Abstract
The importance of HLA-DPB1 matching for the outcome of allogeneic hematologic stem cell (HSC) transplantation is controversial. We have previously identified HLA-DPB1*0901 as a target of cytotoxic T cells mediating in vivo rejection of an HSC allograft. Here we show that HLA-DPB1*0901 encodes a T-cell epitope shared by a subset of DPB1 alleles that determines nonpermissive mismatches for HSC transplantation. Several T-cell clones obtained from the patient at the time of rejection showed HLA-DP restricted recognition of allogeneic targets expressing HLA-DPB1*0901, *1001, *1701, *0301, *1401, and *4501, but not other alleles. Based on these findings, we developed an algorithm for prediction of nonpermissive HLA-DPB1 mismatches. Retrospective evaluation of 118 transplantations showed that the presence of nonpermissive HLA-DPB1 mismatches was correlated with significantly increased hazards of acute grade II to IV graft-versus-host disease (HR = 1.87, P = .046) and transplantation-related mortality (HR = 2.69, P = .027) but not relapse (HR = 0.98, P = .939), as compared with the permissive group. There was also a marked but statistically not significant increase in the hazards of overall mortality (HR = 1.64, P = .1). These data suggest that biologic characterization of in vivo alloreactivity can be a tool for definition of clinically relevant nonpermissive HLA mismatches for unrelated HSC transplantation.
Introduction
Isolation and infusion of high numbers of hematologic precursors in the context of allogeneic hematologic stem cell (HSC) transplantation has made it possible to overcome some of the HLA barriers, increasing the number of patients who can benefit from this therapeutic modality.1 The identification of permissive and nonpermissive HLA mismatches has therefore become one of the main challenges of modern HSC transplantation biology. The DP locus of the HLA class II system was first described in 1978 as a target of alloreactive T-cell responses identified in secondary mixed lymphocyte cultures.2 Due to the scarce availability of specific antisera, the gene products of the HLA-DP locus remained poorly characterized for many years. Only the advent of molecular biology techniques for HLA typing allowed unraveling the full degree of HLA-DP polymorphism, with 20 and 106 HLA-DPA1 and DPB1 alleles, respectively, described to date.3 The functional role of HLA-DP molecules in the immune system is still subject to investigation. A number of autoimmune diseases have been associated with HLA-DP.4-8 HLA-DP–restricted virus,9 bacterial,10 or tumor11 antigen-specific T cells have been described. HLA-DP has also been shown to be capable of presentation of HLA-A1 or HLA-DR3–derived allopeptides for indirect allorecognition by HLA-DP–restricted alloreactive CD4+ T cells.12,13 Recently, it has been demonstrated that the HLA-DPβ polypeptide can induce in vivo alloreactivity leading to acute graft-versus-host disease (aGvHD)14-16 or graft rejection17 after transplantation of allogeneic HSCs. However, the overall clinical significance of HLA-DPB1 mismatches for the clinical outcome of HSC transplantation is still a matter of debate. Retrospective studies initially failed to demonstrate a significant impact of HLA-DPB1 mismatches on the incidence of aGvHD.18,19 In contrast, one early20 and several more recent reports21-24 showed that the incidence of severe aGvHD was increased in HLA-DPB1–mismatched transplantations. Two of these reports suggested a cumulative impact of the number of HLA-DPB1 mismatches on the incidence of aGvHD.22,24 None of these studies demonstrated a correlation between HLA-DPB1 mismatching and transplantation-related mortality (TRM). All retrospective analyses performed to date were based on comparison of HLA-DPB1–matched versus–mismatched donor-recipient pairs, without discrimination according to the nature of these mismatches. This was due to the largely elusive nature of the T-cell epitopes involved in HLA-DP–specific alloreactivity.
In this report, we have used a different approach for evaluating the clinical importance of HLA-DPB1 mismatches. First, we dissected the molecular basis of HSC allograft rejection mediated by alloreactive CD4+ T cells specific for the donor's HLA-DPB1*0901,17 and characterized the cross-recognition pattern of HLA-DPB1 alleles other than *0901 by these T cells. Second, we established an algorithm for permissiveness of HLA-DPB1 mismatches in HSC transplantation, based on the presence or absence of HLA-DPB1 alleles carrying the shared T-cell epitope. This algorithm was used to retrospectively evaluate the clinical outcome of 118 unrelated HSC allografts mismatched only for HLA-DPB1.
Patients, materials, and methods
Cell lines, flow cytometry, and monoclonal antibodies
Most Epstein-Barr virus–transformed B-cell lines (BLCLs) of the Eleventh Histocompatibility Workshop were obtained from the European Collection of Animal Cell Culture. Only BLCLs from patient and donor and BLCLs DKM366 and HSRSL were locally established, and BLCL LG-2 was kindly provided by Dr Thierry Boon (Ludwig Institute for Cancer Research, Brussels, Belgium). For analysis of HLA-DP cell surface expression, BLCLs were stained in 2-color immunofluorescence using the monoclonal antibody (mAb) B7/21,25 a kind gift of Dr S. Tonks (Imperial Cancer Research Fund, London, United Kingdom), and analyzed by fluorescence-activated cell sorting (FACS) scan. For T-cell inhibition studies, the following mAbs were used: W6/32 (HLAclass I), kindly provided by Dr C. Traversari (Molmed-HSR, Milan, Italy), L243 (HLA-DR25 ), and B7/21 (HLA-DP25 ), a generous gift of Dr S. Tonks.
Isolation and maintenance of HLA-DP–specific alloreactive T lymphocytes
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll (Nycomed Pharma, Milan, Italy) gradient centrifugation from a patient at the time of acute clinical rejection of an allogeneic HSC graft. Patient and donor characteristics as well as transplantation conditions have been described in detail elsewhere.17 To generate a panel of clones, PBMCs were plated in limiting dilution conditions in the presence of 600 IU/mL interleukin 2 (IL-2; Chiron Italia, Milan, Italy), irradiated (10 000 rad) allogeneic feeder BLCL LG-2 (3 × 104/well), and irradiated (10 000 rad) donor BLCLs (2 × 104/well). Clones were maintained by bimonthly restimulation with irradiated donor and feeder BLCLs in 24-well plates (Corning, Corning, NY).
Cytotoxicity assays
The cytotoxic activity of effector T lymphocytes was analyzed in a standard 51Cr-release assay as described in detail elsewhere.26 Briefly, 1000 51Cr-labeled (NEN Dupont, Milan, Italy) target BLCLs were incubated for 4 hours with effector cells at various effector-target cell ratios at 37°Cina5% CO2 atmosphere. Subsequently, the supernatant was removed and counted on a γ counter. Percentage of specific lysis was calculated according to the formula: 100 × (51Cr experimental release–spontaneous release)/(maximum release–spontaneous release). Samples were tested in duplicate and the mean value of both duplicates is indicated. For mAb inhibition studies, cytotoxicity assays were performed in the presence of varying dilutions of mAb. Percentage of lysis inhibition was calculated according to the formula: 100 × (51Cr experimental release–51Cr experimental release in the presence of mAb)/(51Cr experimental release).
Retroviral vector-mediated transfer of HLA-DPB1 into BLCLs
Cloning of the full-length cDNA of HLA-DPB1*0901 into a retroviral expression vector LDPB9SΔN and transduction into BLCLs has been described in detail elsewhere.17 HLA-DPB1*4601 was cloned by the same protocol, using cDNA from the BLCL VEC, into the retroviral expression vector LDPB46SΔN. For generation of the retroviral vector LDPB9ΔCYTSΔN expressing the C-terminally truncated form of HLA-DPB1*0901, vector LDPB9SΔN was digested by restriction endonucleases ApaI and XhoI. The ends were blunted using the 5′ polymerase Klenow (Roche, Mannheim, Germany) and the truncated vector was religated.
Cell-mediated inhibition of clonogenic CD34+ cell growth
PBMCs were isolated from leukapheresis products of 2 adult donors, HSRTA, typed as HLA-DPB1*0401, and HSRMIGI, typed as *0901, *1001. CD34+ cells were purified by magnetic beads (Miltenyi Biotec, Bologna, Italy), cultured for 5 days in X-Vivo-10 medium (BioWhittaker, Walkersville, MD) supplemented with 4% fetal bovine serum (Hyclone, South Logan, UT) and the cytokines thrombopoietin (TPO; CellGenix, Freiburg, Germany), fms-like tyrosine kinase-3 ligand (FLT3 ligand; CellGenix), and stem cell factor (SCF; CellGenix), all at 50 ng/mL, and repurified at the end of culture. Clonogenic assays were performed as previously described.27 Briefly, 2.3 × 103 CD34+ cells were incubated for 4 hours at various effector-target cell ratios at 37°C in a 5% CO2 atmosphere with HLA-DP–specific T-cell clones. Subsequently, single-cell suspensions were cultured at a concentration of 103 CD34+ cells/mL in Methocult medium (Stemcell Technologies, Vancouver, BC, Canada). Myeloerythroid colonies consisting of at least 50 cells were counted after 14 days of culture. Percentage of inhibition of clonogenic progenitor growth was calculated according to the formula: 100 × (number of colonies in the absence of alloreactive T cells–number of colonies in the presence of alloreactive T cells)/(number of colonies in the absence of alloreactive T cells).
Patients, HLA typing, and transplantation procedures
For the purpose of this study, only patients with hematologic malignancies were considered. Patients underwent transplantation at 5 different transplantation units between 1995 and 2002: Ospedale S. Martino, Genoa, Italy (n = 36); Hammersmith Hospital, London, United Kingdom (n = 31); Ospedale S. Orsola-Malpighi, Bologna, Italy (n = 18); Ospedale S. Matteo, Pavia, Italy (n = 20); and Ospedale G. Gaslini, Genoa, Italy (n = 13). Patients received unmanipulated bone marrow from an unrelated donor, provided by the National Donor Registries and facilitated through the Italian Bone Marrow Donor Registry or the Anthony Nolan Trust Foundation. Donor-recipient pairs were retrospectively typed at the 4-digit level for loci HLA-A, B, Cw, DRB1/3/4/5, and DQB1 by polymerase chain reaction–sequence-based typing (PCR-SBT),28 or by PCR–sequence-specific priming (PCR-SSP)29,30 and reference strand conformation analysis (RSCA).31 Four-digit HLA-DPB1 typing was performed by PCR-SSP32 and by PCR–sequence-specific oligonucleotide probing (PCR-SSOP).33 A total of 118 pairs matched for all HLA loci studied except for HLA-DPB1 were included in the analysis. All patients received myeloablative therapy, which included total body irradiation (TBI) in 96 cases. In case of lymphoid malignancies, most patients received cyclophosphamide (120 mg/kg) with or without thiotepa (15 mg/kg), whereas most patients with myeloid malignancies were treated with busulfan (16 mg/kg) plus cyclophosphamide (120 mg/kg) with or without melphalan (140 mg/m2). For patients undergoing transplantation in Italy, GvHD prophylaxis consisted of cyclosporine (1-3 mg/kg) and short-course methotrexate (10 mg/m2, 8 mg/m2, and 8 mg/m2 on days 1, 3, and 6, or 15 mg/m2 on day 1 and 10 mg/m2 on days 3, 6, and 11). In 56 cases, pretransplantation antithymocyte globulin (ATG; Sangstat, Lyon, France; 7-15 mg/kg from day –6 today –1, or ATG, Fresenius, Bad Homburg, Germany, 15 mg/kg from days –6 to –2) was also administered. For patients undergoing transplantation at Hammersmith Hospital (London, United Kingdom), GvHD prophylaxis consisted of alemtuzumab (10 mg/kg daily from day –10 to day –6), cyclosporine A (2.5 mg/kg) daily, and methotrexate (8 mg/m2) on days 2, 4, and 8. Patients with acute lymphoblastic leukemia (ALL) or with acute myeloblastic leukemia (AML) in first complete remission, with chronic myeloid leukemia (CML) in first chronic phase, and with myelodysplasia (MDS) characterized by blast percentage less than 5% were considered to belong to the early phase group. Pediatric patients with ALL or with AML in second complete remission were also considered to belong to the early phase group. aGvHD was diagnosed and scored as 0 to IV according to established criteria,34 and monitored for 100 days after transplantation. Time of myeloid engraftment was defined as the first of 3 consecutive days with absolute neutrophil count more than 0.5 × 109/L, and platelet engraftment as the first of 7 consecutive days with an unsupported platelet count more than 50 × 109/L. All patients but one had engraftment.
Statistical analysis
The probability of overall survival at 3 years and of aGvHD at 100 days after transplantation were estimated and represented using the Kaplan-Meier curves, with the relevant SE. Because TRM and the incidence of relapse are known to be competing risks, the cumulative incidence curves for these events were estimated.35 For all 4 end-point parameters (overall survival, TRM, aGvHD, and relapse), log-rank comparisons were carried out between 2 groups (permissive mismatches versus nonpermissive mismatches in graft-versus host [GvH] or host-versus-graft [HvG] direction). The effect of the presence of a permissive or a nonpermissive mismatch in GvH or HvG direction on TRM was evaluated both in univariate and multivariate analyses (including other clinical variables) using a Cox model. Hazard ratio (HR) and confidence interval (CI) were also calculated. All parameters with a P value less than .2 in univariate analysis entered the multivariate model.
Results
Characterization of HLA-DP–specific alloreactive T lymphocytes
We have previously reported on a case of HSC allograft rejection mediated by cytotoxic CD4+ T lymphocytes recognizing the donor's HLA-DPB1*0901.17 Patient and donor were matched by 4-digit typing in HvG direction for all major histocompatibility complex (MHC) loci except for HLA-DP. In particular, the donor carried HLA-DPA1*0201 and DPB1*0901 not shared by the patient (Table 1). Several donor-specific CD4+ T-cell clones were obtained by limiting dilution cloning and 2 of these, designated 501 and 538, are described here in more detail. Both clones specifically recognized BLCLs derived from the donor but not from the patient in an HLA-DP–restricted fashion (Figure 1A-D). Transduction of patient BLCLs with a retroviral vector expressing HLA-DPB1*0901 (LDPB9SΔN) restored recognition by both clones. Transduction with a control vector encoding a truncated form of HLA-DPB1*0901 (LDPB9ΔCYTSΔN), encoding the entire polymorphic region of *0901 but lacking the cytoplasmic tail necessary for cell surface expression of the molecule, had no effect (Figure 1A-B). Transduction of donor BLCLs with the truncated vector did not inhibit recognition of endogenous HLA-DPB1*0901, demonstrating that the presence of truncated *0901 did not interfere with antigen processing of the wild-type molecule (Figure 1A-B). Taken together, these data show that allorecognition of HLA-DPB1*0901 was mediated by the direct and not by the indirect pathway. Recognition of HLA-DPB1*0901 on patient BLCLs that lack the donor's DPA1*0201 (Table 1) demonstrates that the DPα chain was not crucial for formation of the relevant T-cell epitope. In concordance with this notion, the allogeneic BLCL WT100BIS sharing only HLA-DPA1*0201 with the donor (Table 1) was not lysed (Figure 1A-B). Similar data were obtained with 2 additional clones and with a polyclonal T-cell line (data not shown). Clones 501 and 538 were both able to specifically recognize CD34+ cells expressing HLA-DPB1*0901 but not *0401 in cytotoxicity (Figure 2A-B) and clonogenic assays (Figure 2C-D). These data provide in vitro evidence for an involvement of these clones in allograft rejection observed in vivo.
HLA typing and levels of HLA-DP surface expression of patient, donor, and allogeneic BLCLs used as targets for cytotoxicity assays in this study
BLCL . | A* . | B* . | Cw* . | DRB1* . | DQB1* . | DPA1* . | DPB1* . | MFI . |
|---|---|---|---|---|---|---|---|---|
| Patient | 0201, 3001 | 3901, 4901 | 0701, 1203 | 0701, 1601 | 0202, 0502 | 0103 | 0201, 0402 | 1485 |
| Donor | 3001 | 3901, 4901 | 0701, 1203 | 0701, 1601 | 0202, 0502 | 0103, 0201 | 0201, 0901 | 1159 |
| BM21 | 0101 | 4101 | 1701 | 1101 | 0301 | 0201 | 1001 | 1097 |
| HSRSL | 03, 32 | 1402, 3508 | 0802, 1502 | 0102, 1601 | 0501, 0502 | ND | 0201, 1701 | 642 |
| OLL | 3101 | 1501, 1520 | 0102, 0304 | 0802 | 0402 | 0103 | 0301, 0402 | 1171 |
| DKM366 | 0101, 0205 | 3502, 5001 | 0401, 0602 | 0701, 1104 | 0202, 0301 | ND | 0401, 1401 | 670 |
| C212 | ND | ND | ND | 0101, 1301 | 0501, 0603 | 0103, 0201 | 0401, 4501 | 777 |
| WT100BIS | 1101 | 3501 | 0401 | 0101 | 0501 | 0201 | 0101 | ND |
| BEL8-CC | ND | ND | ND | 0101, 0103 | 0501 | 0103 | 0201, 2001 | 854 |
BLCL . | A* . | B* . | Cw* . | DRB1* . | DQB1* . | DPA1* . | DPB1* . | MFI . |
|---|---|---|---|---|---|---|---|---|
| Patient | 0201, 3001 | 3901, 4901 | 0701, 1203 | 0701, 1601 | 0202, 0502 | 0103 | 0201, 0402 | 1485 |
| Donor | 3001 | 3901, 4901 | 0701, 1203 | 0701, 1601 | 0202, 0502 | 0103, 0201 | 0201, 0901 | 1159 |
| BM21 | 0101 | 4101 | 1701 | 1101 | 0301 | 0201 | 1001 | 1097 |
| HSRSL | 03, 32 | 1402, 3508 | 0802, 1502 | 0102, 1601 | 0501, 0502 | ND | 0201, 1701 | 642 |
| OLL | 3101 | 1501, 1520 | 0102, 0304 | 0802 | 0402 | 0103 | 0301, 0402 | 1171 |
| DKM366 | 0101, 0205 | 3502, 5001 | 0401, 0602 | 0701, 1104 | 0202, 0301 | ND | 0401, 1401 | 670 |
| C212 | ND | ND | ND | 0101, 1301 | 0501, 0603 | 0103, 0201 | 0401, 4501 | 777 |
| WT100BIS | 1101 | 3501 | 0401 | 0101 | 0501 | 0201 | 0101 | ND |
| BEL8-CC | ND | ND | ND | 0101, 0103 | 0501 | 0103 | 0201, 2001 | 854 |
MFI indicates mean fluorescence intensity of BLCLs after cell surface staining with the HLA-DP-specific mAb B7/21; ND, not determined.
Characterization of HLA-DP–specific alloreactive CD4+ T cell clones. (A-B) T-cell clones 501 (A) and 538 (B) were used at different effector-target cell ratios in a standard 4-hour cytotoxicity assay. The following BLCLs were used as targets: donor (•); patient (○); patient transduced with retroviral vector LDPB9SΔN (▪); patient transduced with retroviral vector LDPB9ΔCYTSΔN (□); donor transduced with retroviral vector LDPB9ΔCYTSΔN (▴); and WT100BIS (▵). For complete HLA typing of target cells see Table 1. (C-D) T-cell clones 501 (C) and 538 (D) were used at a fixed effector-target cell ratio of 1:1 (clone 501) or 10:1 (clone 538) in mAb inhibition assays of donor BLCL lysis. The mAbs were B7/21 (anti–HLA-DP), L243 (anti–HLA-DR), or W6/32 (anti–HLA class I). All mAbs were used at 1 μg/mL (▪) and mAb B7/21 was also used at 0.3 μg/mL (▦) , 0.1 μg/mL (▨) , or 0.03 μg/mL (□). Specific lysis in the absence of mAbs was 46% and 40% for clones 501 and 538, respectively.
Characterization of HLA-DP–specific alloreactive CD4+ T cell clones. (A-B) T-cell clones 501 (A) and 538 (B) were used at different effector-target cell ratios in a standard 4-hour cytotoxicity assay. The following BLCLs were used as targets: donor (•); patient (○); patient transduced with retroviral vector LDPB9SΔN (▪); patient transduced with retroviral vector LDPB9ΔCYTSΔN (□); donor transduced with retroviral vector LDPB9ΔCYTSΔN (▴); and WT100BIS (▵). For complete HLA typing of target cells see Table 1. (C-D) T-cell clones 501 (C) and 538 (D) were used at a fixed effector-target cell ratio of 1:1 (clone 501) or 10:1 (clone 538) in mAb inhibition assays of donor BLCL lysis. The mAbs were B7/21 (anti–HLA-DP), L243 (anti–HLA-DR), or W6/32 (anti–HLA class I). All mAbs were used at 1 μg/mL (▪) and mAb B7/21 was also used at 0.3 μg/mL (▦) , 0.1 μg/mL (▨) , or 0.03 μg/mL (□). Specific lysis in the absence of mAbs was 46% and 40% for clones 501 and 538, respectively.
Recognition of CD34+ cells by HLA-DP–specific T-cell clones. Clones 501 (A,C) and 538 (B,D) were incubated at different effector-target cell ratios with purified CD34+ cells obtained from 2 healthy adult donors expressing HLA-DPB1*0901,*1001 (•), or *0401 (○). Lysis of CD34+ target cells was measured in a standard 4-hour cytotoxicity assay (A-B). Inhibition of the colony-forming capacity of CD34+ cells was analyzed in a clonogenic assay (C-D).
Recognition of CD34+ cells by HLA-DP–specific T-cell clones. Clones 501 (A,C) and 538 (B,D) were incubated at different effector-target cell ratios with purified CD34+ cells obtained from 2 healthy adult donors expressing HLA-DPB1*0901,*1001 (•), or *0401 (○). Lysis of CD34+ target cells was measured in a standard 4-hour cytotoxicity assay (A-B). Inhibition of the colony-forming capacity of CD34+ cells was analyzed in a clonogenic assay (C-D).
Cross-recognition of allogeneic BLCLs by HLA-DP–specific T lymphocytes
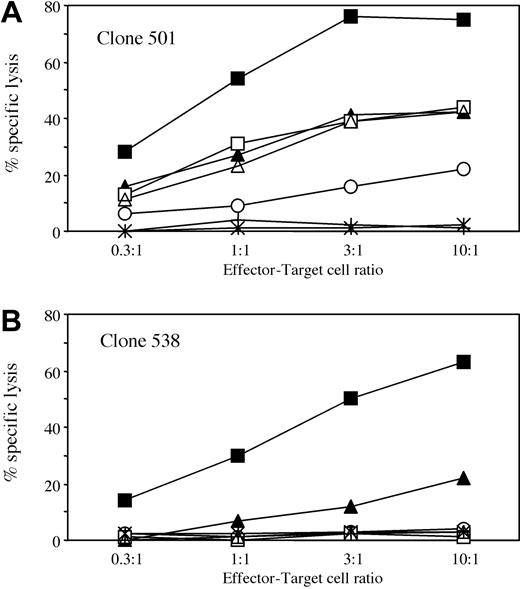
To investigate whether HLA-DPB1*0901 encodes a T-cell epitope shared by other HLA-DPB1 alleles, a panel of allogeneic BLCLs expressing different HLA-DP specificities was tested for recognition by clones 501 and 538. BLCLs expressing HLA-DPB1*1001, *1701, *0301, *1401, and *4501 induced specific, HLA-DP–restricted interferon γ (IFN-γ) release by at least one of the 2 clones (data not shown). In contrast, incubation with a panel of allogeneic BLCLs expressing HLA-DPB1*0101, *0201, *0202, *0401, *0402, *0501, *0601, *1101, *1301, *1501, *1601, *1901, *2001, *2301, or *4601 did not result in specific cytokine release by either of the 2 clones (data not shown). The specificity of HLA-DPB1*0901-specific T-cell clones was further investigated on a selected panel of allogeneic BLCL targets in cytotoxicity assays (Figure 3). Clones 501 and 538 recognized allogeneic BLCLs carrying HLA-DPB1*1001 and, to a lesser extent, *1701. The lower efficiency of recognition of the BLCLs positive for HLA-DPB1*1701 probably reflects heterozygosity for *1701 as well as lower levels of HLA-DP cell surface expression as compared to the BLCLs carrying *1001 (Table 1). Clone 501 but not 538 also showed moderate levels of cytotoxic activity against allogeneic BLCLs expressing HLA-DPB1*0301, *1401, and *4501 (Figure 3A-B). As shown by mAb inhibition studies, lysis of allogeneic BLCLs by both clones was HLA-DP restricted (data not shown). These data were confirmed on at least one additional BLCL expressing HLA-DPB1*1001, *1701, *0301, and *1401 (data not shown). Allogeneic BLCLs expressing HLA-DPB1*2001 or patient BLCLs transduced with *4601 (Figure 3A-B) were not recognized. Two additional clones and the polyclonal T-cell line showed a recognition pattern similar to that of clones 538 and 501, respectively (data not shown).
Cytotoxic activity of clones 501 and 538 against a selected panel of allogeneic BLCLs. Clones 501 (A) and 538 (B) were used at different effector-target cell ratios in a standard 4-hour cytotoxicity assay. The following BLCLs were used as targets: BM21 (HLA-DPB1*1001, ▪); HSRSL (HLA-DPB1*1701, ▴); C212 (HLA-DPB1*4501, □); DKM366 (HLA-DPB1*1401, ○); OLL (HLA-DPB1*0301, ▵); BEL8-CC (HLA-DPB1*2001, stars); or patient BLCLs transduced with retroviral vector LDPB46SΔN (crosses). For complete HLA typing of target cells, see Table 1.
Cytotoxic activity of clones 501 and 538 against a selected panel of allogeneic BLCLs. Clones 501 (A) and 538 (B) were used at different effector-target cell ratios in a standard 4-hour cytotoxicity assay. The following BLCLs were used as targets: BM21 (HLA-DPB1*1001, ▪); HSRSL (HLA-DPB1*1701, ▴); C212 (HLA-DPB1*4501, □); DKM366 (HLA-DPB1*1401, ○); OLL (HLA-DPB1*0301, ▵); BEL8-CC (HLA-DPB1*2001, stars); or patient BLCLs transduced with retroviral vector LDPB46SΔN (crosses). For complete HLA typing of target cells, see Table 1.
Classification of HLA-DPB1 alleles according to their predicted immunogenicity
The observed T-cell reactivity patterns suggest expression, by a defined subset of HLA-DPB1 alleles, of a shared T-cell epitope that determines HLA-DP–specific alloresponses. This epitope is encoded by the HLA-DPB1 alleles that were recognized by all alloreactive T-cell clones studied (represented by clones 501 and 538). These alleles were classified as immunogenic group 1 (Table 2). The epitope is also present, although probably in a conformation with reduced T-cell receptor (TCR) affinity, on the HLA-DPβ chains encoded by the alleles that were recognized by some of the T-cell clones studied (represented by clone 501), but not by others (represented by clone 538). These alleles were classified as intermediately immunogenic group 2 (Table 2). In contrast, the epitope is not encoded by the HLA-DPB1 alleles that were not recognized by any of the T-cell clones studied. These alleles were classified as poorly immunogenic group 3 (Table 2).
Classification of HLA-DPB1 alleles according to their predicted immunogenicity
. | . | Frequencies . | . | . | ||
|---|---|---|---|---|---|---|
| Groups† . | HLA-DPB1* . | White . | Asian . | African . | ||
| 1 | 0901, 1001, 1701 | 0.018 | 0.025 | 0.325 | ||
| 2 | 0301, 1401, 4501 | 0.143 | 0.075 | 0.060 | ||
| 3 | 0101, 0201, 0202, 0401, 0402, 0501, 0601, 1101, 1301, 1501, 1601, 1901, 2001, 2301, 4601 | 0.839 | 0.850 | 0.575 | ||
. | . | Frequencies . | . | . | ||
|---|---|---|---|---|---|---|
| Groups† . | HLA-DPB1* . | White . | Asian . | African . | ||
| 1 | 0901, 1001, 1701 | 0.018 | 0.025 | 0.325 | ||
| 2 | 0301, 1401, 4501 | 0.143 | 0.075 | 0.060 | ||
| 3 | 0101, 0201, 0202, 0401, 0402, 0501, 0601, 1101, 1301, 1501, 1601, 1901, 2001, 2301, 4601 | 0.839 | 0.850 | 0.575 | ||
Based on the T-cell recognition pattern described in this study, HLA-DPB1 alleles are predicted to be immunogenic (group 1), intermediately immunogenic (group 2), or poorly immunogenic (group 3).
The HLA-DPβ chain has been divided into 6 hypervariable regions A-F.36-38 HLA-DPB1 alleles from group 1 share the amino acid sequence of regions A, B, D, and F (Table 3). Alleles from group 2 have substitutions in region D, alone or in combination with regions A or C, as compared to group 1 alleles (Table 3). Alleles from group 3 have substitutions in at least 3 of the 4 regions A, B, D, or F shared by group 1 alleles (Table 3). The 21 HLA-DPB1 alleles assigned to groups 1 to 3 are present with a total frequency of 100%, 95%, and 96% in the white, Asian, and African population, respectively32,39 (Table 2). Interestingly, the combined frequency of the immunogenic group 1 and 2 alleles is over 5 times lower than that of the poorly immunogenic group 3 alleles, in the white and Asian but not in the African population (Table 2).
Amino acid sequence alignment of HLA-DPB1 alleles analyzed in this study
. | . | Polymorphic regions . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | A . | . | . | B . | . | . | C . | . | . | D . | . | E . | F . | . | . | . | |||||||||||||||
| Groups . | HLA-DPB1* . | 8 . | 9 . | 11 . | 33 . | 35 . | 36 . | 55 . | 56 . | 57 . | 65 . | 69 . | 76 . | 84 . | 85 . | 86 . | 87 . | |||||||||||||||
| 1 | 0901 | V | H | L | E | F | V | D | E | D | I | E | V | D | E | A | V | |||||||||||||||
| 1001 | — | — | — | — | — | — | — | — | E | — | — | — | — | — | — | — | ||||||||||||||||
| 1701 | — | — | — | — | — | — | — | — | — | — | — | M | — | — | — | — | ||||||||||||||||
| 2 | 0301 | — | Y | — | — | — | — | — | — | — | L | K | — | — | — | — | — | |||||||||||||||
| 1401 | — | — | — | — | — | — | — | — | — | L | K | — | — | — | — | — | ||||||||||||||||
| 4501 | — | — | — | — | — | — | — | — | E | L | K | — | — | — | — | — | ||||||||||||||||
| 3 | 0101 | — | Y | G | — | Y | A | A | A | E | — | K | — | — | — | — | — | |||||||||||||||
| 0201 | L | F | G | — | — | — | — | — | E | — | — | M | G | G | P | M | ||||||||||||||||
| 0202 | L | F | G | — | L | — | E | A | E | — | — | M | G | G | P | M | ||||||||||||||||
| 0401 | L | F | G | — | — | A | A | A | E | — | K | M | G | G | P | M | ||||||||||||||||
| 0402 | L | F | G | — | — | — | — | — | E | — | K | M | G | G | P | M | ||||||||||||||||
| 0501 | L | F | G | — | L | — | E | A | E | — | K | M | — | — | — | — | ||||||||||||||||
| 0601 | — | Y | — | — | — | — | — | — | — | L | — | M | — | — | — | — | ||||||||||||||||
| 1101 | — | Y | — | Q | Y | A | A | A | E | L | R | M | — | — | — | — | ||||||||||||||||
| 1301 | — | Y | — | — | Y | A | A | A | E | — | — | I | — | — | — | — | ||||||||||||||||
| 1501 | — | Y | G | Q | Y | A | A | A | E | L | R | M | V | G | P | M | ||||||||||||||||
| 1601 | L | F | G | — | — | — | — | — | E | — | — | M | — | — | — | — | ||||||||||||||||
| 1901 | L | F | G | — | — | — | E | A | E | — | — | I | — | — | — | — | ||||||||||||||||
| 2001 | — | Y | — | — | — | — | — | — | — | L | K | M | — | — | — | — | ||||||||||||||||
| 2301 | L | F | G | — | — | — | A | A | E | — | K | M | G | G | P | M | ||||||||||||||||
| 4601 | L | F | G | — | — | — | — | — | — | — | — | M | G | G | P | M | ||||||||||||||||
. | . | Polymorphic regions . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | A . | . | . | B . | . | . | C . | . | . | D . | . | E . | F . | . | . | . | |||||||||||||||
| Groups . | HLA-DPB1* . | 8 . | 9 . | 11 . | 33 . | 35 . | 36 . | 55 . | 56 . | 57 . | 65 . | 69 . | 76 . | 84 . | 85 . | 86 . | 87 . | |||||||||||||||
| 1 | 0901 | V | H | L | E | F | V | D | E | D | I | E | V | D | E | A | V | |||||||||||||||
| 1001 | — | — | — | — | — | — | — | — | E | — | — | — | — | — | — | — | ||||||||||||||||
| 1701 | — | — | — | — | — | — | — | — | — | — | — | M | — | — | — | — | ||||||||||||||||
| 2 | 0301 | — | Y | — | — | — | — | — | — | — | L | K | — | — | — | — | — | |||||||||||||||
| 1401 | — | — | — | — | — | — | — | — | — | L | K | — | — | — | — | — | ||||||||||||||||
| 4501 | — | — | — | — | — | — | — | — | E | L | K | — | — | — | — | — | ||||||||||||||||
| 3 | 0101 | — | Y | G | — | Y | A | A | A | E | — | K | — | — | — | — | — | |||||||||||||||
| 0201 | L | F | G | — | — | — | — | — | E | — | — | M | G | G | P | M | ||||||||||||||||
| 0202 | L | F | G | — | L | — | E | A | E | — | — | M | G | G | P | M | ||||||||||||||||
| 0401 | L | F | G | — | — | A | A | A | E | — | K | M | G | G | P | M | ||||||||||||||||
| 0402 | L | F | G | — | — | — | — | — | E | — | K | M | G | G | P | M | ||||||||||||||||
| 0501 | L | F | G | — | L | — | E | A | E | — | K | M | — | — | — | — | ||||||||||||||||
| 0601 | — | Y | — | — | — | — | — | — | — | L | — | M | — | — | — | — | ||||||||||||||||
| 1101 | — | Y | — | Q | Y | A | A | A | E | L | R | M | — | — | — | — | ||||||||||||||||
| 1301 | — | Y | — | — | Y | A | A | A | E | — | — | I | — | — | — | — | ||||||||||||||||
| 1501 | — | Y | G | Q | Y | A | A | A | E | L | R | M | V | G | P | M | ||||||||||||||||
| 1601 | L | F | G | — | — | — | — | — | E | — | — | M | — | — | — | — | ||||||||||||||||
| 1901 | L | F | G | — | — | — | E | A | E | — | — | I | — | — | — | — | ||||||||||||||||
| 2001 | — | Y | — | — | — | — | — | — | — | L | K | M | — | — | — | — | ||||||||||||||||
| 2301 | L | F | G | — | — | — | A | A | E | — | K | M | G | G | P | M | ||||||||||||||||
| 4601 | L | F | G | — | — | — | — | — | — | — | — | M | G | G | P | M | ||||||||||||||||
An algorithm for permissiveness of HLA-DPB1 mismatches in HSC donor-recipient pairs
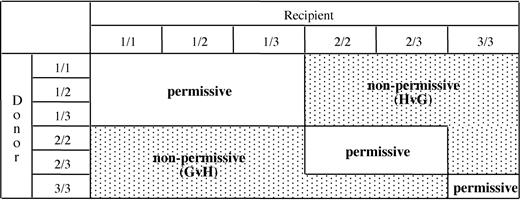
The T-cell epitope shared by the immunogenic HLA-DPB1 alleles of group 1 and 2 was the target of an in vivo immune response resulting in clinical rejection of an HSC allograft.17 This led us to hypothesize that mismatching for group 1 or 2 alleles might have a more general clinical importance for allogeneic HSC transplantation. During thymic education, the T-cell repertoire is shaped by negative selection of potentially autoreactive T cells recognizing immunogenic epitopes present on self-MHC molecules.40-42 Expression of the shared T-cell epitope on self-HLA-DPB1 alleles from group 1 can therefore be predicted to result in clonal deletion of T cells specific for this epitope, that is, T-cell clones similar to clones 501 and 538 from this study. Expression of self-HLA-DPB1 alleles from group 2 is predicted to drive elimination of alloreactive T cells specific for these alleles, including T cells able to recognize both group 1 and group 2 alleles such as clone 501 from this study. In contrast, T cells with selective specificity for group 1 but not group 2 alleles, such as clone 538 from this study, are likely to escape negative thymic selection shaped by self-HLA-DPB1 alleles from group 2. Finally, expression of self-HLA-DPB1 alleles from group 3 is predicted to not induce any negative selection of alloreactive T cells specific for the shared epitope. Based on these considerations, an algorithm for permissiveness of HLA-DPB1 mismatches in allogeneic HSC transplantation was established. According to the donor-recipient combination, this algorithm allows prediction of HLA-DP–specific alloreactivity in GvH or HvG direction, or the absence of alloreactivity (Figure 4). In this model, expression of 2 self-HLA-DPB1 alleles from group 1 is predicted to result in protection from mounting an HLA-DP–specific alloresponse, but in an increased risk of becoming target of such a response. In contrast, expression of 2 self-HLA-DPB1 alleles from group 3 is predicted to have the opposite effect. The functional behavior of 2 self-HLA-DPB1 alleles from group 2 is predicted to vary according to the combination of alleles expressed by the HSC transplantation partner (Figure 4).
An algorithm for permissiveness of HLA-DPB1 mismatches in HSC donor-recipient pairs. Based on the in vitro data from this study, HLA-DPB1 alleles were divided into 3 groups (1-3) according to their predicted immunogenicity (Table 2). The 3 groups of alleles can be present in 6 different combinations in diploid cells. Numbers indicate the group of the first (before the slash) and the second (after the slash) HLA-DPB1 allele of the donor or of the recipient. Classification of HLA-DPB1 mismatches as permissive or nonpermissive in GvH or in HvG direction is indicated for all possible combinations.
An algorithm for permissiveness of HLA-DPB1 mismatches in HSC donor-recipient pairs. Based on the in vitro data from this study, HLA-DPB1 alleles were divided into 3 groups (1-3) according to their predicted immunogenicity (Table 2). The 3 groups of alleles can be present in 6 different combinations in diploid cells. Numbers indicate the group of the first (before the slash) and the second (after the slash) HLA-DPB1 allele of the donor or of the recipient. Classification of HLA-DPB1 mismatches as permissive or nonpermissive in GvH or in HvG direction is indicated for all possible combinations.
Retrospective evaluation of the clinical importance of immunogenic HLA-DPB1 mismatches in unrelated HSC transplantation
An association of HLA-DPB1 mismatches with clinical aGvHD has been reported in several studies.21-24 We therefore sought to determine whether such an association would be evident also using the established algorithm of permissivity. To this end, the clinical outcome of 118 unrelated HSC allografts matched at the 4-digit level for HLA-A, B, Cw, DRB, and DQB1, but mismatched for DPB1, was retrospectively evaluated. Following the algorithm described in Figure 4, HLA-DPB1 mismatches in donor-recipient pairs were classified as permissive (63 pairs) or as nonpermissive (55 pairs). The latter group could be further divided, depending on the vector of the mismatch, in a group displaying a nonpermissive mismatch in the GvH direction (24 pairs), and in a group displaying a nonpermissive mismatch in the HvG direction (31 pairs). Relevant clinical data of the pairs belonging to the 3 groups are listed in Table 4. The probability to develop aGvHD grades II to IV at day 100 after transplantation was significantly higher in the group with nonpermissive mismatches in GvH (44%, SE = 0.11) or HvG direction (42%, SE = 0.09), as compared to the permissive (28%, SE = 0.06) group (P = .046, Figure 5A). The hazards of aGvHD grades II to IV were also significantly increased by the presence of nonpermissive mismatches in GvH or HvG direction (HR = 1.87, 95% CI = 0.99-3.50).
Clinical data of unrelated HSC donor-recipient pairs
. | . | Nonpermissive mismatch . | . | |
|---|---|---|---|---|
. | Permissive mismatch . | GvH direction . | HvG direction . | |
| No. patients | 63 | 24 | 31 | |
| Donor/recipient sex | ||||
| M/M | 19 | 6 | 12 | |
| M/F | 13 | 4 | 7 | |
| F/M | 14 | 8 | 4 | |
| F/F | 17 | 6 | 8 | |
| Age, y (range) | ||||
| Donor | 37 (21-59) | 36 (21-54) | 34 (22-49) | |
| Recipient | 27 (3-55) | 30 (6-54) | 30 (4-59) | |
| Disease | ||||
| CML | 22 | 9 | 18 | |
| CMML | — | 2 | — | |
| AML | 11 | 4 | 5 | |
| ALL | 22 | 6 | 6 | |
| Myeloma | 2 | 1 | 1 | |
| MDS | 5 | 2 | — | |
| NHL | 1 | — | 1 | |
| Early disease phase at transplantation | 39 | 12 | 21 | |
| Time between diagnosis and transplantation, d (range) | 743 (151-4936) | 695 (71-3480) | 810 (104-2392) | |
| Cells infused, 108/kg (range) | 4.4 (0.9-26.1) | 4.8 (1.5-10.9) | 5.3 (1.7-25.7) | |
| TBI conditioning | 53 | 19 | 24 | |
| GvHD prophylaxis | ||||
| Campath | 19 | 3 | 9 | |
| ATG | 32 | 13 | 11 | |
| GvHD grades II-IV | 17 | 10 | 13 | |
| Failure/nonengraftment | — | 1 | — | |
. | . | Nonpermissive mismatch . | . | |
|---|---|---|---|---|
. | Permissive mismatch . | GvH direction . | HvG direction . | |
| No. patients | 63 | 24 | 31 | |
| Donor/recipient sex | ||||
| M/M | 19 | 6 | 12 | |
| M/F | 13 | 4 | 7 | |
| F/M | 14 | 8 | 4 | |
| F/F | 17 | 6 | 8 | |
| Age, y (range) | ||||
| Donor | 37 (21-59) | 36 (21-54) | 34 (22-49) | |
| Recipient | 27 (3-55) | 30 (6-54) | 30 (4-59) | |
| Disease | ||||
| CML | 22 | 9 | 18 | |
| CMML | — | 2 | — | |
| AML | 11 | 4 | 5 | |
| ALL | 22 | 6 | 6 | |
| Myeloma | 2 | 1 | 1 | |
| MDS | 5 | 2 | — | |
| NHL | 1 | — | 1 | |
| Early disease phase at transplantation | 39 | 12 | 21 | |
| Time between diagnosis and transplantation, d (range) | 743 (151-4936) | 695 (71-3480) | 810 (104-2392) | |
| Cells infused, 108/kg (range) | 4.4 (0.9-26.1) | 4.8 (1.5-10.9) | 5.3 (1.7-25.7) | |
| TBI conditioning | 53 | 19 | 24 | |
| GvHD prophylaxis | ||||
| Campath | 19 | 3 | 9 | |
| ATG | 32 | 13 | 11 | |
| GvHD grades II-IV | 17 | 10 | 13 | |
| Failure/nonengraftment | — | 1 | — | |
The pairs were classified as permissive or nonpermissive in GvH or HvG direction, according to the algorithm from Figure 5. CMML indicates chronic myelomonocytic leukemia; NHL, non-Hodgkin lymphoma; and —, not applicable.
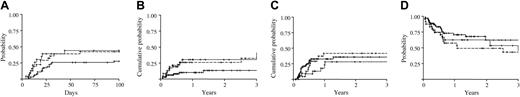
Influence of permissive or nonpermissive HLA-DPB1 mismatches on the clinical outcome of unrelated HSC transplantation. The probability of overall grades II to IV aGvHD (A), TRM (B), relapse (C), or survival (D) was analyzed in 63 pairs with permissive (continuous line) and 24 or 31 pairs with nonpermissive HLA-DPB1 mismatches in GvH (dotted line) or HvG (dashed line) direction, respectively. The probabilities of overall survival and aGvHD were calculated according to Kaplan-Meier estimates; TRM and relapse were analyzed by cumulative incidence curves.
Influence of permissive or nonpermissive HLA-DPB1 mismatches on the clinical outcome of unrelated HSC transplantation. The probability of overall grades II to IV aGvHD (A), TRM (B), relapse (C), or survival (D) was analyzed in 63 pairs with permissive (continuous line) and 24 or 31 pairs with nonpermissive HLA-DPB1 mismatches in GvH (dotted line) or HvG (dashed line) direction, respectively. The probabilities of overall survival and aGvHD were calculated according to Kaplan-Meier estimates; TRM and relapse were analyzed by cumulative incidence curves.
The risks of TRM and relapse, whose occurrences are mutually exclusive, were not analyzed by Kaplan-Meier curves, which are known to overestimate the proportion of patients who develop each event taken separately. Instead, for these 2 parameters, analysis was carried out using the cumulative index rates based on competing risk methods.35 A possible consequence of the increased risk of aGvHD is a higher probability of TRM. In line with this notion, the estimated risk of TRM at 3 years after transplantation was significantly increased in the nonpermissive group (32% and 30% for the GvH and HvG group, respectively, and 13% for the permissive group, P = .021; Figure 5B).
For relapse, the estimated risk at 3 years after transplantation was slightly decreased in the GvH vector (28%, HR = 0.70, 95% CI = 0.28-1.76) and increased in the HvG vector (42%, HR = 1.21, 95% CI = 0.59-2.48), as compared to the permissive group (36%). This resulted in an overall zero effect of nonpermissive mismatches in GvH and HvG direction on relapse (HR = 0.98, 95% CI = 0.516-1.846, P = .939; Figure 5C).
Taken together, all these parameters led to a marked increase in the hazards of mortality for the nonpermissive (HR = 1.64, 95% CI = 0.90-2.64) as compared to the permissive group, although the estimated risk of mortality did not reach statistical significance (P = .100; Figure 5D).
At univariate Cox analysis, the hazards of TRM in the nonpermissive group were significantly increased (HR = 2.69, 95% CI = 1.12-6.43, P = .027 for the nonpermissive GvH and HvG vector as compared to the permissive group). None of the other parameters tested, that is, ATG or alemtuzumab prophylaxis, TBI conditioning, number of cells infused, donor-recipient sex and age, diagnosis (CML versus non-CML), phase of disease, interval between diagnosis and transplantation, and number of HLA-DPB1 mismatches present (1 versus 2), showed a significant impact on the incidence of TRM. A multivariate Cox analysis was then carried out including the 3 groups and all the clinical variables showing a P < .2 (cells infused and recipient age). The HR of nonpermissive as compared to permissive patients did not change adjusting for the clinical variables (HR = 2.64, 95% CI = 1.10-6.35, P = .030), and no parameter other than HLA groups was retained in the final model.
Discussion
In the present study, we have characterized the biologic basis of HSC allograft rejection mediated by T cells specific for the donor's HLA-DPB1*0901.17 These T cells were shown to be dependent on an immunogenic epitope shared by a subset of HLA-DPB1 alleles that encode defined amino acid sequences in the hypervariable regions of the molecule36-38 (Table 3). Mapping of the antigenic epitope to a stretch of amino acids suggests that this sequence might give rise to indirect allorecognition by presentation of an allopeptide bound to self-HLA molecules.12,13 The results of our experiments performed with the truncated form of HLA-DPB1*0901 show that this was not the case for the alloreactive T cells studied here (Figure 1). It is likely that the hypervariable amino acids are not recognized as such but allow a distinct set of peptides to be presented, which in turn give rise to the immunogenic epitope recognized by alloreactive T lymphocytes. In support of this notion, it has been shown that the peptide motif of HLA-DPB1*090110 is profoundly different from that of the poorly immunogenic *020143-45 and *0401.45
The algorithm used in this study for classification of HLA-DPB1 mismatches is based on a hierarchy of immunogenicity that could only be established by functional analysis of alloreactive T lymphocytes. Such a hierarchy could not be detected by any of the conventional approaches to tackling the question of the clinical importance of HLA-DPB1 matching in allogeneic HSC transplantation. These approaches are based on comparison of matched versus mismatched pairs with21,24 or without18-20,22,23 consideration of matching for hypervariable regions of the HLA-DPβ chain. The former strategy has recently been used successfully to establish an algorithm for donor assignment in solid organ transplantation,47 but did not reveal any clinical impact for the outcome of HSC allografts.21,24 The algorithm from the present study is based on the well-established principle of negative thymic selection of T cells specific for self-antigens.40-42 It takes into account that a small number of HLA-DPB1 alleles, classified as intermediately immunogenic group 2, are recognized by some but not all alloreactive T-cell clones studied. The functional behavior of group 2 alleles in terms of immunogenicity or protection thereof is predicted to be dependent on the combination of alleles expressed by stimulator and responder cells (Figure 4). It can be postulated that the amplitude of the alloresponse against group 2 alleles might be lower as compared to alleles from group 1 because it is mediated by only a subset of alloreactive T cells specific for the shared epitope. Similarly, expression of group 2 alleles on responder T cells might reduce the amplitude of the alloresponse against alleles from group 1. These hypothetical considerations were not taken into account in our algorithm. Nevertheless, the evidence obtained in this study from retrospective evaluation of the clinical outcome of allogeneic HSC transplantations suggests the validity of the approach used. In support of this notion, 2 previous case reports have identified 2 HLA-DPB1 alleles of the immunogenic group 1 from this study, *1001 and *1701, as targets of alloreactive T cells mediating clinically manifest aGvHD.16,46 The data from this study suggest that aGvHD may be mediated by HLA-DP–specific T cells in a considerable number of cases. Interestingly, the increased risk of aGvHD was evident independently from the vector of the mismatch, with similar probabilities for the GvH and the HvG groups (Figure 5A). This suggests a dual mechanism for the pathogenesis of aGvHD: first, direct allorecognition of nonpermissive HLA-DPB1 mismatches on host antigen-presenting cells (APCs) by donor-derived T cells, resulting also in a decreased risk of relapse due to a GvL effect mediated by these alloreactive T cells,23,48 and second, release of inflammatory cytokines by host T cells recognizing immunogenic HLA-DPB1 molecules on donor APCs, an indirect pathway for the pathogenesis of GvHD.49,50
The increased risk of aGvHD is probably at least partly responsible for the significantly higher incidence of TRM observed in transplantations with nonpermissive HLA-DPB1 mismatches. To our knowledge, this is the first study to report such a correlation. Despite a marked heterogeneity of the permissive and the nonpermissive groups with regard to patient and donor characteristics, no parameters other than the presence of nonpermissive HLA-DPB1 mismatches were shown to have an impact on TRM in univariate or multivariate analysis. Importantly, the number of HLA-DPB1 mismatches did not have a significant impact on the hazards of TRM (P = .96 in univariate analysis). This makes it unlikely that the observed differences between the permissive and the nonpermissive group are related to the number and not the type of HLA-DPB1 mismatches, as proposed previously by others.22,24
The present study also reports increased hazards of overall survival in pairs with nonpermissive HLA-DPB1 mismatches, which, however, in contrast to aGvHD and TRM, did not reach statistical significance. Overall survival is the result of 2 competing events, TRM and relapse. Whereas nonpermissive HLA-DPB1 mismatches in GvH or HvG direction resulted in a significantly increased risk of TRM, they had opposing effects on relapse, with a decrease in the GvH (28%) and an increase in the HvG group (42%), as compared to permissive pairs (36%). In fact, relapse was the cause of death in 2 (17%) of 12 versus 7 (44%) of 16 patients in the GvH and the HvG group, respectively. This suggests that the lack of statistical significance in the increase of overall mortality in nonpermissive pairs can likely be attributed to the missing influence of relapse in the GvH group. In line with this notion, the cumulative incidence curves of mortality over 3 years markedly diverged between the permissive and the HvG groups, whereas the curve for the GvH group was less divergent (Figure 5D). Taken together, these data suggest that nonpermissive mismatches in HvG direction might have a greater clinical impact than those in GvH direction, because in addition to aGvHD and TRM, they also might worsen the hazards of relapse and therefore of mortality.
The initial failure of retrospective clinical studies to demonstrate a significant clinical impact of HLA-DPB1 disparities on allogeneic HSC transplantation could have been partly due to the obscuring effect of molecular mismatches at HLA class I, undetected by serologic class I typing in early studies.18,19 In fact, 3 more recent reports based on advanced 4-digit typing of all HLA loci demonstrated an increase of aGvHD in HLA-DPB1–mismatched transplantations.22-24 The data from this study show, for the first time, that such retrospective clinical studies can benefit from careful characterization of T lymphocytes mediating in vivo alloreactivity, which can be used as a tool for identification of clinically relevant nonpermissive HLA disparities. This may hold true not only for HLA-DPB1 but also for other MHC loci. In fact, a specific impact of disparity for position 116 of HLA class I alleles on the clinical outcome of allogeneic HSC transplantation has recently been demonstrated.28 The results from this study suggest that matching for HLA-DPB1 might not require complete typing for all 106 alleles known to date, but could be limited to identifying the presence or absence of the immunogenic sequences by a restricted number of allele-specific amplifications or probe hybridizations. Such an approach would reduce the cost and effort of HLA-DPB1 matching while maintaining its potential clinical benefits. Although the allelic frequency of nonpermissive HLA-DPB1 specificities is relatively low (16.1%; Table 2), the resulting phenotypic frequency is approximately 30%, with a considerable probability for at least one of these alleles to occur in either donor or recipient of an HSC transplant. In line with this notion, 55 (47%) of 118 pairs from this study were classified as having a nonpermissive HLA-DPB1 mismatch, suggesting that about 50% of donor searches could directly benefit from the refined selection approach described here. Moreover, knowledge of the innocuousness of mismatches classified as nonpermissive should allow assignment of donors mismatched for such alleles without further searches for a fully matched donor. Therefore, from a practical standpoint, it is likely that most searches can potentially benefit from this selection approach.
Prepublished online as Blood First Edition Paper, October 23, 2003; DOI 10.1182/blood-2003-04-1279.
Supported by grants from the Association “Antonio Castelnuovo,” Cermenate, Italy, the Italian Ministry of Health (grant no. 202), the Associazione Italiana per la Ricerca sul Cancro (AIRC), and the Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Nadia Nobili for technical assistance in screening of the alloreactive T-cell clones, Manuela Battaglia for critical reading of the manuscript, and Richard Szydlo for support in patient data administration. Catia Traversari and Susan Tonks are gratefully acknowledged for providing the mAbs W6/32, and L243 and B7/21, respectively. We are indebted to Tierry Boon for providing the BLCL LG-2.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal