Abstract
Activating mutations in the juxtamembrane domain (FLT3-length mutations, FLT3-LM) and in the protein tyrosine kinase domain (TKD) of FLT3 (FLT3-TKD) represent the most frequent genetic alterations in acute myeloid leukemia (AML) and define a molecular target for therapeutic interventions by protein tyrosine kinase (PTK) inhibitors. We could show that distinct activating FLT3-TKD mutations at position D835 mediate primary resistance to FLT3 PTK inhibitors in FLT3-transformed cell lines. In the presence of increasing concentrations of the FLT3 PTK inhibitor SU5614, we generated inhibitor resistant Ba/F3 FLT3-internal tandem duplication (ITD) cell lines (Ba/F3 FLT3-ITD-R1-R4) that were characterized by a 7- to 26-fold higher IC50 (concentration that inhibits 50%) to SU5614 compared with the parental ITD cells. The molecular characterization of ITD-R1-4 cells demonstrated that specific TKD mutations (D835N and Y842H) on the ITD background were acquired during selection with SU5614. Introduction of these dual ITD-TKD, but not single D835N or Y842H FLT3 mutants, in Ba/F3 cells restored the FLT3 inhibitor resistant phenotype. Our data show that preexisting or acquired mutations in the PTK domain of FLT3 can induce drug resistance to FLT3 PTK inhibitors in vitro. These findings provide a molecular basis for the evaluation of clinical resistance to FLT3 PTK inhibitors in patients with AML.
Introduction
FLT3 (fms-like tyrosine kinase-3) is a receptor tyrosine kinase (RTK),1,2 which is known to play an important role in normal hematopoiesis and leukemogenesis. Most AML blasts express functional FLT3, and both overexpression and activating mutations of FLT3 can be found in patients with acute myeloid leukemia (AML). Between 20% and 25% of patients with AML carry activating FLT3 length mutations (FLT3-LM) consisting of internal tandem duplication (FLT3-ITD) and in some patients also additional insertions in the juxtamembrane domain.3-6 In another 7%, mutations at codons 835/836 in the activation loop of FLT3 can be detected (FLT3-TKD [tyrosine kinase domain] mutations).7-9 Recently, we described a new recurrent FLT3 mutation with transforming potential in patients with AML that is generated by an insertion of 2 amino acids between codons 840 and 841 (FLT3-840GS).10 FLT3-LMs are associated with leukocytosis and poor prognosis in most7,11-14 but not all studies.4,9
FLT3-LM and FLT3-TKD mutants are constitutively autophosphorylated on tyrosine residues, causing activation of signal transducer and activator of transcription 5 (STAT5) and mitogen-activated protein (MAP) kinases.15-19 In addition, transduction of FLT3-ITD and TKD mutants in murine interleukin 3 (IL-3)–dependent cell lines, such as Ba/F3 and 32D induces IL-3–independent growth7,16,17,19,20 in vitro and a myeloproliferative syndrome in the mouse bone marrow transplantation model.21
Although activating FLT3 mutations seem not to be sufficient to cause an AML phenotype, they represent a potential therapeutic target. Targeted inhibition of aberrant kinase signaling can be an effective therapeutic intervention in hematologic malignancies, as evidenced by hematologic and cytogenetic responses in patients with chronic myelogenous leukemia (CML) and CML blast crisis treated with the BCR-ABL kinase inhibitor imatinib mesylate (STI571, Gleevec).22-24 An analogous kinase inhibitor strategy might have therapeutic potential in patients with AML with activating mutations in the FLT3 gene.
In the past decade, many laboratories embarked on projects aimed at generating compounds that specifically inhibit the activity of the signaling cascades triggered by tyrosine kinases. Compounds with selective activity to class III RTK in vitro include AG1295,25 SU5416,26,27 SU5614,15,27 and CT53518.28 Three compounds (CEP-701, SU11248, and PKC412) with in vivo activity are currently evaluated in phase 1 and 2 clinical trials in patients with AML and have shown promising results.29-32
The PTK inhibitor imatinib mesylate has high clinical activity in BCR-ABL–positive CML in chronic phase and blast crisis as well as in BCR-ABL–positive acute lymphoblastic leukemia and gastrointestinal stromal tumors (GISTs) carrying activating KIT mutations.23,33,34 However, small molecule protein tyrosine kinase inhibitors may lead to multifactorial drug resistance.35-37 In certain BCR-ABL–positive cell lines, resistance was associated with amplification of the fusion gene.36-38 Additionally, overexpression of the alpha 1 acid glycoprotein was detected in some resistant cells.39 At least 10 different mutations in the ABL gene that induce imatinib mesylate resistance in vitro were found in patients with BCR-ABL–positive CML and ALL.40-46 Although FLT3 PTK inhibitors have shown promising results in phase 1 and 2 clinical studies in patients with AML, it is unknown whether similar mechanisms of resistance exist for these compounds.
Previously, we and others have demonstrated that the small molecule PTK inhibitor SU5614 is a selective inhibitor of FLT3 that induces growth arrest and apoptosis in FLT3-transformed cells and AML-derived cell lines expressing a constitutively activated FLT3.15,27 It was the aim of this study to analyze mechanisms of primary and secondary resistance to FLT3 PTK inhibitors in vitro. We report here significant differences in the sensitivity of distinct FLT3-TKD mutants that result in primary resistance to SU5614. As a new mechanism of secondary resistance, we identified TKD mutations in FLT3-ITD–transformed hematopoietic cells after prolonged exposure to the PTK inhibitor SU5614 in vitro. Our data demonstrate that distinct molecular alterations in the FLT3 gene define primary and secondary resistance to FLT3 PTK inhibitors. These findings will have a profound affect on the design of clinical studies evaluating FLT3 PTK inhibitors in patients with AML.
Materials and methods
Reagents and cell lines
Low passage murine Ba/F3 cells were obtained from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany) and were maintained in RPMI-1640 medium with 10% fetal bovine serum (FBS) and 10% WEHI-conditioned medium as a source of murine IL-3 when indicated. The PTK inhibitors SU5614, Genistein, and AG1295 were obtained from Calbiochem (Calbiochem-Novabiochem, Bad Soden, Germany). Ara-C (cytosine arabinoside, Cytarabin) was obtained from Cell Pharm GmbH (Hannover, Germany). The PTK inhibitor PKC412 was kindly provided by Novartis Pharma AG (Basel, Switzerland). Final concentrations of dimethyl sulfoxide (DMSO) in growth medium did not exceed 0.1%.
Cell proliferation of Ba/F3 cells
Cells were seeded at a density of 4 × 104/mL growth medium in the presence or absence of IL-3 and inhibitors or Ara-C as indicated. Viable cells were counted at 24, 48, and 72 hours in a standard hemacytometer after staining with trypan blue. The IC50 was defined as the concentration of inhibitor at which 50% of cells were viable compared with cells grown in the absence of inhibitor.
Apoptosis analysis
Assessment of apoptotic cells was carried out by annexin V/7-aminoactinomycin D (7-AAD) staining as recommended by the manufacturer (annexin V-phycoerythrin [PE] apoptosis detection kit; Becton Dickinson, Heidelberg, Germany) using a FacsCalibur flow cytometer (Becton Dickinson, San Jose, CA).
Antibodies
The following antibodies were used: anti-FLT3/flk2 (S18, sc-480; Santa Cruz, Heidelberg, Germany), anti-PY (PY99; Santa Cruz), antiphospho-p42/p44 MAPK, anti-p42/p44 MAPK, antiphospho–STAT5-Tyr694 (all from New England Biolabs, Frankfurt, Germany), anti-STAT5 (sc-835; Santa Cruz), anti–β-actin (A-5441; Sigma).
DNA constructs and vectors
The FLT3-ITD-W51 construct contains a 6-amino-acid (AA) duplicated sequence (REYEDL) inserted between AA601/602 of human FLT3-WT (kindly provided by D.G. Gilliland, Boston, MA).15 All FLT3 constructs were subcloned in the MSCV-IRES-EYFP/EGFP retroviral expression vector (kindly provided by R. K. Humphries, The Terry Fox Laboratory, Vancouver, University of British Columbia, Canada).
In vitro mutagenesis
The FLT3-D835H/Q/V/Y/N and Y842H point mutations were introduced into the full-length human FLT3 wild-type cDNA or the FLT3-ITD-W51 construct using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions as indicated. The correct sequence of all constructs was confirmed by nucleotide sequencing.
Transient transfection of BOSC23 cells and stable transduction of Ba/F3 cells were performed as described previously.15
Expression of CD135 by flow cytometry
Ba/F3 cells were incubated for 30 minutes on ice with a mouse phycoerythrin (PE)–labeled isotype-matched control antibody (Becton Dickinson) or CD135-PE (Becton Dickinson) antibody. Viable cells were analyzed by using a FacsCalibur flow cytometer (Becton Dickinson, San Jose, CA).
Immunoprecipitation and Western blot analysis
Immunoprecipitation and Western blot analysis were performed as described previously.15
Generation of SU5614-resistant Ba/F3-FLT3-ITD cell lines
Ba/F3 cells carrying the FLT3-ITD (W51) mutation were cultured in the presence of increasing concentrations of SU5614 (0.2-0.8 μM) over a period of at least 3 months. Cells were initially cultured in the presence of 0.2 μM SU5614, with culture medium changes every 3 to 4 days, for a total of 27 days. Cells were then cultured in 0.4 μM SU5614 for an additional 35 days, and finally cells were grown in medium containing 0.8 μM SU5614. Then cells were analyzed for drug resistance and were further characterized.
Genomic DNA extraction
Genomic DNA extraction was performed by using the DNeasy Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
RNA isolation and reverse transcription to cDNA
Total RNA was isolated with the RNeasy kit (Qiagen) and reverse transcribed with the Omniscript RT kit (Qiagen) according to the manufacturer's instructions.
PCR for FLT3-ITD and light cycler analysis for FLT3-TKD mutations
Polymerase chain reaction (PCR) and light cycler analysis were performed as described previously.48-50
Results
FLT3-TKD mutants D835Y/V/H/Q induce IL-3–independent growth in Ba/F3 cells
To analyze the transforming potential of FLT3-TKD mutants in IL-3–dependent Ba/F3 cells, we generated FLT3-D835Y/V/H/Q–expressing cell lines essentially as described previously by using the MSCV-IRES-EYFP retroviral expression vector.15 Identical expression levels of all FLT3 constructs were confirmed by flow cytometry and by Western blot after immunoprecipitation (IP) with a FLT3-specific antibody (data not shown). As a positive control we generated Ba/F3 cells expressing the FLT3-ITD (W51) mutant.15
As described previously the FLT3-ITD, but not the FLT3-WT, construct conferred IL-3–independent growth. All FLT3-D835 mutant-transduced Ba/F3 cell lines were able to grow in the absence of IL-3 at a growth rate comparable to that of the FLT3-ITD–expressing cells (data not shown).
FLT3-TKD mutants activate the STAT5 and MAPK signaling pathways
We and others have previously shown that the FLT3-ITD and D835Y mutants induce a constitutive STAT5 and MAPK activation in Ba/F3 or 32D cells.15-19 To characterize the signaling properties of different FLT3-TKD mutants, we analyzed lysates from Ba/F3 cells expressing the FLT3-WT, -ITD, or -D835Y/V/H/Q constructs. By using an antiphosphotyrosine antibody, we could clearly show that the FLT3-D835Y/V/H/Q–transduced Ba/F3 cells express a hyperphosphorylated FLT3 receptor (data not shown). Furthermore, we analyzed the ability of FLT3-TKD mutants to activate STAT5 and MAPK, 2 known downstream targets of FLT3-ITD mutants. For this purpose crude lysates from Ba/F3 cells expressing the FLT3-D835Y/V/H/Q mutants were analyzed by phosphospecific antibodies against STAT5 and MAPK. All FLT3-D835 mutants induced a strong constitutive activation of STAT5 and MAPK at levels comparable to the FLT3-ITD construct (data not shown).
These data clearly demonstrate that all FLT3-TKD mutants analyzed in our study have transforming potential and induce a hyperphosphorylation of FLT3. STAT5 and MAPK represent common downstream targets of the constitutively active FLT3-ITD and FLT3-TKD mutant receptors.
FLT3-TKD mutants differ significantly in their sensitivity to the growth inhibitory activity of the FLT3 PTK inhibitor SU5614
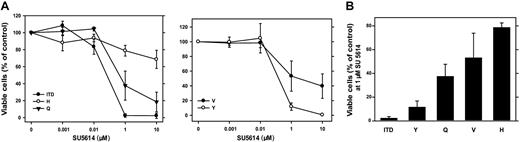
The PTK inhibitor SU5614 has selective inhibitory activity against class III RTK, including FLT3, and inhibits the growth of FLT3-ITD– and FLT3-D835Y–carrying Ba/F3 cells.15,27 To analyze the sensitivity of different FLT3-D835 mutants to SU5614, we cultured FLT3-D835Y/V/H/Q–expressing Ba/F3 cells in the presence of different concentrations of SU5614 for 72 hours. As shown in Figure 1A, FLT3-TKD mutants differ significantly in their sensitivity to the inhibitor. FLT3-D835Y and FLT3-ITD cells show a similar and high sensitivity to SU5614 (IC50 = 0.2 μM and 0.1 μM, respectively).
FLT3-TKD mutants differ significantly in their sensitivity to the growth inhibitory activity of the FLT3 PTK inhibitor SU5614. (A) Dose-response curves of the inhibitory activity of SU5614 in Ba/F3 FLT3-ITD and FLT3-TKD cells after 72 hours of incubation. Ba/F3 cells expressing different FLT3 constructs were seeded at a density of 4 × 104 cells/mL in the absence or presence of different concentrations of SU5614. Viable cells were counted after 72 hours by trypan blue exclusion. The growth of cells that were incubated without inhibitor was defined as 100%. Values represent means and standard errors from 3 independent experiments. (B) Sensitivity of FLT3-ITD and TKD mutants to SU5614. Ba/F3 cells were seeded at a density of 4 × 104 cells/mL in the presence of 1 μM SU5614 and counted after 72 hours. Values represent means and standard errors from 3 independent experiments.
FLT3-TKD mutants differ significantly in their sensitivity to the growth inhibitory activity of the FLT3 PTK inhibitor SU5614. (A) Dose-response curves of the inhibitory activity of SU5614 in Ba/F3 FLT3-ITD and FLT3-TKD cells after 72 hours of incubation. Ba/F3 cells expressing different FLT3 constructs were seeded at a density of 4 × 104 cells/mL in the absence or presence of different concentrations of SU5614. Viable cells were counted after 72 hours by trypan blue exclusion. The growth of cells that were incubated without inhibitor was defined as 100%. Values represent means and standard errors from 3 independent experiments. (B) Sensitivity of FLT3-ITD and TKD mutants to SU5614. Ba/F3 cells were seeded at a density of 4 × 104 cells/mL in the presence of 1 μM SU5614 and counted after 72 hours. Values represent means and standard errors from 3 independent experiments.
In contrast, the FLT3-D835Q and D835V mutants were significantly less sensitive to the inhibitor (IC50 = 0.4 and 1 μM, respectively), and the FLT3-D835H was the most resistant mutant (IC50 > 10 μM). At concentrations of 1 μM inhibitor FLT3-ITD cells showed a viability of 3% compared with 10% (FLT3-D835Y), 38% (FLT3-D835Q), 52% (FLT3-D835V), and 78% (FLT3-D835H) after 72 hours of incubation (Figure 1B).
These data demonstrate that FLT3-TKD mutants differ significantly in their sensitivity to the FLT3 PTK inhibitor SU5614 in the following order: D835Y > D835Q > D835V > D835H.
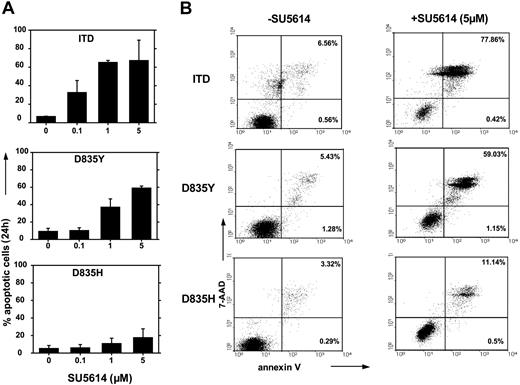
SU5614 induces apoptosis in FLT3-ITD– and FLT3-D835Y–transformed cells but not in FLT3-D835H–transformed cells
To further characterize the mechanisms of primary resistance of the FLT3-D835H mutant to SU5614, we analyzed induction of apoptosis after inhibitor treatment of FLT3-ITD, -D835Y–, and -D835H–transformed Ba/F3 cells with SU5614. Figure 2A (upper panel) shows that FLT3-ITD–transformed cells underwent rapid apoptotic cell death after exposure to increasing concentrations of SU5614 after 24 hours as measured by the expression of annexin V/7-AAD. FLT3-D835Y cells show the same level of sensitivity to SU5614 (IC50 = 0.1 μM) (Figure 2A, middle panel) but not the FLT3-D835H cells (Figure 2A, bottom panel).
SU5614 induces apoptosis in FLT3-ITD and FLT3-D835Q/V/Y cells but not in FLT3-D835H cells. (A-B) Ba/F3 cells transduced with the FLT3-ITD, FLT3-D835Y, or FLT3-D835H constructs were incubated with different concentrations of SU5614 for 24 hours and were analyzed by flow cytometry after staining with annexin V-PE and 7-AAD. Representative dot plots from 1 of 3 independently performed experiments are shown. Values represent means and standard deviations from 3 independent experiments. The numbers in dot-plot quadrants (B) show the percentages of annexin V+/7-AAD- (lower right quadrants) and annexin V+/7-AAD+ (upper right quadrants).
SU5614 induces apoptosis in FLT3-ITD and FLT3-D835Q/V/Y cells but not in FLT3-D835H cells. (A-B) Ba/F3 cells transduced with the FLT3-ITD, FLT3-D835Y, or FLT3-D835H constructs were incubated with different concentrations of SU5614 for 24 hours and were analyzed by flow cytometry after staining with annexin V-PE and 7-AAD. Representative dot plots from 1 of 3 independently performed experiments are shown. Values represent means and standard deviations from 3 independent experiments. The numbers in dot-plot quadrants (B) show the percentages of annexin V+/7-AAD- (lower right quadrants) and annexin V+/7-AAD+ (upper right quadrants).
In the presence of 5 μM SU5614, FLT3-D835H cells show only 18% apoptotic cells compared with FLT3-ITD (63%) and FLT3-D835Y cells (60%). A representative experiment is shown in Figure 2B and clearly demonstrates that SU5614 at a concentration of 5 μM induces apoptosis in 11.6% of D835H (lowest panel) cells compared with 60.2% and 78.3% in FLT3-D835Y (middle panel) and FLT3-ITD (upper panel) expressing Ba/F3 cells, respectively.
FLT3 PTK inhibitor SU5614 down-regulates autophosphorylation of the FLT3-ITD but not the FLT3-D835H receptor mutant
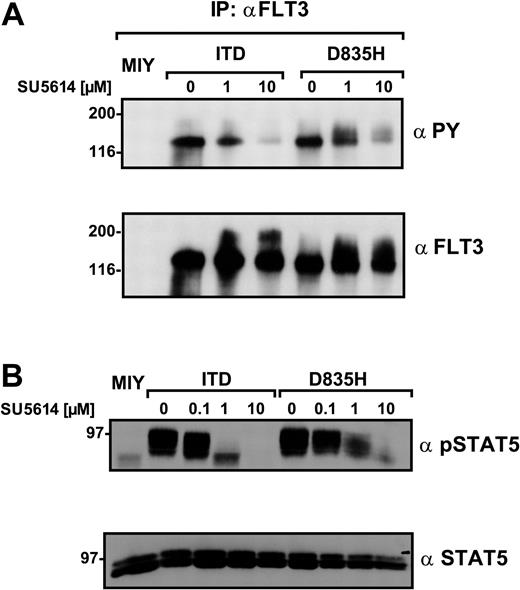
It was recently shown that SU5614 inhibits phosphorylation of the FLT3-ITD receptor and down-regulates STAT5 and MAPK phosphorylation.15,27 To confirm the resistance of FLT3-D835H cells to SU5614 on the level of individual signaling pathways, we analyzed the activation of FLT3, MAPK, and STAT5 after incubation with SU5614.
For this purpose, 293T cells were transiently transfected with either the FLT3-ITD or the D835H construct. As shown in Figure 3A, SU5614 induced an efficient dephosphorylation of the FLT3-ITD receptor at a concentration of 1 μM (38.5% of control, defined as the FLT3 phosphorylation in the absence of SU5614 compared with basal levels of FLT3 measured by densitometry). In contrast, the D835H mutant was still strongly hyperphosphorylated (92.7%) in the presence of 1 μM SU5614. At concentrations of 10 μM SU5614, the FLT3-ITD construct was almost completely dephosphorylated (7.4%), whereas a significant autophosphorylation was still detectable in FLT3-D835H–transfected cells (46.4%).
The FLT3 PTK inhibitor SU5614 down-regulates autophosphorylation of the FLT3-ITD but not FLT3-D835H receptor mutants. (A) 293T cells were transiently transfected with empty vector (MIY), FLT3-ITD, and FLT3-D835H constructs. Forty-four hours after transfection the cells were incubated with SU5614 as indicated for 4 hours and lysed. FLT3 was immunoprecipitated with a polyclonal anti-FLT3 antibody. Tyrosine phosphorylation of FLT3 was determined by Western blot analysis with the use of a monoclonal anti-PY antibody, and identical loading was confirmed by reblotting with a polyclonal anti-FLT3 antibody. (B) The expression and phosphorylation of STAT5 in Ba/F3 MIY-, FLT3-ITD–, and FLT3-D835H–expressing cells treated with 0.1, 1, and 10 μM SU5614 or untreated cells was determined by Western blot analysis in crude lysates with the use of polyclonal pSTAT5 and anti-STAT5 antibodies.
The FLT3 PTK inhibitor SU5614 down-regulates autophosphorylation of the FLT3-ITD but not FLT3-D835H receptor mutants. (A) 293T cells were transiently transfected with empty vector (MIY), FLT3-ITD, and FLT3-D835H constructs. Forty-four hours after transfection the cells were incubated with SU5614 as indicated for 4 hours and lysed. FLT3 was immunoprecipitated with a polyclonal anti-FLT3 antibody. Tyrosine phosphorylation of FLT3 was determined by Western blot analysis with the use of a monoclonal anti-PY antibody, and identical loading was confirmed by reblotting with a polyclonal anti-FLT3 antibody. (B) The expression and phosphorylation of STAT5 in Ba/F3 MIY-, FLT3-ITD–, and FLT3-D835H–expressing cells treated with 0.1, 1, and 10 μM SU5614 or untreated cells was determined by Western blot analysis in crude lysates with the use of polyclonal pSTAT5 and anti-STAT5 antibodies.
We further analyzed the effects of SU5614 on the activation of 2 important downstream signaling pathways activated by FLT3-LM/TKD mutants. In the presence of 1 μM SU5614, STAT5 phosphorylation in Ba/F3 FLT3-ITD cells was almost completely inhibited (Figure 3B). In contrast, in Ba/F3 FLT3-D835H–expressing cells STAT5 was only slightly dephosphorylated in the presence of this concentration of inhibitor. The incubation of the cells with 10 μM SU5614 produced an almost complete inhibition of STAT5 phosphorylation in both Ba/F3 FLT3-ITD– and D835H-transduced cell lines. These results confirm at a molecular level the different sensitivity of FLT3-TKD mutants to FLT3 PTK inhibitors.
Generation of SU5614-resistant Ba/F3 FLT3-ITD cells
FLT3 PTK inhibitors are now being evaluated in clinical phase 1 and 2 studies in patients with AML (described in “Introduction”), but it is unknown to what extent clinical resistance will develop and influence the clinical efficiency of these inhibitors.
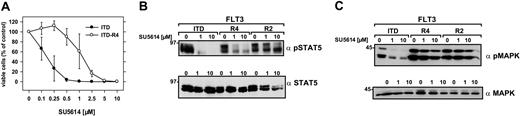
To address this important issue, FLT3-ITD-W51–expressing polyclonal Ba/F3 cells were cultivated either in the presence of increasing concentrations of SU5614 (0.2-0.8 μM) or in the absence of inhibitor (control cells) for a time period of 3 months. After a cultivation period of 2 weeks at a concentration of 0.2 μM SU5614, 2 independent cell lines were obtained that were each split in 2 sublines (ITD-R1+R2 and ITD-R3+R4, respectively). In contrast to the control cell line all 4 cell lines (ITD-R1-R4) growing in the presence of inhibitor developed a partial resistance to SU5614 and showed an IC50, which was at least 6.5-fold higher than the IC50 of control Ba/F3 FLT3-ITD cells (Tables 1, 2). A representative example is shown in Figure 4A: Ba/F3 FLT3-ITD-R4 cells were cultured for 72 hours in the presence of increasing concentrations of SU5614. The IC50 for SU5614 was 0.2 μM in the control ITD cell line and 1.3 μM in ITD-R4 cells. At high concentrations of 10 μM inhibitor, the ITD-R4 line was still sensitive to the growth inhibitory activity of SU5614, showing that the proliferation of these cells was still dependent on the presence of the activating FLT3 mutant.
Functional and molecular characterization of Ba/F3 FLT3-ITD-R1-4 cells
. | IC50 . | . | FLT3 expression . | . | . | ||
|---|---|---|---|---|---|---|---|
| Cell line . | SU5614, μM . | AraC, μg/mL . | FACS, MCF . | WB, % of ITD native . | FLTc-TKD mutation . | ||
| ITD native | 0.2 | 0.06 | 14.1 | 100 | — | ||
| ITD-R1 | 4.5 | 0.03 | 50.9 | 725 | Y842H | ||
| ITD-R2 | 5.2 | 0.08 | 41.2 | 918 | Y842H | ||
| ITD-R3 | 2.1 | ND | 63.3 | 1016 | D835N | ||
| ITD-R4 | 1.3 | 0.04 | 54.4 | 1285 | D835N | ||
. | IC50 . | . | FLT3 expression . | . | . | ||
|---|---|---|---|---|---|---|---|
| Cell line . | SU5614, μM . | AraC, μg/mL . | FACS, MCF . | WB, % of ITD native . | FLTc-TKD mutation . | ||
| ITD native | 0.2 | 0.06 | 14.1 | 100 | — | ||
| ITD-R1 | 4.5 | 0.03 | 50.9 | 725 | Y842H | ||
| ITD-R2 | 5.2 | 0.08 | 41.2 | 918 | Y842H | ||
| ITD-R3 | 2.1 | ND | 63.3 | 1016 | D835N | ||
| ITD-R4 | 1.3 | 0.04 | 54.4 | 1285 | D835N | ||
The IC50 of SU5614 and Ara-C in ITD-R1-4 cells was determined as described in Figure 4. FLT3 expression was measured by FACS, and the mean channel fluorescence (MCF) was calculated. The expression of FLT3 (WB) was determined by Western blot analysis with the use of an anti-FLT3 antibody. The blot was stripped and reprobed with an anti–β-actin antibody. The results were quantified by densitometry according to the FLT3–β-actin ratio of parental cells that was set to 100%. FLT3-TKD mutation indicates the presence (Y842H and D835N) or absence (—) of point mutations in TKD. ND indicates not done.
SU5614-resistant cell lines are sensitive to PKC412
. | SU5614 . | . | PKC412 . | . | ||
|---|---|---|---|---|---|---|
. | IC50, μM . | Fold . | IC50, nM . | Fold . | ||
| ITD native | 0.2 | 1.0 | 7.5 | 1.0 | ||
| ITD-R1 | 4.5 | 23 | ND | ND | ||
| ITD-R2 | 5.2 | 26 | 8.7 | 1.2 | ||
| ITD-R3 | 2.1 | 11 | ND | ND | ||
| ITD-R4 | 1.3 | 6.5 | 17 | 2.3 | ||
. | SU5614 . | . | PKC412 . | . | ||
|---|---|---|---|---|---|---|
. | IC50, μM . | Fold . | IC50, nM . | Fold . | ||
| ITD native | 0.2 | 1.0 | 7.5 | 1.0 | ||
| ITD-R1 | 4.5 | 23 | ND | ND | ||
| ITD-R2 | 5.2 | 26 | 8.7 | 1.2 | ||
| ITD-R3 | 2.1 | 11 | ND | ND | ||
| ITD-R4 | 1.3 | 6.5 | 17 | 2.3 | ||
The IC50 of SU5614 in Ba/F3 FLT3-ITD-R2 and -R4 cells was determined as described in Figure 4. The IC50 of PKC412 was determined by MTT assays as described previously.15 Fold indicates the ratio of the IC50 of parental Ba/F3 FLT3-ITD cells to IC50 of R2 and R4 cell lines, respectively. All values represent means of 3 independent experiments.
Resistance of FLT3-ITD-R4 cell line to SU5614 and activation of STAT5 and MAPK in FLT3-ITD-R1-4 cells in response to the inhibitor. (A) Ba/F3 FLT3-ITD-R4 cells were seeded at a density of 4 × 104 cells/mL in the absence or presence of different concentrations of SU5614 and viable cells were counted after 72 hours by trypan blue exclusion. Values represent means and standard errors from 3 independent experiments. (B-C) The phosphorylation status of STAT5 and MAPK in extracts of Ba/F3 FLT3-ITD and FLT3-ITD-R4/2 cells treated with 1 and 10 μM SU5614 for 4 hours was determined by Western blot analysis by using the polyclonal anti-pSTAT5 (B) and anti-pMAPK antibodies (C). Expression of STAT5 and MAPK in the same lysates was analyzed by immunoblotting with polyclonal anti-STAT5 and anti-MAPK antibodies.
Resistance of FLT3-ITD-R4 cell line to SU5614 and activation of STAT5 and MAPK in FLT3-ITD-R1-4 cells in response to the inhibitor. (A) Ba/F3 FLT3-ITD-R4 cells were seeded at a density of 4 × 104 cells/mL in the absence or presence of different concentrations of SU5614 and viable cells were counted after 72 hours by trypan blue exclusion. Values represent means and standard errors from 3 independent experiments. (B-C) The phosphorylation status of STAT5 and MAPK in extracts of Ba/F3 FLT3-ITD and FLT3-ITD-R4/2 cells treated with 1 and 10 μM SU5614 for 4 hours was determined by Western blot analysis by using the polyclonal anti-pSTAT5 (B) and anti-pMAPK antibodies (C). Expression of STAT5 and MAPK in the same lysates was analyzed by immunoblotting with polyclonal anti-STAT5 and anti-MAPK antibodies.
Activation of STAT5 and MAPK in FLT3-ITD-R1-4 cell lines in response to SU5614
To confirm the resistance to SU5614 at a molecular level, we analyzed 2 important downstream targets of FLT3: STAT5 and MAPK. For this purpose FLT3-ITD-R2 and -R4 cell lines were incubated with different concentrations of SU5614 (0, 1, and 10 μM) for 4 hours. As shown in Figure 4 (B-C), SU5614 efficiently induces dephosphorylation of STAT5 and MAPK at a concentration of 1 μM in Ba/F3 FLT3-ITD cells. In contrast to the parental cell line, STAT5 and MAPK were still phosphorylated in the FLT3-ITD-R2 and -R4 cells even in the presence of 10 μM SU5614. These results are in good agreement with the IC50 of these cell lines in cell proliferation assays, showing a 4-fold higher IC50 of ITD-R2 compared with ITD-R4 cells (Table 1).
Ba/F3 FLT3-ITD-R1-4 cells are resistant to the FLT3 PTK inhibitor AG1295 but not to PKC412, Genistein, and Ara-C
To further characterize the SU5614-resistant cell lines and to analyze whether these cells developed a specific resistance to FLT3 PTK inhibitors, we cultivated FLT3-ITD-R1-4 cells in the presence of several different PTK inhibitors. We evaluated PKC41251 and AG1295,25 which have inhibitory activity to class III PTK, as well as the broad-spectrum PTK inhibitor Genistein.52 Our data show that the R2 and R4 cell lines were almost as sensitive to PKC412 as parental Ba/F3 FLT3-ITD cell lines (IC50 = 8.7 nM, 17 nM, and 7.5 nM, respectively; Table 2).
Furthermore, we investigated whether the combination of different FLT3 PTK inhibitors might be superior to single agents. For this purpose we incubated Ba/F3 FLT3-ITD parental cells with either SU5614 or PKC412 alone or in combination. Our results clearly show that a combination of inhibitors has additive, but not synergistic, activity (Table 3).
Combination of 2 selective FLT3 PTK inhibitors has additive cytotoxic activity in Ba/F3 FLT3-ITD cells
. | . | Ci values at . | . | . | ||
|---|---|---|---|---|---|---|
. | IC50 . | IC50 . | IC75 . | IC90 . | ||
| SU5614, μM | 0.17 | NA | NA | NA | ||
| PKC412, nM | 7.3 | NA | NA | NA | ||
| SU5614, μM + PKC412, nM | 0.11 + 2.5 | 1.0 | 1.0 | 1.1 | ||
. | . | Ci values at . | . | . | ||
|---|---|---|---|---|---|---|
. | IC50 . | IC50 . | IC75 . | IC90 . | ||
| SU5614, μM | 0.17 | NA | NA | NA | ||
| PKC412, nM | 7.3 | NA | NA | NA | ||
| SU5614, μM + PKC412, nM | 0.11 + 2.5 | 1.0 | 1.0 | 1.1 | ||
IC50 values were determined as described in Table 2 and Figure 4. To assess combined drug effects, SU5614 and PKC412 were added simultaneously to cells in culture medium. The combination indices were evaluated by solving the equation Ci=[(D)1/(Dx)1] + [(D)2/(Dx)2] as described previously.47 (Dx)1 is the concentration of drug 1 required to produce an x percent effect of that drug alone. (D)1 is the concentration of drug 1 required to produce the same x percent effect in combination with (D)2. A combination index (Ci) less than 1 indicates synergy, a value more than 1 indicates antagonism, and a value of 1 indicates an additive effect. All values represent means of 3 independent experiments. NA indicates not applicable.
In addition, we could demonstrate that all SU5614-resistant cell lines were still sensitive to Genistein (data not shown). In contrast, the ITD-R1-4 cell lines developed almost complete resistance to the FLT3-selective PTK inhibitor, AG1295 (data not shown), which might be explained by structure similarity of AG1295 and SU5614.
Next, we investigated whether the ITD-R1-4 cells developed an unspecific resistance to apoptotic cell death induced by cytotoxic drugs. For this purpose we exposed these cells to different concentrations of Ara-C (cytosine arabinoside, 0-5 μg/mL), a deoxycytidine analog that is the most effective cytotoxic agent in the treatment of AML. We found that FLT3-ITD-R1-4 mutants are sensitive to Ara-C at a level comparable to that seen in the parental FLT3-ITD line (data not shown, summarized in Table 1).
These data clearly show that the ITD-R1-4 cell lines are resistant to the growth inhibitor activity of the FLT3 PTK inhibitor AG1295 but not to unselective PTK inhibitors or the cytotoxic agent Ara-C.
Structurally unrelated and selective FLT3 PTK inhibitors like PKC412 were able to overcome SU5614 resistance and showed additive activity in combination with SU5614 in FLT3-ITD–transformed cells.
Expression of FLT3 protein in resistant cell lines
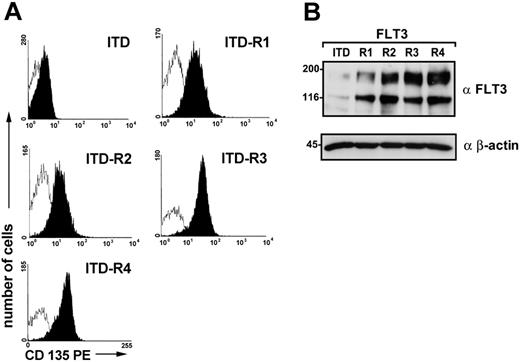
Overexpression of the target kinase is a well-known mechanism of PTK inhibitor resistance.36-38 To further evaluate the mechanisms of SU5614 resistance in the ITD-R1-4 cell lines, we analyzed FLT3 surface expression by flow cytometry with the use of a PE-labeled monoclonal anti-FLT3 antibody. All resistant cell lines expressed significantly higher levels of FLT3 compared with the parental FLT3-ITD cell line (Figure 5A; Table 1).
Expression of FLT3 in SU5614-resistant and parental FLT3-ITD cell lines. (A) The expression of FLT3 in Ba/F3 FLT3-ITD-R1-4 was analyzed by fluorescence activated cell sorting (FACS) analysis with the use of an anti-CD135 PE-labeled antibody compared with parental FLT3-ITD cells. Open histograms represent isotype control (PE-labeled control antibody); filled histograms show fluorescence intensity of CD135. (B) Lysates were prepared from these cells, and FLT3 expression was determined by immunoblotting with the use of a polyclonal anti-FLT3 antibody. Identical protein loading in all lanes was confirmed by immunoblotting with the use of an anti–β-actin antibody.
Expression of FLT3 in SU5614-resistant and parental FLT3-ITD cell lines. (A) The expression of FLT3 in Ba/F3 FLT3-ITD-R1-4 was analyzed by fluorescence activated cell sorting (FACS) analysis with the use of an anti-CD135 PE-labeled antibody compared with parental FLT3-ITD cells. Open histograms represent isotype control (PE-labeled control antibody); filled histograms show fluorescence intensity of CD135. (B) Lysates were prepared from these cells, and FLT3 expression was determined by immunoblotting with the use of a polyclonal anti-FLT3 antibody. Identical protein loading in all lanes was confirmed by immunoblotting with the use of an anti–β-actin antibody.
These data were confirmed by Western blot analyses using a polyclonal antibody directed against the interkinase domain of FLT3 (Figure 5B; Table 1). FLT3 expression was significantly higher in ITD-R4 (1285%), ITD-R3 (1016%), ITD-R2 (918%), and ITD-R1 cells (725%) compared with FLT3 levels in the parental FLT3-ITD cell line (100%).
SU5614-resistant ITD-R1-4 cell lines acquired distinct FLT3-TKD mutations
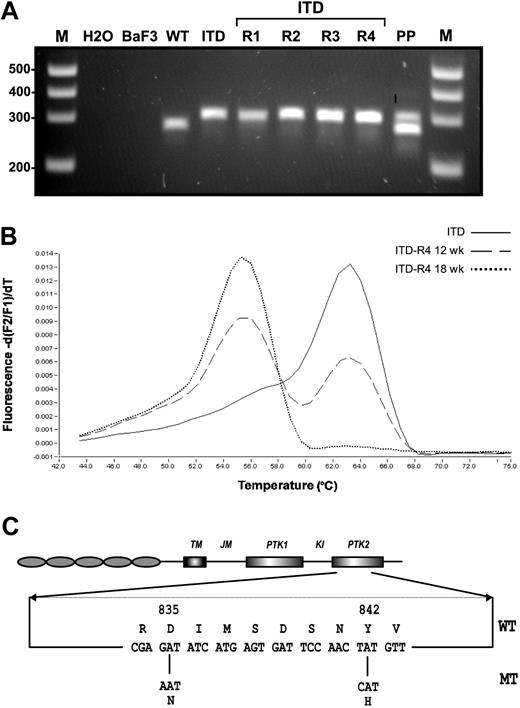
Another mechanism of clinical resistance to PTK inhibitors represents the acquisition of distinct mutations in the adenosine triphosphate (ATP) binding site and the kinase activation loop.40-45 We hypothesized that similar mechanisms might also be responsible for resistance to FLT3 PTK inhibitors in SU5614-resistant FLT3-ITD cell lines. For this purpose we analyzed the known hot spot regions for activating mutations in the FLT3 gene found in patients with AML: the juxtamembrane (JM) region and the activation loop of the kinase domain. The JM region was screened for mutations by using the previously published primers 11F and 12R and subsequent visualization of the PCR products in agarose gels.48,49 The ITD-R1-4 lines expressed the parental ITD (W51) mutation, as shown by an identical size of the PCR product compared with the control cell line (Figure 6A).
Molecular characterization of SU5614-resistant cells. (A) The juxtamembrane region of FLT3 was amplified by PCR with the use of the published primer pair 11F and 12R,48,49 and PCR products were separated by agarose gel electrophoresis and viewed under UV illumination after ethidium bromide staining. M indicates DNA molecular weight marker; H2O, water control; Ba/F3, native Ba/F3 cells; WT, Ba/F3 FLT3-WT; ITD, Ba/F3 FLT3-ITD; ITD-R1-4, Ba/F3 FLT3-ITD-R1-4; PP, positive control (patient with AML carrying a FLT3-LM). (B) Detection of FLT3-TKD mutations was performed by melting curve analysis after amplification of a 244-bp fragment by real-time PCR as described previously.50 In the presence of the FLT3-TKD wild-type sequence or the FLT3 TKD mutant DNA the fluorescence peak is observed at 63°C and 55°C, respectively. (C) The structural domains of the FLT3 protein with the position of the TKD mutation found in Ba/F3 FLT3-ITD-R1-4 cells (D835N and Y842H) are shown.
Molecular characterization of SU5614-resistant cells. (A) The juxtamembrane region of FLT3 was amplified by PCR with the use of the published primer pair 11F and 12R,48,49 and PCR products were separated by agarose gel electrophoresis and viewed under UV illumination after ethidium bromide staining. M indicates DNA molecular weight marker; H2O, water control; Ba/F3, native Ba/F3 cells; WT, Ba/F3 FLT3-WT; ITD, Ba/F3 FLT3-ITD; ITD-R1-4, Ba/F3 FLT3-ITD-R1-4; PP, positive control (patient with AML carrying a FLT3-LM). (B) Detection of FLT3-TKD mutations was performed by melting curve analysis after amplification of a 244-bp fragment by real-time PCR as described previously.50 In the presence of the FLT3-TKD wild-type sequence or the FLT3 TKD mutant DNA the fluorescence peak is observed at 63°C and 55°C, respectively. (C) The structural domains of the FLT3 protein with the position of the TKD mutation found in Ba/F3 FLT3-ITD-R1-4 cells (D835N and Y842H) are shown.
To screen for mutations in the TKD domain, we used melting curve analyses after amplification of a 244–base pair fragment by real-time PCR as described previously.50,53 The design of the hybridization probe allows detection of the point mutations and insertions/deletions ranging from amino acid 825 to 839 in the TKD domain of FLT3. The real-time PCR analysis demonstrated abnormal melting curves for the ITD-R3 and -R4 mutants by using either cDNA or genomic DNA as a template (Figure 6B). Nucleotide sequencing of genomic DNA from these cells confirmed a single and identical G → A nucleotide (nt) exchange in both the ITD-R3 and ITD-R4 cell lines, resulting in a D835N mutation (Figure 6C). Sequential analysis of the FLT3-ITD-R4 cell line at 12 weeks and 18 weeks during selection with SU5614 indicated that the ratio of D835N/D835 DNA continuously increased over time. The melting curve analyses shown in Figure 6B clearly demonstrate that the D835N mutant allele accounted for approximately 50% of DNA after 12 weeks of selection and increased to 100% of total DNA after continuous exposure to SU5614 for another 6 weeks. These results were confirmed by direct nucleotide sequencing of the DNA templates used for melting curve analysis (data not shown).
After the identification of a D835N mutant in ITD-R3 and -R4 cells, we sequenced the entire TKD domain of the ITD-R1 and -R2 cell lines to screen for further mutations outside the region that was covered by the hybridization primers used in the light cycler analysis. Direct nucleotide sequencing clearly demonstrated that both the ITD-R1 and -R2 cells carried a T → C nucleotide substitution in codon 842 of FLT3, resulting in a Y842H mutation (Figure 6C).
Taken together, these data clearly indicate that distinct mutations in the PTK domain of FLT3 are acquired in Ba/F3 FLT3-ITD cells during selection with the FLT3 PTK inhibitor SU5614.
FLT3-ITD/TKD dual mutation restores the SU5614-resistant phenotype in FLT3-ITD-R1-4 cells
To confirm that the D835N and Y842H TKD mutations are responsible for the resistance to SU5614 in ITD-R1/2 and ITD-R3/4 cells, we introduced either the D835N or the Y842H mutation in the FLT3-WT and the FLT3-ITD constructs. Then Ba/F3 cell lines expressing the FLT3-D835N, Y842H, or the FLT3-ITD mutant alone were generated. In addition, the FLT3 dual mutant FLT3-ITD/D835N and FLT3-ITD/Y842H Ba/F3 cell lines were established. All FLT3 mutants conferred IL-3–independent growth to Ba/F3 cell lines (data not shown, summarized in Table 4).
Characterization of Ba/F3 cells expressing FLT3-ITD/TKD dual mutants
Ba/F3 cell line . | IC50 of SU5614, μM . | IL-3–independent growth . |
|---|---|---|
| ITD (W51) | 0.2 | + |
| D835N | 0.1 | + |
| ITD-D835N | 1.0 | + |
| Y842H | 0.2 | + |
| ITD-Y842H | 1.1 | + |
Ba/F3 cell line . | IC50 of SU5614, μM . | IL-3–independent growth . |
|---|---|---|
| ITD (W51) | 0.2 | + |
| D835N | 0.1 | + |
| ITD-D835N | 1.0 | + |
| Y842H | 0.2 | + |
| ITD-Y842H | 1.1 | + |
The IC50 of SU5614 in Ba/F3 cell lines expressing either the D835N or Y842H or the FLT3-ITD mutant alone and the FLT3-ITD/D835N, FLT3-ITD/Y842H dual mutants were determined as described in Figure 7. + indicates IL-3–independent growth.
Further, we characterized the activation and signaling properties of the FLT3-Y842H receptor mutant that has not been described in clinical samples so far. In Western blot analyses, the FLT3-Y842H receptor was hyperphosphorylated on tyrosine compared with the FLT3-WT receptor and was also able to activate STAT5 and MAPK (data not shown).
As shown in Figure 7A and Table 4, the IC50 of FLT3-D835N cells to SU5614 did not significantly differ from the IC50 of FLT3-ITD cells. In contrast, the FLT3-ITD/D835N dual mutant cells were partially resistant to SU5614 at levels comparable to the original ITD-R4 cell line (IC50 = 1.0 and 1.3 μM, respectively). Similar results were obtained when FLT3-Y842H and FLT3-ITD/Y842H cell lines were analyzed (Table 4).
The FLT3-ITD/D835N dual mutant restores the SU5614-resistant phenotype in Ba/F3 cells. (A) The Ba/F3 FLT3-ITD–, FLT3-D835N–, and FLT3-ITD/D835N–expressing cells were seeded at a density of 4 × 104 cells/mL in the absence or presence of different concentrations of SU5614, and viable cells were counted after 72 hours by trypan blue exclusion. Values represent means and standard deviations from 3 independent experiments. (B-C) MIG FLT3-ITD (GFP+)– and MIY FLT3-ITD/D835N (YFP+)–expressing Ba/F3 cells were mixed in a ratio of 10:1, and the percentage of GFP+ and YFP+ cells was measured every 3 to 4 days for a time period of 2 weeks by FACS analysis in the presence (B) or absence (C) of 0.2 μM SU5614.
The FLT3-ITD/D835N dual mutant restores the SU5614-resistant phenotype in Ba/F3 cells. (A) The Ba/F3 FLT3-ITD–, FLT3-D835N–, and FLT3-ITD/D835N–expressing cells were seeded at a density of 4 × 104 cells/mL in the absence or presence of different concentrations of SU5614, and viable cells were counted after 72 hours by trypan blue exclusion. Values represent means and standard deviations from 3 independent experiments. (B-C) MIG FLT3-ITD (GFP+)– and MIY FLT3-ITD/D835N (YFP+)–expressing Ba/F3 cells were mixed in a ratio of 10:1, and the percentage of GFP+ and YFP+ cells was measured every 3 to 4 days for a time period of 2 weeks by FACS analysis in the presence (B) or absence (C) of 0.2 μM SU5614.
We further analyzed whether the dual FLT3 ITD/TKD mutant provides a competitive growth advantage to Ba/F3 cells in the absence or presence of 0.2 μM SU5614 compared with the parental ITD cell line. For this purpose, we mixed MIG FLT3-ITD (GFP+)– and MIY FLT3-ITD/D835N (YFP+)–expressing Ba/F3 cells in a ratio of 10:1 and measured the percentage of GFP+ and YFP+ cells every 3 to 4 days for a time period of 2 weeks by FACS analysis (Figure 7B-C).
We could show that over a period of 14 days the FLT3-ITD/D835N dual-mutant cells had a substantial competitive growth advantage in the presence, but not in the absence, of SU5614. The percentage of FLT3-ITD-D835N-YFP–positive cells in the presence of SU5614 increased from 10% (day 0) to 88% (day 14), whereas the percentage of dual-mutant YFP-positive cells cultured in the absence of SU5614 increased only to 22%.
These data clearly show that the FLT3-ITD/TKD dual mutant restores the ITD-R1-4 phenotype and that the acquisition of specific TKD mutations in the FLT3-ITD gene represents a new mechanism of resistance to FLT3 PTK inhibitors in vitro.
Discussion
We and others have previously shown that selective FLT3 PTK inhibitors, like SU5614, AG1295, PKC412, or CEP701, can induce growth arrest and apoptosis in FLT3-transformed cell lines and in primary AML blasts.15,25,27,28,51 These promising in vitro results have stimulated phase 1 and 2 clinical studies evaluating the efficacy of FLT3 PTK inhibitors in patients with AML.29,31,32
In contrast to the well-studied mechanisms of drug resistance found in patients treated with the PTK inhibitor imatinib mesylate, little is known about primary and secondary mechanisms of resistance to FLT3 PTK inhibitors. In the present study we could show that certain activating point mutations found in the PTK domain of FLT3 in patients with AML significantly alter the sensitivity to the FLT3 PTK inhibitor SU5614. As a mechanism of secondary resistance, we identified that FLT3-ITD–transformed cell lines acquire distinct mutations in the TKD domain after prolonged cultivation in the presence of FLT3 PTK inhibitors. Reconstruction of these FLT3 dual mutants confirmed that the presence of a certain TKD mutant on the genetic background of the ITD mutation is responsible for the FLT3-PTK inhibitor-resistant phenotype. These data provide genetic evidence that preexisting and acquired mutations in the TKD domain of FLT3 are sufficient to induce drug resistance to PTK inhibitors.
Genetically heterogeneous mutations in the TKD domain of FLT3 have been found in 7% to 8% of patients with AML and include point mutations at codon D835 and also insertions/deletions in codons 835/836.7-9 To analyze the sensitivity of these mutants to FLT3 PTK inhibitors, we generated Ba/F3 cell lines expressing the most frequent FLT3-TKD mutants, namely D835Y/V/H. All FLT3-TKD mutants confer IL-3–independent growth to Ba/F3 cells and activate similar signal transduction pathways (STAT5 and MAPK) compared with the FLT3-ITD construct. Importantly, these FLT3-TKD mutants differed significantly in their sensitivity to the growth inhibitory and apoptosis-inducing activity of the FLT3 PTK inhibitor SU5614 in the following order: FLT3-ITD = FLT3-D835 Y > V > H.
AML-specific mutations in the TKD domain of PTKs occur in conserved amino acids at codons 835/836 and 816 in FLT3 and KIT, respectively.7,8,54 It has been hypothesized that these mutations change the conformation of the activation loop (A-loop), thereby allowing spontaneous opening of the catalytic pocket and ligand-independent activation of the kinase.55,56 The conformational change in the A-loop of KIT by the D816V mutation is probably also responsible for the resistance of this mutant to the PTK inhibitor imatinib mesylate.54,57 On the basis of these findings, the observation of decreased SU5614 sensitivity of certain FLT3-TKD mutants, eg, D835H, might also be attributable to the decreased affinity of the inhibitor for the catalytic pocket of FLT3. Recently, Grundler et al58 have analyzed the sensitivity of mutants of FLT3 at codon 835/836 to the FLT3 PTK inhibitors SU5614, PKC412, and AG1295. Those researchers found in accordance with our results that distinct FLT3-TKD mutations lead to an altered sensitivity to PTK inhibitors.58
These findings have important clinical implications for studies evaluating the efficacy of FLT3 PTK inhibitors in patients with FLT3-TKD mutation-positive AML. FLT3-TKD mutations can change the affinity of the kinase to inhibitors and might, therefore, result in clinical resistance. Treatment of patients with AML carrying TKD mutations will, therefore, require a preclinical sensitivity analysis to select an appropriate inhibitor.
We characterized 4 sublines of the parental FLT3-ITD–transformed Ba/F3 cell line after selection with SU5614. All cell lines acquired a partial resistance to the FLT3 PTK inhibitor SU5614 but not to other apoptosis-inducing agents like Ara-C, and showed an IC50 that was 7- to 25-fold higher compared with that of control cells. Importantly, the ITD-R1-4 lines were completely sensitive to SU5614 at high concentrations of 10 μM inhibitor, showing that the proliferation of all cell lines was still dependent on the presence of the FLT3 mutant. By using the structurally different FLT3 PTK inhibitor PKC412 we could demonstrate that the SU5614-resistant cell lines were almost as sensitive to inhibition as the Ba/F3-FLT3-ITD cells (Table 2). Thus, our results show that cell lines harboring a FLT3 mutation that was acquired by pretreatment with a FLT3 inhibitor might still be sensitive to treatment with a structurally unrelated compound.
The upfront combination therapy with PTK inhibitors in patients with AML cannot be supported so far from our observations, because the combination of 2 inhibitors did not show any significant synergism in FLT3-LM–transformed cells. It still remains unclear if concomitant application of structurally unrelated inhibitors could prevent AML cells from developing drug resistance.
Our detailed molecular analysis showed that each of the ITD-R1-4 cell lines acquired a specific TKD mutation, resulting either in a D835N or Y842H FLT3 mutant. In sequential analyses performed in the ITD-R4 cell line during selection with SU5614, the ratio of mutant/wild-type DNA increased over time, showing a substantial competitive growth advantage of the ITD-D835N dual-mutant–expressing cell population.
Y842 of FLT3 is also a highly conserved amino acid in class III RTK, such as in the colony-stimulating factor-1 receptor (CSF1R) and KIT,2 but mutations of this position have not been described in patients with AML so far. Further studies are necessary to clarify whether conserved amino acids other than D835/I836 in the TKD domain of FLT3 are mutated in AML.
Clinical samples obtained from patients with AML usually carry either the LM or the TKD mutation.3,4,9 However, in some patients both mutations can be detected, and preliminary data suggest that these patients might have an even worse clinical prognosis compared with patients with a single LM or TKD mutation.59,60 In a recent study of 979 patients, Thiede et al9 found an additional TKD mutation in 17 of 200 (8.5%) patients with AML carrying a FLT3-LM. Further analyses revealed that 40% of the LM and TKD mutations occurred on the same allele, showing that FLT3-LM/TKD dual mutants spontaneously arise in vivo.9 Although the dual FLT3 LM/TKD mutant is infrequently found at diagnosis in patients with AML, our in vitro data suggest that these mutants might develop during treatment with FLT3 PTK inhibitors.
To prove that the acquisition of the TKD mutation is responsible for the FLT3 PTK inhibitor resistance in the ITD-R1-4 cells, we generated Ba/F3 cells expressing either the FLT3-D835N/Y842H mutant alone or on the background of the ITD mutation. Although both TKD mutants alone induced IL-3–independent growth in Ba/F3 cells, these mutants were equally sensitive to SU5614 as the FLT3-ITD construct. In contrast, when introduced on the ITD background, both the D835N and the Y842H mutation restored the SU5614-resistant phenotype in Ba/F3 cells.
Although the FLT3-D835N and the Y842H mutants have transforming activity, these constructs alone did not mediate resistance to SU5614. The explanation for this phenomenon is not obvious, but one has to assume that only the combination of the ITD and the D835N/Y842H mutation changes the conformation of the catalytic domain in a way which decreases its affinity for the FLT3 inhibitor. Structural analyses of the Ephrin-receptors have shown that both the activation loop and the JM region directly interact with the PTK domain by the formation of autoinhibitory loops.55,56 To what extent the exact amino acid sequence and the site of insertion of the FLT3-LM will influence the resistance to a specific FLT3 PTK inhibitor is unknown.
Although the FLT3-ITD/TKD dual mutant restores the SU5614-resistant phenotype when introduced in FLT3-ITD–transformed Ba/F3 cells, we cannot completely rule out that other mechanisms might contribute to the phenotype of ITD-R1-4 cells. Such mechanisms of resistance include altered metabolism of the drug, induction of drug efflux, eg, by ATP-binding cassette (ABC) transporters or overexpression of the target kinase. Recently, Weisberg et al51 have generated a PKC412-resistant Ba/F3 cell line that was selected with increasing inhibitor concentrations and showed overexpression of FLT3 as a potential mechanism of drug resistance. Although the FLT3-ITD/TKD dual-mutant cells generated by transduction of Ba/F3 cells with the FLT3 cDNA carrying both mutations resembled the Ba/F3 FLT3-ITD-R1-4 phenotype, the IC50 of SU5614 in ITD-R1-4 cells was still 2 to 5 times higher (Tables 1,4). It is possible that the FLT3 overexpression found in the ITD-R1-4 cells might contribute to the enhanced SU5614 resistance in these cells compared with the dual ITD/TKD mutant Ba/F3 cells. As shown in patients with CML developing clinical resistance during imatinib mesylate treatment, several mechanisms of resistance can occur in parallel, and overexpression of the target kinase might contribute to drug resistance in vivo.
Taken together, mutations in the TKD domain of FLT3 can induce primary and secondary resistance to FLT3 PTK inhibitors. These findings have a profound impact on clinical studies evaluating FLT3 PTK inhibitors in patients with AML and should be considered when clinical resistance to these compounds occurs.
Prepublished online as Blood First Edition Paper, November 6, 2003; DOI 10.1182/blood-2003-05-1653.
Supported by a grant from the Deutsche Krebshilfe (grant 10-1562) and the Deutsche Forschungsgemeinschaft (DFG Sp 556/1-3).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Ralf Dirschinger for helpful discussions and Susan King and Stefan Bohlander for critical reading of the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal