged hD1D2, BSA-N9A was first coupled to the biosensor surface and then used as an anchor for the Ca2+-dependent, high-affinity binding of CaM-tagged hD1D2. Prior to affinity measurements, the purified scFv was gel filtered (Superose 12; Amersham Biosciences) in HBS to remove the minor portion of dimeric scFv.35 Affinity of the scFvs was calculated by using proprietary FastFit software (ThermoLabsystems). For epitope mapping studies the responses at equilibrium were used.
Flow cytometry
Flow cytometry was performed as previously described33 by using purified scFv at 20 μg/mL. For the inhibition assays, scFv (20 μg/mL) was preincubated with hD1D2-His (1-300 μg/mL), in total volume of 150 μL for 15 minutes at room temperature prior to addition to platelets.
Aggregometry and perfusion
Aggregometry was performed in autologous plasma with an adjusted platelet count of 200 × 109/L from fresh citrate anticoagulated blood at 37°C as previously described.36 To study the effect of scFvs on aggregation, purified scFv was added 5 minutes before the induction of platelet aggregation by the addition of either collagen type I fibers (Ethicon, Somerville, NJ), adenosine 5′-diphosphate (ADP; Sigma), U46619 (Sigma), thrombin receptor–activating peptide (TRAP; Sigma), or epinephrine (EPI; Sigma). Perfusion assays were performed as previously described.37
Modelling
With the use of PSI-BLAST at the National Center for Biotechnology Information, hD1D2 and mD1D2 were aligned to the Brookhaven Protein Databank to identify suitable templates for modeling. This search returned 1B6U, 1G0X, 1NKR, 2DL2, and 2DL1 as suitable templates that were then used with MODELLER38 to construct models of hD1D2 and mD1D2 without subjective intervention. A similar approach for the scFvs identified 1AQK, 1MCW, 2CDO, and 2MCG as suitable templates for modeling. Images of protein surfaces were generated by using GRASP39 with electrostatic potential going from blue (potential of +13 kT/e) to red (–8 kT/e). Electrostatic potential was calculated with Poisson-Boltzmann solver within GRASP, using a uniform dielectric constant of 80 for the solvent, 2 for the protein interior, and an ionic strength of zero. A probe radius of 1.4 Å was used. Here, K and R have a single-positive and D and E a single-negative charge. All other residues are neutral. The mutation of K59 to E was introduced into hD1D2 with the program O40 by using the most preferred side-chain conformation from the rotomer database.41
Results
Human and murine D1D2 bind CRP with different affinities
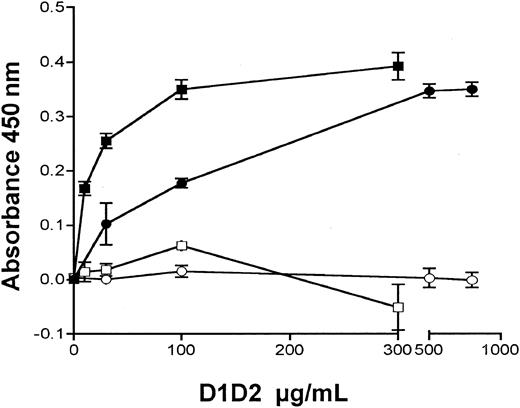
A protein sequence alignment of human and murine GPVI with KIR2DL1-3 and ILT2 shows 39% to 45% identity at the amino acid level. Numerous structural features, including the 2 entire Ig-like domains (residues 1-185 or D1D2), are preserved (Figure 1A-B). We generated pure, soluble forms of hD1D2 and mD1D2 fused to CaM, a versatile affinity tag.43 When hD1D2 and mD1D2 were allowed to bind to wells coated with CRP or the negative control GPP10, 2 observations were made. First, both mD1D2 and hD1D2 bound specifically to CRP (Figure 2). This observation confirms that the recombinant proteins and platelet GPVI have the same specificity.24,25 Second, hD1D2 binds to CRP with a one log higher affinity than mD1D2 when estimated by median effective concentration (EC50) values of 3.4 ± 0.8 μg/mL for hD1D2 (n = 8) and of 156.3 ± 25.8 μg/mL for mD1D2 (n = 3).
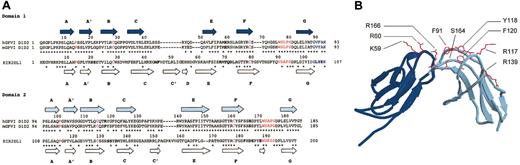
Predicted secondary structure of Ig-like domains of GPVI, compared with a KIR. (A) Alignment of hD1D2 and mD1D2 to KIR2DL1 (pdb code 1NKR42 ). Residue numbers are shown above each sequence, either hD1D2 (identical to mD1D2) or KIR2DL1. Identity between sequences is indicated by a star, either below mD1D2 (conserved in human and murine D1D2) or below KIR2DL1 (identity with those conserved residues in D1D2). Predicted β-strands of D1D2 are shown as arrows above hD1D2 (dark blue for D1, light blue for D2). Actual β-strands in 1NKR are shown as white arrows below KIR2DL1. Gaps produced in D1D2 by alignment to KIR2DL1, or the converse, are indicated by hyphens. Cysteines are shown in purple, conserved structural motifs in red, the interdomain linker in blue, and potential N-glycosylation sites in green. Bold type in D1D2 indicates the sites of human-to-murine substitutions. Bold type in KIR2DL1 indicates the sites of interaction with HLA-peptide complex.21 (B) Ribbon diagram of hD1D2 model showing the relative positions of residues in hD1D2 that were mutated.
Predicted secondary structure of Ig-like domains of GPVI, compared with a KIR. (A) Alignment of hD1D2 and mD1D2 to KIR2DL1 (pdb code 1NKR42 ). Residue numbers are shown above each sequence, either hD1D2 (identical to mD1D2) or KIR2DL1. Identity between sequences is indicated by a star, either below mD1D2 (conserved in human and murine D1D2) or below KIR2DL1 (identity with those conserved residues in D1D2). Predicted β-strands of D1D2 are shown as arrows above hD1D2 (dark blue for D1, light blue for D2). Actual β-strands in 1NKR are shown as white arrows below KIR2DL1. Gaps produced in D1D2 by alignment to KIR2DL1, or the converse, are indicated by hyphens. Cysteines are shown in purple, conserved structural motifs in red, the interdomain linker in blue, and potential N-glycosylation sites in green. Bold type in D1D2 indicates the sites of human-to-murine substitutions. Bold type in KIR2DL1 indicates the sites of interaction with HLA-peptide complex.21 (B) Ribbon diagram of hD1D2 model showing the relative positions of residues in hD1D2 that were mutated.
Binding of D1D2 to collagenlike peptides measured in a plate assay. Binding of hD1D2 to CRP (▪) or GPP10 (□). Binding of mD1D2 to CRP (•) or GPP10 (○). Mean absorbance ± SD is shown for triplicate wells in a single experiment, representative of at least 3 independent experiments in each case.
Binding of D1D2 to collagenlike peptides measured in a plate assay. Binding of hD1D2 to CRP (▪) or GPP10 (□). Binding of mD1D2 to CRP (•) or GPP10 (○). Mean absorbance ± SD is shown for triplicate wells in a single experiment, representative of at least 3 independent experiments in each case.
Homology modeling of the Ig-like domains of GPVI
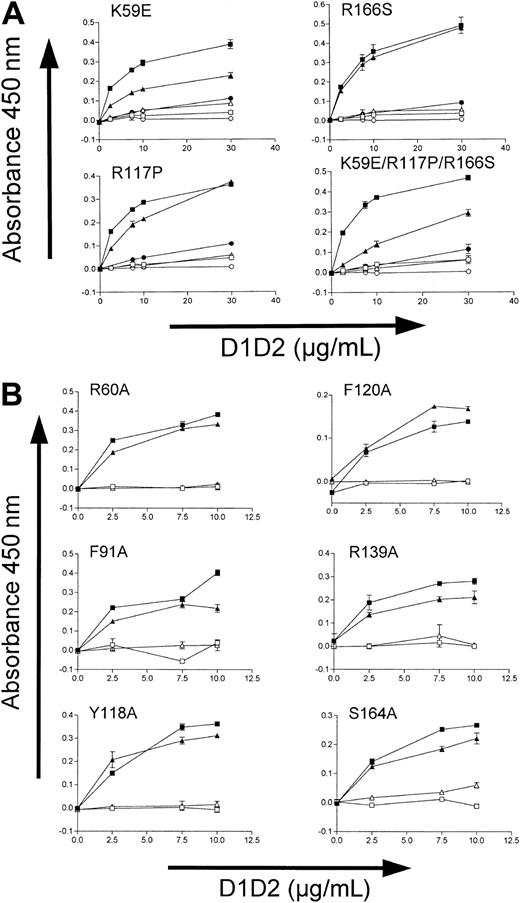
The difference in affinity of mD1D2 and hD1D2 for CRP (Figure 2) prompted us to examine the molecular models of the Ig-like domains for surface-exposed residues that might be responsible for the difference in affinity. The alignment of mD1D2 to hD1D2 is ungapped with 78% identity between species (calculated according to Jandrot-Perrus et al9 ). This revealed a number of interesting residue differences between hD1D2 and mD1D2 (Figure 1A-B). Alignment with KIR2DL1, in which the ligand-binding residues are marked (Figure 1A; taken from Fan et al21 ), highlights the corresponding apical loops in D1D2. We postulated that differences in the amino acid sequence of the apical loops are the basis for differences in binding affinity for CRP between hD1D2 and mD1D2. We identified 9 major surface-exposed residues in the apical loops (Figure 1A). For 3 of these (59, 117, and 166) there were major differences in the nature of the residue between species (Figure 1A-B). Ten mutant GPVI molecules were expressed. Four molecules were the result of the replacement of human residues by their murine counterpart (K59E, R117P, R166S, and a “triple mutant” with all 3) and, for the remaining 6, the residues in positions R60, F91, Y118, F120, R139, and S164 were replaced by alanine.
K59E substitution reduces binding to CRP and 10B12
Consistent with the behavior of the wild-type proteins, all mutants clearly demonstrated higher levels of binding to CRP than to GPP10 (Figure 3). The binding to CRP of 3 of the 4 human-to-mouse mutants was significantly reduced (Figure 3A). The most profound reduction of binding was observed with the triple mutant (K59E, R117P, R166S), and the only other one with major reduction of binding was K59E. The R117P replacement had only a minor effect. For all but one of the 6 alanine mutants (R60, F91, Y118, F120, S164) binding to CRP was unaltered, and the R139A replacement caused only a minor reduction of binding (Figure 3B).
Binding of hD1D2 mutants to collagenlike peptides in a plate assay. (A) Binding of the 4 human-to-murine D1D2 mutants to CRP (▴) or GPP10 (▵), compared with the binding of wild-type hD1D2 to CRP (▪) or to GPP10 (□), and of wild-type mD1D2 to CRP (•) or to GPP10 (○). (B) Binding of hD1D2 (wild type, ▪) and the 6 hD1D2 “alanine” mutants (▴) to CRP or to GPP10 (□ and ▵ for wild type and mutants, respectively) at a range of concentrations of hD1D2. Mean ± SD for 3 separate experiments.
Binding of hD1D2 mutants to collagenlike peptides in a plate assay. (A) Binding of the 4 human-to-murine D1D2 mutants to CRP (▴) or GPP10 (▵), compared with the binding of wild-type hD1D2 to CRP (▪) or to GPP10 (□), and of wild-type mD1D2 to CRP (•) or to GPP10 (○). (B) Binding of hD1D2 (wild type, ▪) and the 6 hD1D2 “alanine” mutants (▴) to CRP or to GPP10 (□ and ▵ for wild type and mutants, respectively) at a range of concentrations of hD1D2. Mean ± SD for 3 separate experiments.
Human phage antibodies against different epitopes on GPVI
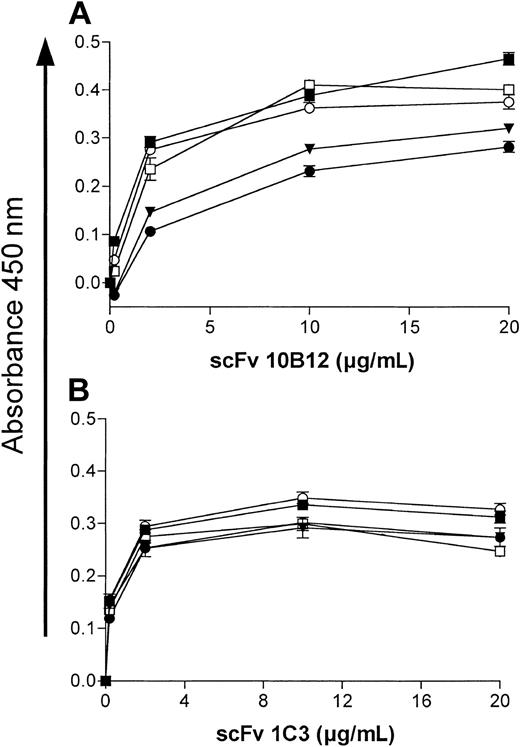
Two human scFvs specific for GPVI were obtained by selecting 2 phage display libraries with hD1D2. By ELISA, both antibodies (10B12 and 1C3) showed specific binding to hD1D2 but not to a set of unrelated protein antigens (Figure 4A). scFv 1C3 did not bind the isolated domains of hGPVI but did bind to mD1D2, indicating that this antibody recognizes an epitope that is conserved between the 2 species (Figure 4B). Some reactivity between 10B12 and hD1 was observed by ELISA, but there was no reactivity with mD1D2 (Figure 4B). By flow cytometry, specific binding of 10B12 and 1C3 to human platelets (Figure 4C) but not to leucocytes and erythrocytes (data not shown) was observed. Preincubating the scFvs with recombinant hD1D2 inhibited antibody binding in a dose-dependent manner (Figure 4D). The 2 antibodies produced additive and noncompetitive binding to platelets in flow cytometry (data not shown) and to hD1D2 in a biosensor assay (Figure 4E). In the latter experiment the response obtained at saturating levels of antibody was similar for both (462 and 448 arc.sec–1 for 1C3 and 10B12, respectively). Interpretation of the biosensor data on the basis of respective molecular masses of antibody and hD1D2 indicates equimolar binding (734 arc.sec–1 for hD1D2). Further biosensor analysis was conducted to determine the affinity of each scFv. hD1D2 was immobilized by way of the CaM tag, and the Kon for each monomeric scFv over a concentration range from 0.5 to 8.2 μM was determined (data not shown). Dissociation constants (Kd) of 5.4 × 10–7 M for 1C3 and 7.9 × 10–7 M for 10B12 were obtained.
Binding of scFvs to GPVI in ELISA, flow cytometry, and biosensor assays. Bacterial culture supernatants of clones 10B12 and 1C3 were added to wells prepared as follows: (A) Wells were directly coated with a panel of antigens (hD1D2 and irrelevant scFv were both CaM-tagged, lysozyme, thyroglobulin, IV Ig = intravenous immunoglobulin G, soluble collagen type I, streptavidin) or none (plate), before blocking with BSA. (B) Wells were precoated with BSA-N9A, and then buffer alone (plate) or buffer containing CaM-tagged antigens (hD1D2, irrelevant scFv, hD1, hD2, mD1D2) was added, capturing them to the solid phase. (A-B) The mean ± SD is shown for triplicate wells in a single experiment, representative of at least 3 independent experiments. (C) Fluorescence histograms of fresh human platelets incubated with scFv 1C3 (white area) or scFv 10B12 (light gray area) at saturating concentrations, or secondary antibody only (dark gray area). (D) scFvs 10B12 (▪), 1C3 (▴), or control scFv (anti-GPIIbIIIa; ○) at 20 μg/mL were preincubated with increasing concentrations of hexahistidine-tagged hD1D2 (hD1D2-His) before binding of scFvs to platelets was measured by flow cytometry. Each line in panel C and each point in panel D are from a single experiment, representative of 2 identical experiments. (E) Resonant mirror sensorgram of the binding of purified scFvs to hD1D2. The immobilization of hD1D2 (arrow 1) is followed by a brief wash to remove excess hD1D2. A saturating amount of scFv 1C3 (5 μL of 4.2 mg/mL) (arrow 2) was then added. Saturation was confirmed by the addition of a second aliquot of 1C3 (arrow 3). Without washing, scFv 10B12 was then added (arrows 4 and 5 mark the addition of 5 μL each of 4.5 mg/mL). The response in arc.sec–1 immediately prior to each addition was subtracted from the response at equilibrium afterward. The results are presented in the text. This is representative of 2 independent experiments.
Binding of scFvs to GPVI in ELISA, flow cytometry, and biosensor assays. Bacterial culture supernatants of clones 10B12 and 1C3 were added to wells prepared as follows: (A) Wells were directly coated with a panel of antigens (hD1D2 and irrelevant scFv were both CaM-tagged, lysozyme, thyroglobulin, IV Ig = intravenous immunoglobulin G, soluble collagen type I, streptavidin) or none (plate), before blocking with BSA. (B) Wells were precoated with BSA-N9A, and then buffer alone (plate) or buffer containing CaM-tagged antigens (hD1D2, irrelevant scFv, hD1, hD2, mD1D2) was added, capturing them to the solid phase. (A-B) The mean ± SD is shown for triplicate wells in a single experiment, representative of at least 3 independent experiments. (C) Fluorescence histograms of fresh human platelets incubated with scFv 1C3 (white area) or scFv 10B12 (light gray area) at saturating concentrations, or secondary antibody only (dark gray area). (D) scFvs 10B12 (▪), 1C3 (▴), or control scFv (anti-GPIIbIIIa; ○) at 20 μg/mL were preincubated with increasing concentrations of hexahistidine-tagged hD1D2 (hD1D2-His) before binding of scFvs to platelets was measured by flow cytometry. Each line in panel C and each point in panel D are from a single experiment, representative of 2 identical experiments. (E) Resonant mirror sensorgram of the binding of purified scFvs to hD1D2. The immobilization of hD1D2 (arrow 1) is followed by a brief wash to remove excess hD1D2. A saturating amount of scFv 1C3 (5 μL of 4.2 mg/mL) (arrow 2) was then added. Saturation was confirmed by the addition of a second aliquot of 1C3 (arrow 3). Without washing, scFv 10B12 was then added (arrows 4 and 5 mark the addition of 5 μL each of 4.5 mg/mL). The response in arc.sec–1 immediately prior to each addition was subtracted from the response at equilibrium afterward. The results are presented in the text. This is representative of 2 independent experiments.
10B12 specifically blocks GPVI interaction with CRP and collagen
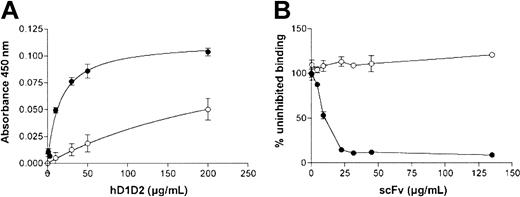
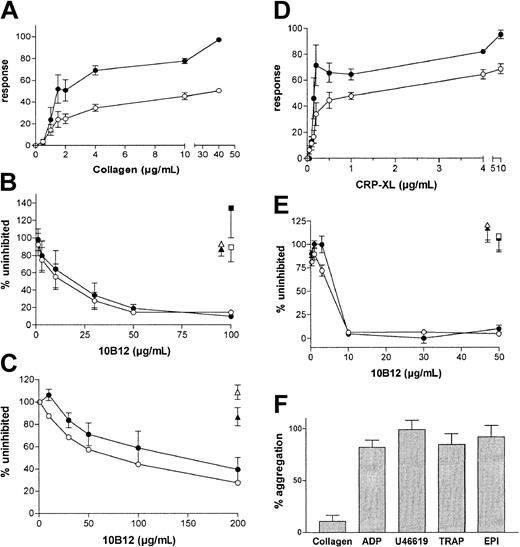
With the ligand-binding assay, we observed that 10B12 dose-dependently inhibits the binding of hD1D2 to CRP with an IC50 of 14 ± 3 μg/mL (n = 3), whereas 1C3 does not (data not shown). In addition, the binding of hD1D2 to collagen fibers (Figure 5A) was completely inhibited by 10B12 (Figure 5B). The effects of scFvs on the response of human platelets to collagen and to CRP-XL were then tested by platelet aggregometry. First, the dose-response to collagen and CRP-XL was determined with platelets from 3 individuals (Figure 6A,D). Subsequently, the ability of scFv 10B12 to inhibit aggregation was tested at a 4-μg/mL concentration of agonist (Figure 6B,E). When a higher concentration of collagen fibers was used (10 μg/mL), complete inhibition was not obtained with platelets from all individuals (Figure 6C). At a concentration of 200 μg/mL 10B12, the response was 28% ± 4% (slope) and 40% ± 11% (extent) of aggregation without scFv (means ± SEM, n = 8). The inhibitory effect of 10B12 on CRP-XL–mediated aggregation occurs rather dramatically between 5 and 10 μg/mL antibody (0.2-0.4 μM; Figure 6E). This finding is close to the Kd of 10B12 and consistent with a large increase in receptor occupancy at that concentration. When compared with CRP-XL, a smoother profile of inhibition was observed when collagen was used to induce aggregation (Figure 6B-C). In contrast to its inhibitory effect on CRP-XL and collagen-mediated aggregation, 10B12 did not block aggregation induced by non-GPVI agonists (Figure 6F).
Binding of hD1D2 to collagen and inhibition by scFv 10B12. (A) Binding of hD1D2 to collagen I fibers (•) or GPP10 (○). Mean absorbance ± SD is shown for triplicate wells in a single experiment, representative of at least 3 independent experiments. (B) Binding of hD1D2 (10 μg/mL) to collagen fibers, in the presence of scFv 10B12 (•) or 2D4 (anti-HLA-A2; ○). The binding in the presence of scFv is shown as a percentage of the signal obtained in the absence of scFv. Mean ± SD is shown for triplicate wells in a single experiment, representative of at least 3 independent experiments.
Binding of hD1D2 to collagen and inhibition by scFv 10B12. (A) Binding of hD1D2 to collagen I fibers (•) or GPP10 (○). Mean absorbance ± SD is shown for triplicate wells in a single experiment, representative of at least 3 independent experiments. (B) Binding of hD1D2 (10 μg/mL) to collagen fibers, in the presence of scFv 10B12 (•) or 2D4 (anti-HLA-A2; ○). The binding in the presence of scFv is shown as a percentage of the signal obtained in the absence of scFv. Mean ± SD is shown for triplicate wells in a single experiment, representative of at least 3 independent experiments.
Platelet aggregation induced by collagen and CRP-XL is selectively inhibited by scFv 10B12. (A) Aggregation in response to increasing doses of collagen I fibers. (B) Effect of scFvs on response to 4 μg/mL collagen I fibers or (C) 10 μg/mL collagen I fibers. (D) Aggregation in response to increasing doses of CRP-XL. (E) Effects of scFvs on response to 4 μg/mL CRP-XL. (B,C,E) scFvs are shown by 10B12 (circles), 2D4 (anti-HLA A2; triangles), and 1C3 (squares, in panels B and E only). (F) Effect of scFv 10B12 on response to moderate doses of various agonists; collagen I fibers 1 μg/mL, ADP 5 μM, U46619 5 μM, EPI 4 μM, TRAP 10 μM. (A-E) The maximal rate of aggregation or “slope” is shown in open symbols, whereas the extent of aggregation is shown in closed symbols. (B,C,E,F) The response with scFv 10B12 is shown as a percentage of the response without scFv. All points or bars represent the mean ± SEM of measurements from at least 3 separate donors.
Platelet aggregation induced by collagen and CRP-XL is selectively inhibited by scFv 10B12. (A) Aggregation in response to increasing doses of collagen I fibers. (B) Effect of scFvs on response to 4 μg/mL collagen I fibers or (C) 10 μg/mL collagen I fibers. (D) Aggregation in response to increasing doses of CRP-XL. (E) Effects of scFvs on response to 4 μg/mL CRP-XL. (B,C,E) scFvs are shown by 10B12 (circles), 2D4 (anti-HLA A2; triangles), and 1C3 (squares, in panels B and E only). (F) Effect of scFv 10B12 on response to moderate doses of various agonists; collagen I fibers 1 μg/mL, ADP 5 μM, U46619 5 μM, EPI 4 μM, TRAP 10 μM. (A-E) The maximal rate of aggregation or “slope” is shown in open symbols, whereas the extent of aggregation is shown in closed symbols. (B,C,E,F) The response with scFv 10B12 is shown as a percentage of the response without scFv. All points or bars represent the mean ± SEM of measurements from at least 3 separate donors.
In a perfusion system designed to more closely represent vascular blood flow, anticoagulated whole blood was treated with scFvs and perfused at low (300s–1) or high (1600s–1) shear rate over a surface coated with soluble type III collagen. Here, 10B12 inhibited thrombus formation in a dose-dependent fashion, whereas 1C3 and a non-GPVI control scFv did not (Figure 7A-B). The dose of antibody required to achieve 50% reduction in thrombus formation was similar at each shear rate.
Effects of scFvs on thrombus formation in whole blood. (A) Anticoagulated whole blood, preincubated with scFvs 10B12 (circles), 1C3 (squares), or a control 2D4 (anti-HLA A2; triangles) was passed over collagen III–coated coverslips at low (300s–1; open symbols) or high shear (1600s–1; closed symbols). Data are expressed as a percentage of surface coverage in the absence of scFv at each shear rate and represent the mean ± SD for 3 separate donors. (B) Micrographs of surface coverage obtained at high shear in the presence of 0, 3, or 50 μg/mL scFv 10B12. Individual platelets are seen as the smaller dots in the right-hand boxes.
Effects of scFvs on thrombus formation in whole blood. (A) Anticoagulated whole blood, preincubated with scFvs 10B12 (circles), 1C3 (squares), or a control 2D4 (anti-HLA A2; triangles) was passed over collagen III–coated coverslips at low (300s–1; open symbols) or high shear (1600s–1; closed symbols). Data are expressed as a percentage of surface coverage in the absence of scFv at each shear rate and represent the mean ± SD for 3 separate donors. (B) Micrographs of surface coverage obtained at high shear in the presence of 0, 3, or 50 μg/mL scFv 10B12. Individual platelets are seen as the smaller dots in the right-hand boxes.
Residue K59 is critical to 10B12 binding
scFv 10B12 but not 1C3 shows some binding to hD1 (Figure 4B) and both antibodies bind different epitopes (Figure 4E). To obtain more information on the epitopes recognized by each antibody, reactivity was tested with all 10 mutant hD1D2 proteins. Binding to the alanine mutants by ELISA was identical to that of wild-type hD1D2 (data not shown). However, binding of 10B12 to the `triple mutant' and to the K59E mutant was strongly reduced (Figure 8A). Not unexpectedly, the binding of 1C3 to these 2 mutants was unaltered (Figure 8B). In other words, the substitution of a lysine (K) for a glutamate (E) at position 59 of hD1D2 not only reduced the affinity for CRP but also reduced the binding of the inhibitory clone 10B12.
Binding of scFvs 10B12 and 1C3 to hD1D2 and hD1D2 mutants by ELISA. (A) Binding of 10B12 to hD1D2 (▪), hD1D2-117P (□), hD1D2-166S (○), hD1D2-59E (•), and triple mutant (▾). (B) Binding of 1C3 to the antigens as represented in panel A. Points represent mean ± SD of triplicate wells in 1 experiment, representative of 2 identical experiments.
Binding of scFvs 10B12 and 1C3 to hD1D2 and hD1D2 mutants by ELISA. (A) Binding of 10B12 to hD1D2 (▪), hD1D2-117P (□), hD1D2-166S (○), hD1D2-59E (•), and triple mutant (▾). (B) Binding of 1C3 to the antigens as represented in panel A. Points represent mean ± SD of triplicate wells in 1 experiment, representative of 2 identical experiments.
The third loop (H3) of the VH domain of 10B12 is highly acidic
The amino acid sequence of the VH and VL domains of 1C3 and 10B12 were derived from the nucleotide sequences. These sequences have been submitted to the GenBank database under accession numbers 10B12VH, AF539528; 10B12Vκ, AF539529; 1C3VH, AF539530; and 1C3Vλ, AF539531. A number of features of the variable domain of the heavy chain (VH) of 10B12 are noteworthy. First, the VH gene encoding the 10B12 VH domain is in near germ line configuration and has a 97.8% sequence homology with the VH3-30 gene. Second, as a consequence of mutations in the VH gene, highly conserved residues at positions 93 and 94 were mutated (A to T and K to D, respectively). This is of particular interest because the latter is involved in the formation of a conserved salt-bridge in the third hypervariable loop (H3) of the VH domain.44 Third, although the H3 loop is of average length, its sequence is highly acidic with 3 aspartates in addition to the one at position 94 and a glutamate at position 98 (sequence of the H3 loop, C92TDGWAEMA100T100aTDDAF100fD101IWG104 and Figure 9A,E). The sequence of the Vκ domain of clone 10B12 has a 87.6% sequence homology with the VκI gene L12a. The L1 loop of the Vκ domain contains an additional acidic residue at position 27 (Figure 9A,E). For 1C3, the VH domain is 91.3% homologous to the VH5-51 gene of the VH5 family, and the Vλ domain is encoded by a hybrid of 2 Vλ genes (1b and 2a2). This hybrid has probably arisen by PCR crossover during creation of the VL gene repertoires.
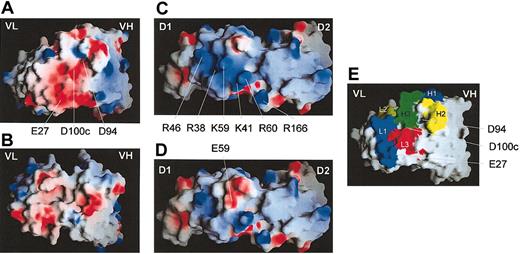
Modelling of scFvs, hD1D2, and hD1D2-59E. (A) The antigen-binding site of scFv 10B12. (B) The antigen-binding site of scFv 1C3; VH, variable domain of the heavy chain, VL domain, variable domain of the light chain. (C) The apical surface of hD1D2. (D) The apical surface of hD1D2-59E. (E) The antigen binding loops H1-3 and L1-3 (according to Chothia et al45 ) of scFv 10B12. Residues pertinent to the discussion are labeled. Numbering of residues in scFv 10B12 are according to Kabat et al.46 Images were produced using the GRASP program39 and are colored for surface charge (red = negative, blue = positive) as described in the text.
Modelling of scFvs, hD1D2, and hD1D2-59E. (A) The antigen-binding site of scFv 10B12. (B) The antigen-binding site of scFv 1C3; VH, variable domain of the heavy chain, VL domain, variable domain of the light chain. (C) The apical surface of hD1D2. (D) The apical surface of hD1D2-59E. (E) The antigen binding loops H1-3 and L1-3 (according to Chothia et al45 ) of scFv 10B12. Residues pertinent to the discussion are labeled. Numbering of residues in scFv 10B12 are according to Kabat et al.46 Images were produced using the GRASP program39 and are colored for surface charge (red = negative, blue = positive) as described in the text.
Modelling of the 10B12 interface with hD1D2
Molecular models of 10B12 and 1C3 and wild-type hD1D2 and the hD1D2-59E mutant were generated to gain further insight into the epitopes recognized by both scFvs (Figure 9A-D). For 10B12, the highly acidic H3 loop forms part of a negatively charged region in the antigen-binding surface of the antibody (Figure 9A,E). This surface has both charge and shape complementarity with a basic multilobed patch on the apical surface of hD1D2 (Figure 9C). The modeling suggests that this lobed patch is at the center of the 10B12 epitope. The basic patch on hD1D2 appears to be formed by R38, K41, R46, K59, and R60 (Figures 1A and 9C). The K59E mutation reduces the basic nature of the patch, whereas the surrounding features are preserved (Figure 9D), possibly explaining the reduced binding of 10B12 to hD1D2-59E. From ELISA data, 10B12 reacts with hD1 but not with hD2 (Figure 4B). However, this reactivity is considerably reduced compared with hD1D2, suggesting that residues outside D1 also contribute to the 10B12 epitope. The model generated is compatible with this observation, as the footprint of the antibody would cover the interdomain hinge and extend to the apical surface of D2. Further biochemical analysis is required to verify the proposed model of the GPVI-antibody interface.
Discussion
GPVI is the major platelet signaling receptor for collagen.8,47,48 The studies described here were designed to better define the interaction between collagen and GPVI, by exploiting the unique feature that GPVI specifically recognizes the synthetic collagen-related peptide (CRP). This ligand is thought to interact with the primary binding site for collagen on GPVI.26,27 When testing human and murine recombinant forms of D1D2 for their ability to bind to CRP, we observed a surprising difference in the strength of binding (Figure 2). That the affinity of hGPVI for CRP is at least a log higher than that of mGPVI has not been observed before in platelet aggregation tests with CRP-XL. There are several reasons why this difference has not been observed when studying platelets. First, the membrane organization of GPVI in platelets of mice and humans may be different. For example, the cytoplasmic domain of hGPVI contains signaling motifs to bind CaM and kinases,18,19 which are incomplete in mGPVI (full sequences in Jandrot-Perrus et al9 ). Second, in humans, minor differences in the sequence of GPVI have a significant effect on the expression level of GPVI and on the responsiveness of platelets to CRP-XL.36 Third, our study of CRP and D1D2 binding is based on the use of monomeric components. CRP is a solution of monomeric triple helices, whereas a cross-linked form is traditionally used to activate platelets in suspension.24 The recombinant D1D2 molecules are monomeric, whereas cellular GPVI can cluster and may operate as a dimer.49
It was hypothesized that the difference in strength of binding was due to residue differences in the CRP binding site between hD1D2 and mD1D2. Co-crystallography of KIR with HLA-C20 revealed that the recognition of the HLA α helices is by residues at the apical surface of the KIRs. Clemetson et al6 suggested in their GPVI cloning study that a similar surface of GPVI could be used for the recognition of the helical collagen structure. When comparing the sequence of the apical surfaces of hD1D2 and mD1D2, a number of critical differences were apparent, in particular at residues 59, 117, and 166. Each of these murine residues (E59, P117, S166) was transferred into the human backbone, and 6 alanine substitutions at residues 60, 91, 118, 120, 139, and 164 were performed. The effect of each mutation on CRP binding was observed (Figure 3). The K59E mutation was the only one that caused a profound reduction in CRP binding. In our model of GPVI, the K59E mutation produces localized changes at the apical surface of the first domain (Figure 9C-D), suggesting that this surface is required for CRP binding. The reversal of the charge at residue 59 from positive to negative may in part explain the difference in affinity of human and murine D1D2 for CRP. However, a molecular mechanism for CRP binding on the basis of these observations cannot be proposed.
In a parallel approach antibody V gene phage display technology was used. With the use of a classic phage selection method and a novel one with CRP as competitor, 2 human scFvs (clones 1C3 and 10B12) specific for GPVI were obtained. The latter was selected by its ability to displace hD1D2 from CRP. This resulted in its property as an scFv to selectively block the hD1D2-collagen interaction (Figure 5), as well as CRP-XL and collagen evoked platelet activation in plasma and in flowing whole blood (Figures 6, 7). Platelet aggregation induced by CRP was more sensitive to inhibition by 10B12 than was aggregation induced by collagen fibers (Figure 6). This finding concurs with the work of others using the JAQ-1 antibody to block GPVI on murine platelets.26 Thrombus formation on a collagen surface was inhibited by 10B12, confirming the importance of GPVI in this process and supporting previous studies on human platelets.50,51 In contrast, scFv 1C3, although having a similar affinity for hD1D2, was unable to produce inhibition. That an scFv like 10B12, with a moderate, submicromolar affinity, can effectively inhibit collagen-mediated platelet activation is compatible with the recent observation that the affinity of recombinant dimeric GPVI for fibrous collagen is 5.8 × 10–7 M.49
By using mD1D2, hD1D2, and hD1D2 mutants as antigens for the scFvs in ELISA and biosensor assays, we were able to attribute the functional differences of the scFvs to separate epitopes. First, both antibodies bind GPVI noncompetitively (Figure 4E). 10B12 retains some binding to the first domain on its own, but 1C3 does not (Figure 4B). Second, the noninhibitory scFv 1C3 does not bind the first domain but reacts with an epitope in part conserved between human and mouse. Finally, the binding of 10B12 of both the K59E and the `triple' mutant is strongly reduced (Figure 8A), whereas none of the other 8 mutations have a significant effect on antibody binding (data not shown).
The model of the GPVI-10B12 interface supports the notion that the first domain makes an important contribution to the epitope recognized by 10B12. Reliable modeling was possible as GPVI has a 39% to 45% sequence identity to the KIRs, and structural data of several antibodies were available to model the binding sites of both GPVI antibodies. From primary sequence analysis of the V domains, the presence of an unusual acidic sequence in the H3 loop of 10B12 was surprising. The H3 itself has 4 aspartates and one glutamate but no basic residues. In particular, the aspartate at the carboxy-terminal end of framework 3 at position 94 is in place of arginine or lysine that is more typical for this position. The basic residue at position 94 is required to form a salt bridge with D101 and stabilize this loop.44 These factors combined make it more likely that the H3 loop would be unusually flexible and require considerable charge neutralization by the antigen. The model of the binding interface between hD1D2 and 10B12 supports this assumption. It reveals a patch of basic residues on the GPVI surface (Figure 9C) that would offer a suitable docking site for the acidic H3 loop (Figure 9A,E). The basic patch would be disrupted by a charge reversal at position 59 (Figure 9D), compatible with the detrimental effect of the K59E mutation on 10B12 binding. This model supports the notion as K59 extends from the loop joining strands E-F in D1, near the interdomain linker (Figure 1B), and linker residues would face Vκ domain residues (Figure 9E). Therefore, the 10B12 epitope maps to the apical surface of the first domain and the interdomain residues of GPVI. Compared with IgG antibodies the inhibitory scFv has a relatively small footprint. Therefore, it is postulated that the 10B12 epitope overlaps with the CRP binding site to a large degree.
Another inhibitory human scFv (clone A10) has recently been reported.52 Whether the epitope recognized by this antibody is the same as the 10B12 epitope is not known, as no epitope mapping studies were performed with A10. Direct binding studies like the ones we have performed with 10B12 and 1C3 will be needed to answer this question.
To conclude, there is converging evidence to localize the CRP/primary collagen-binding surface of GPVI to the area where the first domain meets the interdomain linker. Structural studies of GPVI, either on its own or combined with 10B12 or CRP should permit the precise identification of residues underlying these interactions and lead to the development of GPVI antagonists. Such studies are currently in progress in our laboratory.
Prepublished online as Blood First Edition Paper, September 22, 2003; DOI 10.1182/blood-2003-01-0308.
Supported by the British Heart Foundation (P.A.S.), the Medical Research Council (R.W.F., C.G.K., T.R.D., A.B., and E.W.), and the National Blood Service R&D (L.J.-K., S.F.G., and W.H.O.).
P.A.S. and L.J.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank L. Morton, D. Onley, I. Harmer, P. Siljander, V. Dhanaraj (now sadly deceased), and C. Chothia for helpful discussions and Prof J. P. Allain for editorial help.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal