Abstract
The hypothesis that increased ADAMTS13 (a disintegrin and metalloprotease with thrombospondin repeats) activity or increased susceptibility of von Willebrand factor (VWF) to proteolysis by ADAMTS13 may underlie type I von Willebrand disease (VWD) in some patients was investigated. Plasma from 4 patients with type I VWD was cryoprecipitated. ADAMTS13 activity in the VWF-poor cryodepleted fraction was assessed by incubation with purified VWF; susceptibility to proteolysis of the VWF in the VWF-rich cryoprecipitate was assessed by incubation with a normal, group O cryodepleted plasma. ADAMTS13 activity was similar in all 4 type I VWD cryodepleted plasmas and comparable to a normal control plasma. In contrast, the VWF of one patient showed increased susceptibility to proteolysis by ADAMTS13. Investigation of additional family members indicated that increased susceptibility was heritable, but it did not track uniquely with type I VWD. Sequence analysis of VWF exon 28 indicated that increased susceptibility to proteolysis tracked with the “G” allele of the A/G polymorphism at position 24/1282, encoding the amino acid polymorphism Tyr/Cys1584 (“G” = Cys1584). A prospective study of 200 individuals yielded 2 Tyr/Cys1584 heterozygotes; for both, plasma VWF showed increased susceptibility to proteolysis. The finding that an amino acid polymorphism in VWF may influence susceptibility to ADAMTS13 has potentially significant implications in diverse areas. (Blood. 2004;103:941-947)
Introduction
The plasma glycoprotein von Willebrand factor (VWF) circulates as a series of homopolymers (multimers) of increasing size, ranging from 500 kDa to over 20 000 kDa.1 Each VWF monomer within a multimer is a potential target for proteolysis by the VWF metalloprotease ADAMTS13 (a disintegrin and metalloprotease with thrombospondin repeats), which proteolyses VWF at the peptide bond Tyr1605-Met1606.2,3 This bond occurs in the A2 domain of VWF (amino acid residues 1480-1682) and is flanked by domains that bind platelet glycoprotein Ib (the A1 domain, residues 1260-1479)4 and collagen (the A3 domain, residues 1683-1874)5 (numbering from the start methionine of pre-pro-VWF6 ).
The degree of polymerization of plasma VWF appears to be regulated by this specific proteolysis. A deficiency or dysfunction of ADAMTS13 results in the presence of high-molecular-weight (HMW) and ultra-HMW multimers in the circulation and is the molecular basis for thrombotic thrombocytopenic purpura (TTP).3 Mutations in the ADAMTS13 gene, located on chromosome 9 at q34,7 which lead to a dysfunctional or inactive molecule have been characterized in hereditary TTP,7 whereas autoantibodies against ADAMTS13 have been demonstrated in acquired TTP.8-10
Measurement of ADAMTS13 activity for the diagnosis of TTP is typically done indirectly, for example, using a Western blotting approach to assess the increase in the 176-kDa dimer released on proteolysis,10 or using a collagen-binding assay,11 or VWF multimer analysis.12 Comparison of the VWF collagen-binding capacity (VWF:CB) or multimer profile at the start of the incubation with that at the end gives a measure of VWF proteolysis, the VWF:CB or HMW multimers decreasing as proteolysis proceeds. These approaches are used in many investigations involving the measurement of ADAMTS13 activity.
The VWF:CB is also used in the diagnosis of von Willebrand disease (VWD); an aberrant ratio of VWF:CB to VWF antigen (VWF:Ag; < 0.7) is suggestive of a qualitative VWF defect.13 A second phenotypic assay used in the diagnosis of VWD measures the functional ability of VWF to bind platelets—the ristocetin cofactor agglutination activity (VWF:RCo). An aberrant ratio of VWF:RCo to VWF:Ag (< 0.7) is also suggestive of a qualitative VWF defect.13 Both the VWF:CB and the VWF:RCo are dependent on the multimeric size of VWF and may decrease as a result of amino acid substitutions that destabilize VWF, causing its increased proteolysis, as observed in type IIA VWD.2,14
Although type IIA VWD represents a pathologic state arising from increased susceptibility of VWF to proteolysis, a pathologic state associated with elevated ADAMTS13 activity has not yet been described. The recent finding that blood group may influence the susceptibility of VWF to proteolysis by ADAMTS13 and that, where susceptibility is highest VWF level is lowest (in group O), leads to the possibility that elevated ADAMTS13 activity may be associated with decreased circulating VWF.15 This has been proposed as a possible mechanism for type I VWD.15 An alternative mechanism would be increased susceptibility of VWF itself to proteolysis by ADAMTS13. These possibilities were explored in the present study, which revealed an amino acid polymorphism in VWF that correlates with increased susceptibility to proteolysis by ADAMTS13. Increased susceptibility to proteolysis was not uniquely associated with type I VWD but was also found among healthy control subjects.
Patients and methods
Patients
Four unrelated patients were tested. All patients had a personal history of mucosal bleeding and decreased VWF:Ag, VWF:RCo, and VWF:CB tested on at least 3 separate occasions. The ratios of VWF:Ag/VWF:RCo and VWF:Ag/VWF:CB were more than 0.7, except for patient M for whom borderline or marginally discordant ratios (> 0.6) were sometimes obtained. The VWF multimer profile was normal in all cases.
Local cohort
Citrated whole blood that was in excess of that required from routine blood tests of 200 individuals was anonymizd and tested in accordance with Medical Research Council (MRC) guidelines (http://www.mrc.ac.uk/txt/pdf-tissue_guide_fin.pdf “Human tissue and biological samples for use in research”). Samples from the hemostasis and thrombosis clinics were excluded.
Phenotypic analysis
VWF multimer analysis was done using sodium dodecyl sulfate (SDS)-agarose gel electrophoresis as previously described.16 VWF:Ag17 and the VWF:CB15 were measured using enzyme-linked immunosorbent assay (ELISA), and VWF:RCo was measured using platelet aggregrometry.18 Factor VIII activity (FVIII:C) was measured using a 1-stage clotting assay.
Genotypic analysis
Genomic DNA (gDNA) was extracted using rapid lysis.19 The relevant region of exon 28 of the VWF gene was amplified by the polymerase chain reaction (PCR) using primers K2A and K1B (Table 1).
Primers used for genetic investigations
Primer . | Sequence (5′-3′) . | Position . |
|---|---|---|
| K1A | TCT GTG GGA ATA TGG AAG TCA TTG | 24/111-24/134 |
| K1B | AAG CCA GGA TTA GAA CCC GAG TCG | 24/1692-24/1666 |
| K2A | AAC TCC ATG GTT CTG GAT GTG GCG TTC | 24/1008-24/1034 |
| N1A | ACT GTA GGG CTC AGA AGT GTC CAC | 24/138-24/161 |
| N1B | TCG TAT CTT GGC AGA TGC ATG TAG | 24/1669-24/1645 |
Primer . | Sequence (5′-3′) . | Position . |
|---|---|---|
| K1A | TCT GTG GGA ATA TGG AAG TCA TTG | 24/111-24/134 |
| K1B | AAG CCA GGA TTA GAA CCC GAG TCG | 24/1692-24/1666 |
| K2A | AAC TCC ATG GTT CTG GAT GTG GCG TTC | 24/1008-24/1034 |
| N1A | ACT GTA GGG CTC AGA AGT GTC CAC | 24/138-24/161 |
| N1B | TCG TAT CTT GGC AGA TGC ATG TAG | 24/1669-24/1645 |
Amplification was done using AmpliTaq DNA polymerase (PE-Applied Biosystems, Warrington, United Kingdom) in the buffer supplied by the manufacturer supplemented with 1.5 mM MgCl2. K2A and K1B were used at 0.2 μM. The PCR comprised 35 cycles of 94°C for 30 seconds, 65°C for 1 minute, and 72°C for 1 minute.
The Tyr/Cys1584 amino acid polymorphism results from an A/G single nucleotide polymorphism (SNP) at nucleotide 24/1282 (http://www.shef.ac.uk/VWF/polymorphisms/ex28.html), which affects a KpnI restriction site (GGTACC, A = KpnI+ and encodes tyrosine, G = KpnI- and encodes cysteine). Thus, the 682-base pair (bp) K2A-K1B PCR product was genotyped at codon 1584 by digestion with KpnI; KpnI+ yielded fragments of 276 + 406 bp, for KpnI- the PCR product remained undigested at 682 bp.
Nucleotide sequence analysis
DNA extracted by rapid lysis was amplified using nested PCR: outer primers K1A and K1B (Table 1), product size 1579 bp, inner primers N1A and N1B (Table 1), product size 1531 bp.
Amplification was as described (see “Genotypic analysis”) using 30 cycles of 94°C for 30 seconds, 62°C for 1 minute, 72°C for 3 minutes, with the addition of 5 s/cycle to the extension step.
The nested N1A + N1B product, containing exon 28 of the VWF gene plus flanking sequences, was purified using the HighPure kit (Boehringer Mannheim, Nottingham, United Kingdom) and then sequenced using BigDye (PE-Applied Biosystems) under conditions recommended by the manufacturer. Sequencing products were electrophoresed on an Applied Biosystems 3777 Genetic Analyzer.
Primers K1A, K2A, and K1B contain 3′ mismatches against the VWF pseudogene (Table 1). These, together with the stringent hybridization temperatures used in the respective PCRs, minimize the risk of obtaining contaminating products from the VWF pseudogene. Amplification products were tested for such contamination using nucleotide sequence analysis and no evidence for the presence of pseudogene sequence was found.
Preparation of cryoprecipitate and cryodepleted plasma
An aliquot of citrated plasma (3 mL) was defrosted from -20°C to 0°C on ice in a cold room (4°C) overnight; 750 μL was then decanted into a microcentrifuge tube on ice and the cryoprecipitate harvested by centrifugation at 3000g, 4°C for 20 minutes. Half of the supernatant (cryodepleted plasma) was decanted and stored for subsequent use; the residual supernatant was thoroughly drained from the cryoprecipitate and discarded. The cryoprecipitate was resuspended in 50 μL T buffer (5 mM Tris [tris(hydroxymethyl)aminomethane]-HCl, pH 8.0) at 37°C and stored for subsequent use.
ADAMTS13 reactions
These were done using urea dialysis as previously described.3,15 Briefly, cryodepleted supernatant (the source of ADAMTS13) and resuspended cryoprecipitate (the source of plasma VWF) were mixed in final reaction conditions of 20 mM Tris-HCl, 150 mM NaCl, 10 mM BaCl2, pH 8.0. Within a given experiment, an equal volume of cryodepleted plasma was added to all incubations to ensure an equivalent addition of ADAMTS13. Resuspended cryoprecipitate was added to a give a final concentration of VWF of 0.45 U/mL. The mixture was transferred to microdialysis chambers and dialyzed against Tris-HCl (5 mM), urea (1 M), pH 8.0, at 37°C for the desired time interval. In some incubations, ristocetin was added to a final concentration of 1.5 mg/L. Ristocetin has previously been shown to increase the rate of VWF proteolysis by ADAMTS13.15
To assess the susceptibility of test VWF to proteolysis, the source of ADAMTS13 was cryodepleted plasma from a hemostatically healthy individual of group O. The VWF content of the cryodepleted plasma was shown to be negligible on multimer analysis and less than 1 U/mL using ELISA.
To assess ADAMTS13 activity in test cryodepleted plasma, the VWF used was a gel-filtration purified, HMW fraction obtained as previously described.15 The VWF was added to a final concentration of 0.6 U/mL.
Proteolysis was assessed using either multimer analysis or VWF:CB. In the case of multimer analysis, gels were assessed by visual inspection independently by 2 individuals, and scanning densitometry was used to provide an objective visual read out of the multimer profiles. Scanning densitometry was done using a Bio-Rad model GS-700 Imaging Densitometer (Bio-Rad, Hertfordshire, United Kingdom) and scans were analyzed using Bio-Rad Quantity One software (version 4.2.1).
Comparison of VWF proteolysis for homozygous Tyr/Tyr1584 and heterozygous Tyr/Cys1584 individuals
ADAMTS13 incubations (8 hours, without ristocetin) were done in triplicate using cryoprecipitate prepared from 20 randomly selected individuals homozygous for Tyr1584 and from the 4 Tyr/Cys heterozygous individuals. Proteolysis was quantified using the VWF:CB assay and was expressed as the “percent loss of VWF:CB” (decrease in VWF:CB between T = 0 hours and T = 8 hours expressed as a percentage of the VWF:CB at T = 0 hours). For each sample, the mean percent loss of VWF:CB for the triplicate assays was calculated.
Statistical analysis of data
The mean percent loss of VWF:CB was calculated for the group of 20 Tyr/Tyr1584 homozygotes and for the group of 4 Tyr/Cys1584 heterozygotes. The means were compared using an unpaired t test and the 95% confidence interval for the difference between the means was calculated using standard formulas.
Results
ADAMTS13 activity in plasma from patients with type I VWD
Multimer analysis showed that the cryodepleted plasmas from all 4 patients with type I VWD caused comparable proteolysis of a purified, HMW VWF fraction and this was similar to the proteolysis obtained using cryodepleted plasma from a healthy control individual (data not shown). This result was consistent for proteolysis over an 8-hour interval in the absence of ristocetin and a 4-hour interval in the presence of ristocetin. Thus, ADAMTS13 activity was similar in plasma samples from the 4 patients with type I VWD and a healthy control.
VWF susceptibility to proteolysis
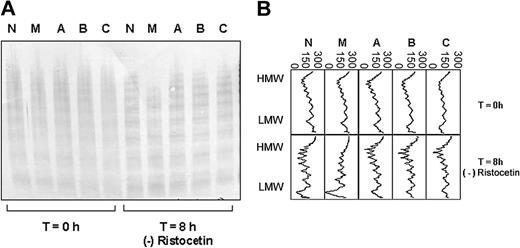
The plasma VWF of 1 of the 4 patients with type I VWD (patient M, female) showed increased susceptibility to proteolysis by ADAMTS13 (Figure 1A-B). This was shown to be reproducible for 2 further samples from this patient, taken on independent occasions (data not shown). Increased susceptibility to proteolysis was observed both in the presence and absence of ristocetin (Figure 3A-B). The plasma VWF of the remaining 3 patients showed similar proteolysis to the normal control plasma, both in the presence (Figure 1A) and absence (not shown) of ristocetin.
ADAMTS13 proteolysis of VWF from 4 unrelated patients with type I VWD compared with a healthy control. (A) Multimer analysis showing the multimer profile of the VWF in each cryoprecipitate before (T = 0 hours) and after (T = 8 hours) incubation with cryodepleted plasma containing ADAMTS13. M, A, B, and C indicate patients with type I VWD; N represents a healthy control. (B) Densitometric scans of the lanes in A. Individual panels correspond with individual lanes. At T = 8 hours, the loss of HMW multimers, gain of LMW multimers, and quantitative increase in the triplet bands are apparent as proteolysis proceeds. The horizontal axis is optical density: 0 = black, 300 = white.
ADAMTS13 proteolysis of VWF from 4 unrelated patients with type I VWD compared with a healthy control. (A) Multimer analysis showing the multimer profile of the VWF in each cryoprecipitate before (T = 0 hours) and after (T = 8 hours) incubation with cryodepleted plasma containing ADAMTS13. M, A, B, and C indicate patients with type I VWD; N represents a healthy control. (B) Densitometric scans of the lanes in A. Individual panels correspond with individual lanes. At T = 8 hours, the loss of HMW multimers, gain of LMW multimers, and quantitative increase in the triplet bands are apparent as proteolysis proceeds. The horizontal axis is optical density: 0 = black, 300 = white.
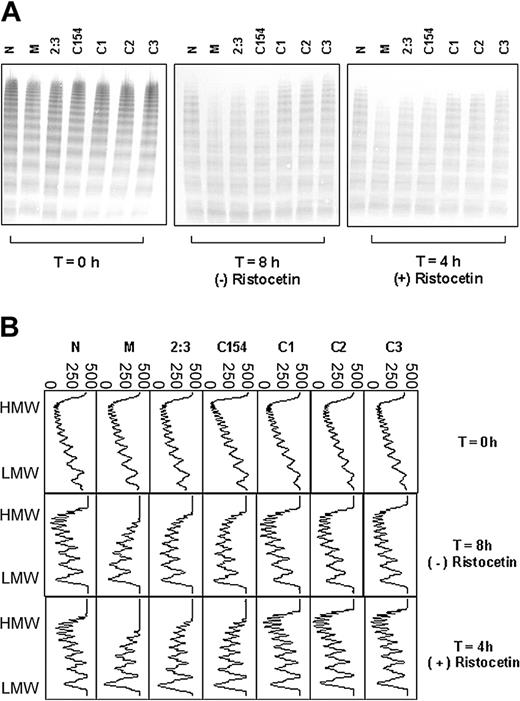
Proteolysis of VWF from Tyr/Cys1584 heterozygotes and randomly selected Tyr/Tyr homozygotes. (A) Multimer analysis of each cryoprecipitate at the start (T = 0 hours) of incubation with cryodepleted plasma as a source of ADAMTS13, and after further incubation in the presence (4 hours) and absence (8 hours) of ristocetin. M represents patient M, 2:3 represents the sister of patient M, C154 and C1-3 represent individuals from the random cohort of 200 people and N represents a healthy control. The extent of proteolysis is greater for M, 2:3, and C154 compared with the other samples tested. (B) Densitometric scans of the lanes in panel A. Each panel corresponds with an individual lane. The multimer profiles of all samples are similar at T = 0 hours; however, the extent of proteolysis for the heterozygotes M, 2:3, and C154 is greater than for the homozygotes C1, C2, and C3 after incubation in the presence or absence of ristocetin. In all cases, proteolysis is accompanied by a quantitative increase in the triplet bands. Horizontal axis is optical density: 0 = black, 500 = white.
Proteolysis of VWF from Tyr/Cys1584 heterozygotes and randomly selected Tyr/Tyr homozygotes. (A) Multimer analysis of each cryoprecipitate at the start (T = 0 hours) of incubation with cryodepleted plasma as a source of ADAMTS13, and after further incubation in the presence (4 hours) and absence (8 hours) of ristocetin. M represents patient M, 2:3 represents the sister of patient M, C154 and C1-3 represent individuals from the random cohort of 200 people and N represents a healthy control. The extent of proteolysis is greater for M, 2:3, and C154 compared with the other samples tested. (B) Densitometric scans of the lanes in panel A. Each panel corresponds with an individual lane. The multimer profiles of all samples are similar at T = 0 hours; however, the extent of proteolysis for the heterozygotes M, 2:3, and C154 is greater than for the homozygotes C1, C2, and C3 after incubation in the presence or absence of ristocetin. In all cases, proteolysis is accompanied by a quantitative increase in the triplet bands. Horizontal axis is optical density: 0 = black, 500 = white.
Control incubations lacking cryodepleted plasma, or urea, or barium ions gave negligible proteolysis (data not shown). This, together with the quantitative increase in the triplet bands (a characteristic feature of ADAMTS13-catalyzed proteolysis) shown by the densitometric scans (Figures 1B, 2C, and 3B), supports the observed degradation of VWF to be due to proteolysis by ADAMTS13. Neither nonspecific proteolysis nor VWF depolymerase (thrombospondin-1) activity would yield an increase in the triplet bands and show a requirement for these specific reaction conditions.
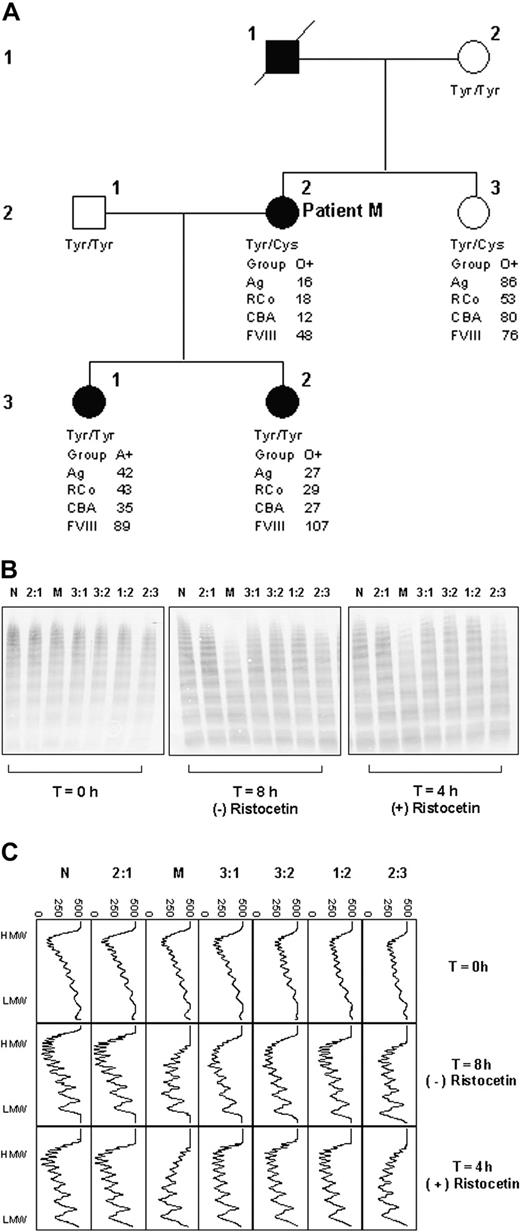
Family study. (A) Pedigree of patient M. □ indicates unaffected male; ▪, affected male; ○, unaffected female; •, affected female; / indicates deceased. Tyr and Cys, respectively, represent tyrosine and cysteine at residue 1584 in VWF, determined by genotype analysis. (B) Multimer analysis showing proteolysis of VWF from each family member. T = 0 hours shows the multimer profile of the VWF in each cryoprecipitate before incubation with cryodepeleted plasma containing ADAMTS13. Incubations were done for fixed time intervals according to the presence (4 hours) and absence (8 hours) of ristocetin. The extent of proteolysis is greater in the case of lanes M and 2:3 both in the presence and absence of ristocetin. (C) Densitometric scans of the lanes in panel B. Each panel corresponds with one lane. In both the presence and absence of ristocetin, the plasma VWF from patient M and sibling 2:3 shows greater proteolysis than that of the other individuals tested. In all cases, proteolysis is accompanied by a quantitative increase in the triplet bands. The horizontal axis is optical density: 0 = black, 500 = white.
Family study. (A) Pedigree of patient M. □ indicates unaffected male; ▪, affected male; ○, unaffected female; •, affected female; / indicates deceased. Tyr and Cys, respectively, represent tyrosine and cysteine at residue 1584 in VWF, determined by genotype analysis. (B) Multimer analysis showing proteolysis of VWF from each family member. T = 0 hours shows the multimer profile of the VWF in each cryoprecipitate before incubation with cryodepeleted plasma containing ADAMTS13. Incubations were done for fixed time intervals according to the presence (4 hours) and absence (8 hours) of ristocetin. The extent of proteolysis is greater in the case of lanes M and 2:3 both in the presence and absence of ristocetin. (C) Densitometric scans of the lanes in panel B. Each panel corresponds with one lane. In both the presence and absence of ristocetin, the plasma VWF from patient M and sibling 2:3 shows greater proteolysis than that of the other individuals tested. In all cases, proteolysis is accompanied by a quantitative increase in the triplet bands. The horizontal axis is optical density: 0 = black, 500 = white.
Family study
Susceptibility of VWF to proteolysis was subsequently investigated in further members of the family of patient M (Figure 2A-C). Whereas both daughters of patient M (3:1 and 3:2; Figure 2A) have type I VWD, their VWF level is higher than that of patient M and their bleeding tendency is clinically less severe. Their VWF did not show increased susceptibility to proteolysis (Figure 2B-C). However, the VWF of patient M's sister (2:3; Figure 2A) did show increased susceptibility to proteolysis (Figure 2B-C), although the sister does not have type I VWD. These observations indicated that increased susceptibility to proteolysis is a heritable trait; however, it did not track with the type I VWD in this family.
Nucleotide sequence analysis
Exon 28 of the VWF gene in patient M did not contain any mutations; however, heterozygosity was observed at 3 nucleotide positions: 24/672, 24/1171, and 24/1282 (numbering according to Mancuso et al20 ). Two of these are polymorphisms reported in the international VWF database (http://www.shef.ac.uk/VWF/polymorphisms/ex28.html): 24/672 is a G/A SNP that results in the amino acid polymorphism Ala/Thr1381; 24/1282 is an A/G SNP that results in the amino acid polymorphism Tyr/Cys1584. At position 24/1171, patient M possessed both a C and a T nucleotide; however, this is a silent change (Thr/Thr1547); it is not reported in the international VWF database.
VWF susceptibility to proteolysis correlates with the Tyr/Cys1584 polymorphism
Of the 2 amino acid polymorphisms for which patient M was heterozygous, Ala/Thr1381 was not considered unusual; Ala/Thr is observed in many proteins. However, Tyr/Cys1584 was considered unusual on the basis of the nonconservative nature of the amino acids involved and the absence/presence of a sulfydryl group.
Genotypic analysis of patient M's family showed that both patient M and her sister were heterozygous for Tyr/Cys1584, whereas all other family members were homozygous for tyrosine (Figure 2A).
To exclude the possibility that the effect was familial and not necessarily related to the Tyr/Cys1584 polymorphism, a cohort of 200 individuals from the local population were genotyped at this locus: 2 heterozygous individuals were found (C154 and C188); the remaining 198 were homozygous Tyr/Tyr. For both heterozygotes, the plasma VWF showed an increased susceptibility to proteolysis by ADAMTS13 and this was comparable to that observed for individual 2:3, the sister of patient M (Figure 3A-B; data not shown for C188 the analysis of which was done independently at a later date).
In contrast, the plasma VWF of 3 individuals selected randomly from those homozygous for tyrosine at position 1584 showed comparable proteolysis to the healthy control (Figure 3A-B).
Comparison of VWF proteolysis for homozygous Tyr/Tyr1584 and heterozygous Tyr/Cys1584 individuals
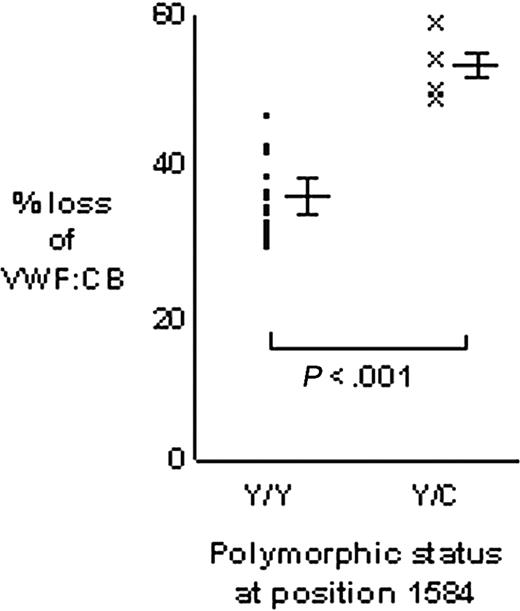
The VWF:CB was used to quantify the proteolysis of VWF from Tyr/Tyr1584 (n = 20) and Tyr/Cys1584 (n = 4) individuals (Figure 4). The mean percent loss of VWF:CB for homozygous VWF was 35.1% ± 4.6% and for heterozygous VWF it was 53.4% ± 4.6% (P < .001). The 95% confidence interval for the difference between the means was 18.3% ± 5.2%, indicating that heterozygous VWF showed between 13.1% and 23.5% greater loss of VWF:CB than homozygous VWF.
Comparison of VWF proteolysis for homozygous Tyr/Tyr1584 and heterozygous Tyr/Cys1584 individuals. Proteolysis of VWF from 20 Tyr/Tyr homozygous individuals was measured in triplicate and the mean of each triplicate plotted (•). Similarly, the mean of triplicate proteolysis assays for heterozygous Tyr/Cys VWF was plotted (x). Major bars to the right of each data set indicate the mean; minor bars above and below indicate 1 SD each side of the mean. Proteolysis was expressed as the percent loss of VWF:CB (the loss of VWF:CB as a percentage of the T = 0 hours value).
Comparison of VWF proteolysis for homozygous Tyr/Tyr1584 and heterozygous Tyr/Cys1584 individuals. Proteolysis of VWF from 20 Tyr/Tyr homozygous individuals was measured in triplicate and the mean of each triplicate plotted (•). Similarly, the mean of triplicate proteolysis assays for heterozygous Tyr/Cys VWF was plotted (x). Major bars to the right of each data set indicate the mean; minor bars above and below indicate 1 SD each side of the mean. Proteolysis was expressed as the percent loss of VWF:CB (the loss of VWF:CB as a percentage of the T = 0 hours value).
Comparison of exon 28 sequences between individuals who show increased VWF proteolysis and those who do not
Exon 28 of the VWF gene contains 15 identified SNPs, of which 4 are silent and 11 result in amino acid polymorphisms (Table 2).
Polymorphisms within exon 28 of the VWF gene
Polymorphism . | . | . | . | |
|---|---|---|---|---|
| Nucleotide . | Position . | Amino acid . | Position . | |
| A/G | 24/223 | Asn/Ser | 1231 (468) | |
| G/T | 24/295 | Leu/Leu | 1287 (524) | |
| G/A | 24/542 | Pro/Pro | 1337 (574) | |
| C/T | 24/661 | Ala/Val | 1377 (614) | |
| A/G | 24/669 | Ile/Val | 1380 (617) | |
| G/A | 24/672 | Ala/Thr | 1381 (618) | |
| G/A | 24/727 | Arg/His | 1399 (638) | |
| A/G | 24/835 | Asn/Ser | 1435 (672) | |
| G/C | 24/945 | Asp/His | 1472 (709) | |
| C/T | 24/1048 | Ser/Leu | 1506 (743) | |
| A/C | 24/1196 | Ala/Ala | 1555 (792) | |
| G/T | 24/1224 | Val/Leu | 1565 (802) | |
| A/G | 24/1282 | Tyr/Cys | 1584 (821) | |
| C/A | 24/1332 | Pro/Thr | 1601 (838) | |
| T/A | 24/1577 | Pro/Pro | 1682 (919) | |
| C/T | 24/1171 | Thr/Thr | 1547 (784) | |
Polymorphism . | . | . | . | |
|---|---|---|---|---|
| Nucleotide . | Position . | Amino acid . | Position . | |
| A/G | 24/223 | Asn/Ser | 1231 (468) | |
| G/T | 24/295 | Leu/Leu | 1287 (524) | |
| G/A | 24/542 | Pro/Pro | 1337 (574) | |
| C/T | 24/661 | Ala/Val | 1377 (614) | |
| A/G | 24/669 | Ile/Val | 1380 (617) | |
| G/A | 24/672 | Ala/Thr | 1381 (618) | |
| G/A | 24/727 | Arg/His | 1399 (638) | |
| A/G | 24/835 | Asn/Ser | 1435 (672) | |
| G/C | 24/945 | Asp/His | 1472 (709) | |
| C/T | 24/1048 | Ser/Leu | 1506 (743) | |
| A/C | 24/1196 | Ala/Ala | 1555 (792) | |
| G/T | 24/1224 | Val/Leu | 1565 (802) | |
| A/G | 24/1282 | Tyr/Cys | 1584 (821) | |
| C/A | 24/1332 | Pro/Thr | 1601 (838) | |
| T/A | 24/1577 | Pro/Pro | 1682 (919) | |
| C/T | 24/1171 | Thr/Thr | 1547 (784) | |
Nucleotide numbering according to Mancuso et al.20 Amino acid positions that are not in parentheses are from the start methionine of pre—pro-VWF; amino acid positions in parentheses are from residue 1 of the mature VWF subunit. The numbering schemes can be respectively interconverted by the subtraction or addition of 763. With the exception of the silent C/T 24/1171 SNP revealed in the present study, all other polymorphisms are reported in the international VWF database (http://www.shef.ac.uk/vwf/polymorphisms/ex28.html).
Comparison of exon 28 sequences between 7 individuals, of whom 4 showed increased VWF proteolysis (patient M, sister 2:3, C154, and C188) and of whom 3 did not, showed that all individuals shared identical genotypes at 13 SNP positions, whereas at the 2 remaining positions (Ala/Thr1381 and Tyr/Cys1584) the 2 groups differed. Heterozygosity for Tyr/Cys1584 was common to the 4 with increased VWF proteolysis, compared with homozygosity for tyrosine in the 3 with normal proteolysis. At position Ala/Thr1381, Ala/Ala, Ala/Thr, and Thr/Thr were observed and did not correlate with increased susceptibility to proteolysis.
Phenotypic data for Tyr/Cys1584 VWF and Tyr/Tyr1584 VWF
Individuals homozygous for Tyr/Tyr1584 showed VWF:Ag, VWF:RCo, and VWF:CB within the normal range, and concordant ratios for VWF:RCo/VWF:Ag and VWF:CB/VWF:Ag (Table 3).
Phenotypic parameters for Tyr/Tyr1584 and Tyr/Cys1584 individuals
ID . | Position 1584 . | Blood group . | VWF:Ag, % . | VWF:RCo, % . | RCo/Ag ratio . | VWF:CB, % . | CBA/Ag ratio . | FVIII:C, % . |
|---|---|---|---|---|---|---|---|---|
| C1 | Tyr/Tyr | A | 99 | 145 | 1.47 | 79 | 0.80 | 88 |
| C2 | Tyr/Tyr | O | 72 | 123 | 1.71 | 57 | 0.79 | 86 |
| C3 | Tyr/Tyr | A | 94 | 133 | 1.41 | 76 | 0.81 | 97 |
| 2:3 | Tyr/Cys | O | 86 | 53 | 0.62 | 80 | 0.93 | 76 |
| C154 | Tyr/Cys | O | 34 | — | — | 18 | 0.53 | 40 |
| C188 | Tyr/Cys | A | 309* | 464 | 1.50 | 262 | 0.84 | 192 |
| M* | Tyr/Cys | O | 13 | < 10 | > 0.76 | — | — | 37 |
| 16 | 18 | 1.13 | 12 | 0.75 | 48 | |||
| 13 | < 10 | > 0.77 | — | — | 27 | |||
| 21 | 13 | 0.61 | 9 | 0.42 | 48 |
ID . | Position 1584 . | Blood group . | VWF:Ag, % . | VWF:RCo, % . | RCo/Ag ratio . | VWF:CB, % . | CBA/Ag ratio . | FVIII:C, % . |
|---|---|---|---|---|---|---|---|---|
| C1 | Tyr/Tyr | A | 99 | 145 | 1.47 | 79 | 0.80 | 88 |
| C2 | Tyr/Tyr | O | 72 | 123 | 1.71 | 57 | 0.79 | 86 |
| C3 | Tyr/Tyr | A | 94 | 133 | 1.41 | 76 | 0.81 | 97 |
| 2:3 | Tyr/Cys | O | 86 | 53 | 0.62 | 80 | 0.93 | 76 |
| C154 | Tyr/Cys | O | 34 | — | — | 18 | 0.53 | 40 |
| C188 | Tyr/Cys | A | 309* | 464 | 1.50 | 262 | 0.84 | 192 |
| M* | Tyr/Cys | O | 13 | < 10 | > 0.76 | — | — | 37 |
| 16 | 18 | 1.13 | 12 | 0.75 | 48 | |||
| 13 | < 10 | > 0.77 | — | — | 27 | |||
| 21 | 13 | 0.61 | 9 | 0.42 | 48 |
C1-3 indicate individuals from a cohort of 200 random subjects; M indicates patient M; 2:3 indicates the sister of patient M; C154 and C188 indicate the 2 Tyr/Cys 1584 heterozygous individuals from the cohort of 200 random subjects;—indicates not determined.
Data are shown for testing on 4 independent occasions.
Individuals heterozygous for Tyr/Cys1584 showed variable results. The sister of patient M (individual 2:3; Table 3) gave normal phenotypic parameters with concordant ratios. C154 gave parameters suggestive of a qualitative and quantitative VWF defect that was not detected on multimer analysis, and C188 gave an abnormally high VWF:Ag, VWF:RCo, and VWF:CB possibly caused by an acute-phase response; however, the ratios were concordant.
Patient M gave normal or marginally discordant ratios for VWF:RCo/VWF:Ag and VWF:CB/VWF:Ag when tested on independent occasions (Table 3). In some instances, the parameters were consistent with type I VWD, and in others they suggested borderline type IIM VWD.
Discussion
The amino acid polymorphism Tyr/Cys1584 is located within the A2 domain of VWF, the domain in which the Tyr1605-Met1606 peptide bond cleaved by ADAMTS13 occurs.2,3 The polymorphic site is 22 residues N-terminal to the ADAMTS13 site and involves a nonconservative amino acid variation that is associated with the absence/presence of a sulfydryl group. These observations support a causal role for the polymorphism in increasing the susceptibility of VWF to proteolysis by ADAMTS13; however definitive evidence, for example, using expression studies, is required to confirm causality. The possibility that the effect may be due to an amino acid polymorphism elsewhere in VWF and in linkage disequilibrium with Tyr/Cys1584 cannot be excluded at this stage.
The increase in proteolysis is associated with cysteine at position 1584 and is between 13% and 23% greater than that observed for tyrosine at this position. Increased susceptibility of VWF to proteolysis has potentially significant implications in diverse areas.
In vivo, it may have an impact on the efficiency of primary hemostasis. Recent findings suggest that ultra-HMW VWF is present on the luminal surface of endothelial cells and is rapidly proteolysed in situ by ADAMTS13 when platelets bind.22 It is possible that increased susceptibility to proteolysis may affect the length of VWF at the site of the evolving primary hemostatic plug as platelets bind and thereby influence VWF efficacy. The data in the present study show that the increased susceptibility to proteolysis associated with Tyr/Cys1584 occurs both in the presence and absence of ristocetin in vitro. Ristocetin has been shown to increase the rate of proteolysis of VWF by ADAMTS1315 ; thus the increased proteolysis associated with Tyr/Cys1584 is evident even in the presence of that brought about by ristocetin. A possible implication of these in vitro data are that in vivo, the increased susceptibility of VWF to proteolysis associated with Tyr/Cys1584 may be a property of VWF both before and after incorporation into the soft clot. This, however, remains to be proven.
Thus, although some heterozygous individuals may give normal VWF parameters in static, in vitro assays, the significance of increased susceptibility to proteolysis may lie in its impact on the dynamic situation in vivo at the time of hemostatic challenge. It remains to be seen whether the effect is indeed clinically relevant in the context of normal VWF parameters or in the context of type I VWD.
Increased susceptibility of VWF to proteolysis may result in a change in the distribution of multimers in the plasma. This may not be detected in phenotypic assays such as multimer analysis, VWF:RCo and VWF:CB despite the fact that in vivo, at the site of formation of the soft clot, primary hemostasis may be significantly affected.
The long-established relationship between blood group and VWF levels23 may reflect increased proteolysis and clearance of VWF in group O individuals compared with non-O individuals.15 In combination with the effect of blood group, the increased proteolysis of VWF associated with the presence of cysteine at position 1584 may result in phenotypic parameters consistent with VWD. The data in the present study indicate that heterozygous individuals may show concordant VWF:Ag, VWF:RCo, and VWF:CB, in which case, if the overall VWF:Ag is low, type I VWD would result. However, the present data also show that discordant phenotypic parameters may be obtained, suggestive of type IIM VWD.
In families affected by VWD (type I or type II) and in families affected by TTP due to low levels (but not the absence) of ADAMTS13, increased susceptibility of VWF to proteolysis may influence the penetrance of the disorder. Thus, in VWD, increased proteolysis may exacerbate the bleeding tendency, whereas in TTP it may moderate the thrombotic tendency. Increased susceptibility of VWF to ADAMTS13 cleavage may, in part, explain the variable phenotype of families with VWD.
If Tyr/Cys1584 is directly responsible for the increased susceptibility of VWF to proteolysis, it may not be the only amino acid polymorphism of VWF to exert such an effect. Of the 15 SNPs reported within exon 28 of the VWF gene (http://www.shef.ac.uk/VWF/polymorphisms/ex28.html), 11 encode amino acid polymorphisms, some of which may similarly influence proteolysis. In addition, amino acid polymorphisms elsewhere in VWF may influence proteolysis. We have investigated one high-frequency polymorphism in exon 28 (His/Asp 1472) (http://www.shef.ac.uk/VWF/polymorphisms/ex28.html) and found that it does not exert an effect (D.J.B. and P.W.C., manuscript in preparation); therefore, clearly not all nonconservative amino acid polymorphisms encoded within exon 28 are relevant to proteolysis. A knowledge of which polymorphisms do and do not affect VWF proteolysis would be valuable in the context of VWD and TTP.
The possibility that one or more amino acid polymorphisms in VWF may influence its susceptibility to proteolysis forces the recent findings that blood group may influence susceptibility to be revisited.15 In that recent study, VWF of each blood group was purified from the pooled plasma of 4 individuals. According to the amino acid phenotype of each VWF in the pool, the VWF may have had an intrinsic susceptibility to proteolysis, independent of blood group.
Studies of VWF half-life in type I VWD have reached varying conclusions, with a normal half-life reported by some24 and a shortened half-life reported by others.25,26 It is not yet known whether a shortened half-life reflects increased catabolism in some patients, but if so, this may arise from increased proteolysis followed by clearance or increased clearance without increased proteolysis. Increased proteolysis arising from homozygosity or compound heterozygosity for relevant amino acid polymorphisms represents a novel candidate mechanism for type I VWD. Further investigations are clearly needed to explore this possibility.
Determination of the frequencies for the Tyr/Cys1584 polymorphism to statistical significance requires a large-scale population study. In the present study, 2 heterozygotes were detected among 200 randomly chosen individuals from the local (Welsh) population; therefore, the polymorphism may not be uncommon and it would be reasonable to anticipate its occurrence in some VWD and TTP families. It is present in the type I VWD family investigated in the present study and interestingly, 2 of the 3 affected individuals studied (the daughters of patient M) are homozygous for Tyr1584 and have higher VWF levels and a clinically less severe bleeding tendency than the third affected individual (patient M) who is heterozygous for tyrosine and cysteine. The effect of the Tyr/Cys1584 polymorphism in patient M appears to change her phenotype from type I VWD, as seen in other family members, to a borderline type IIM.
It would be valuable to establish the phenotypic parameters for Tyr/Tyr, Tyr/Cys, and Cys/Cys individuals according to blood group. Homozygotes for Cys1584 would be extremely rare; however, they are potentially of the greatest interest because, just as for Tyr1584 homozygotes, all multimers are built up from identical monomers, and, therefore, there should not be a preferential loss of HMW multimers. In comparison, in heterozygotes, HMW multimers, which by chance contain a greater number of the more susceptible VWF monomers, will suffer greater proteolysis than multimers of corresponding size that happen to contain the less susceptible monomers. Clearly, from the heterozygotes observed in the present study this does not result in a VWD type IIA profile. The paradox of increased susceptibility to proteolysis but a normal multimer profile for heterozygotes may reflect the following: The increased susceptibility to proteolysis brought about by the cysteine variant may be many fold less than that induced by VWD type IIA substitutions. Thus, there may well be a loss in the HMW range, but this may be minor in comparison to the effect seen in VWD type IIA and may be insufficient to be revealed on multimer analysis. Additionally, multimers containing predominantly tyrosine monomers may be released normally from the cell, whereas those possessing predominantly cysteine monomers may be retained. The snapshot of in vivo plasma VWF given by multimer analysis may therefore appear normal, whereas the static, in vitro ADAMTS13 assays, in which there is no replenishment or removal of VWF, unveil the presence of multimers possessing cysteine monomers. Differences between individuals in VWF synthesis, cellular release, catabolism/clearance, and ADAMTS13 levels may cause considerable variation in the overall impact of the polymorphism on the VWF multimer profile and in vitro parameters.
The prospect that one or more amino acid polymorphisms in VWF increases its susceptibility to proteolysis therefore has implications in diverse areas. A fuller picture can only be obtained with further detailed research; however, at the very least, the data presented here reveal that in some individuals, VWF is more susceptible to proteolysis than in others and that this is an intrinsic property of VWF, independent of blood group and ADAMTS13 levels. This novel insight into VWF biochemistry opens new avenues of exploration into the role of VWF in disease.
Prepublished online as Blood First Edition Paper, October 2, 2003; DOI 10.1182/blood-2003-05-1505.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Steve Bowley, Nicky McCartney, Noel Murrin, and Lee Hathaway for their help in generating some of the phenotypic data.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal