Abstract
DNA methylation plays critical roles in the development and differentiation of mammalian cells, and its dysregulation has been implicated in oncogenesis. This study was designed to determine whether DNA hypomethylation-associated aberrant gene expression is involved in adult T-cell leukemia (ATL) leukemogenesis. We isolated hypomethylated DNA regions of ATL cells compared with peripheral blood mononuclear cells from a carrier by a methylated CpG-island amplification/representational difference analysis method. The DNA regions identified contained MEL1, CACNA1H, and Nogo receptor genes. Sequencing using sodium bisulfite-treated genomic DNAs revealed the decreased methylated CpG sites, confirming that this method detected hypomethylated DNA regions. Moreover, these hypomethylated genes were aberrantly transcribed. Among them, MEL1S, an alternatively spliced form of MEL1 lacking the PR (positive regulatory domain I binding factor 1 and retinoblastoma-interacting zinc finger protein) domain, was frequently transcribed in ATL cells, and the transcriptional initiation sites were identified upstream from exons 4 and 6. Transfection of MEL1S into CTLL-2 cells conferred resistance against transforming growth factor β (TGF-β), suggesting that aberrant expression of MEL1S was associated with dysregulation of TGF-β-mediated signaling. Although Tax renders cells resistant to TGF-β, Tax could not be produced in most fresh ATL cells, in which MEL1S might be responsible for TGF-β resistance. Our results suggest that aberrant gene expression associated with DNA hypomethylation is implicated in leukemogenesis of ATL. (Blood. 2004;103:2753-2760)
Introduction
Adult T-cell leukemia (ATL) is a neoplastic disease of CD4+ T lymphocytes that is etiologically associated with human T-cell leukemia virus type I (HTLV-I).1-6 After transmission, HTLV-I increases the number of infected cells through its viral proteins, including Tax. Tax encoded by the pX region between env and the 3′ long terminal repeat (LTR) is an oncoprotein with pleiotropic actions,7,8 including transactivation of the nuclear factor kappaB (NFκB), serum responsive factor (SRF), and cyclic adenosine monophosphate (cAMP) response element-binding protein (CREB) pathways; transrepression of genes, such as DNA polymerase β, lck, and p18 genes; and functional inactivation of p53, p16, and mitotic arrest-defective 1 (MAD1).8,9 With these actions, Tax promotes the proliferation and inhibits apoptosis of infected cells, resulting in their clonal expansion and increased proviral load.
The cumulative risk of developing ATL after HTLV-I infection was estimated to be 6% in men and 2% in women.10 In addition, there is a long latent period before the onset of ATL, suggesting that additional factors other than viral proteins are implicated in leukemogenesis. Although somatic changes of genes, such as mutation of p5311 or deletion of p16,12 were reported in ATL, they were not so frequent in ATL cells and were predominantly observed in aggressive forms of ATL, such as acute and lymphoma types. This suggested that these genetic changes were implicated in the progression of ATL. In addition, it has been reported that DNA methylation of the p16 gene silenced its expression, indicating that epigenetic changes were also implicated in leukemogenesis.13
Epigenetic changes include dysregulated DNA methylation in cancer cells, which consists of hypermethylation and hypomethylation of DNA.14 Hypermethylation is frequently associated with gene silencing when hypermethylation occurs in the promoter region of genes.15 The same mechanism is considered to regulate the transcriptional silencing of various tumor suppressor genes, including p16,16 p15,17 hMLH1,18 BRCA1,19 and GSTP1 genes.20 From the perspective of the whole genome, genome-wide hypomethylation has been reported in cancer cells,21-23 and demethylation due to decreased expression of DNA methyltransferase has been shown to be associated with oncogenesis.24 Two mechanisms by which DNA hypomethylation is associated with oncogenesis have been considered: (1) DNA hypomethylation directly induces genetic instability, and (2) aberrant transcription is linked with DNA hypomethylation. In the immunodeficiency, centromeric instability, and facial anomalies (ICF) syndrome, the underlying abnormalities are mutations of both alleles of the gene that encodes DNA methyltransferase 3B (Dnmt3B).25,26 Chromosomal instability has also been demonstrated in mice with decreased expression of Dnmt1.24 Taken together, these findings suggest that undermethylation of the CpG sites is closely linked with organization and stabilization of chromatin structures. Although DNA hypomethylation in cancer cells was frequently observed in both highly and moderately repetitive DNA sequences,14 it was also detected in transcriptional units of single-copy genes. Such genes included MAGE-1,27 MDR1,28 and HOX11.29
The present study was designed to determine the DNA methylation status in ATL and whether DNA hypomethylation-associated aberrant gene expression is involved in ATL leukemogenesis. We report the isolation of hypomethylated DNA regions from ATL cells compared with cells in the carrier state by the methylated CpG-island amplification/representational difference analysis (MCA/RDA) method and identification of hypomethylated and aberrantly expressed genes in ATL cells.
Materials and methods
Cell lines and antibody
HTLV-I-transformed cell lines and ATL-derived cell lines used in this study were as follows: ATL-43T, ATL-55T, ED, and ATL-2 were cell lines derived from leukemic clones,13 and MT-4 and ATL-35T were derived from nonleukemic clones. Hut78, Jurkat, SupT1, and Kit225 were T-cell lines not associated with HTLV-I. The human embryonic kidney cell line, 293, was used as a control. Rabbit serum anti-MDS1/EVI1-like gene 1 (anti-MEL1) DNA binding domain 2 (nucleotides [nt's] 2613-3152) was prepared as described previously.30
MCA/RDA
MCA/RDA was performed as described previously.31 For preparation of MCA/RDA amplicons, genomic DNA extracted from peripheral blood mononuclear cells of an HTLV-I carrier was used as a tester and that from acute ATL cells was used as a driver. Five micrograms of DNA was digested with 110 units of SmaI overnight. The DNA was then digested with 20 units of XmaI overnight. DNA fragments were then precipitated with ethanol. The RMCA polymerase chain reaction (PCR) adaptor was prepared by incubation of 2 oligonucleotides RMCA24 (5′-CCACCGCCATCCGAGCCTTTCTGC-3′) and RMCA12 (5′-CCGGGCAGAAAG-3′) at 65°C for 2 minutes, followed by cooling to room temperature. DNA (0.5 μg) was ligated to 0.5 nmol of RMCA adaptor using T4 DNA ligase. PCR was performed using 2 μL of each of ligation mix as a template in a 100-μL volume containing 100 pmol RMCA24 primer, 2 units of ExTaq (Takara, Shiga, Japan), 2 mM MgCl2, 16 mM NH4 (SO4)2, 10 μg/mL bovine serum albumin (BSA), and 5% vol/vol dimethyl sulfoxide (DMSO). The reaction mixture was incubated at 72°C for 5 minutes and 95°C for 3 minutes. Samples were then subjected to 25 cycles amplification consisting of 1 minute at 95°C and 3 minutes at 77°C in a thermal cycler (Perkin Elmer Applied Biosystems, Norwalk, CT). The final extension time was 10 minutes. All restriction enzymes and T4 DNA ligase were from New England Biolabs (Beverly, MA). In the RDA step, 500 ng and 100 ng of ligation mixture were used for the first and second competitive hybridization, respectively. To eliminate the digested adaptor, CHROMA SPIN Column (BD Biosciences Clontech, Palo Alto, CA) was used. Primers used for the first and second rounds of RDA were as follows: JMCA24, 5′-GTGAGGGTCGGATCTGGATGGCTC-3′; JMCA12, 5′CCGGGAGCCAGC-3′; NMCA24, 5′-GTTAGCGGACACAGGGCGGGTCAC-3′; and NMCA12, 5′-CCGGGTGACCCG. After the second round of competitive hybridization, the PCR products were subcloned into pCR-XL-TOPO (Invitrogen, Carlsbad, CA), and then the insert of each clone was amplified with M13 forward (M13F) (5′-CCCAGTCACGACGTTGTAAAACGA-3′), and M13 reverse (M13R) (5′-AGCGGATAACAATTTCACACAGGA-3′) primers. Sequence analysis was carried out using automated DNA sequencers (Perkin Elmer Applied Biosystems). Sequence homologies were identified using the BLAST program of the National Center for Biotechnology Information available at http://www.ncbi.nlm.nih.gov:80/BLAST/.
Southern blot analysis
To confirm that the MCA/RDA method isolated hypomethylated DNA fragments, we performed Southern blot analysis using the isolated DNA fragments as probes. Tester and driver MCA amplicons (500 ng each) were separated by electrophoresis in an agarose gel and transferred to a positive charge nylon membrane (Hybond-N; Amersham Biosciences, Piscataway, NJ). Filters were hybridized with alkaline phosphatase-labeled probes and then washed using Alphos Direct Labeling and Detection with CDP-Star (Amersham Biosciences). Filters were then exposed to film (Medical X-ray film; Kodak, Rochester, NY) for a few seconds and analyzed.
Direct sequencing after sodium bisulfite treatment
One microgram of genomic DNA (10 μL) was denatured by the addition of an equal volume of 0.6 N NaOH for 15 minutes and then 208 μL of 3.6 M sodium bisulfite and 12 μL of 10 mM hydroxyquinone were added. This mixture was incubated at 55°C for 16 hours to convert cytosine to uracil. Treated genomic DNA was subsequently purified using the Wizard clean-up system (Promega, Madison, WI), precipitated with ethanol, and resuspended in 100 μL of dH2O. Sodium bisulfite-treated genomic DNAs (50 ng) were amplified with the specific primers to isolated DNA regions, and then PCR products were subcloned into plasmid DNA. Sequences of 10 clones of each were determined using Big Dye Terminator (Perkin Elmer Applied Biosystems) with an ABI 377 autosequencer.
Synthesis of cDNA and semiquantitative reverse transcriptase (RT)-PCR
Transcripts of MEL, CACNA1H, and Nogo receptor genes were quantified using semiquantitative RT-PCR as described previously.32 Total RNA was extracted from each sample using TRIZOL (Invitrogen), and cDNA was synthesized from 1 to 5 μg of total RNAs by the Superscript Preamplification System (Invitrogen) according to the protocol recommended by the manufacturer. PCR was performed using 1 μL of reverse transcriptase reaction sample mixed with 50 μL of PCR reaction buffer containing 0.2 mM each of deoxynucleotide triphosphate, 2 mM MgCl2, 1.25 units of ExTaq polymerase (Takara), and 0.5 μM of each primer using the hot start technique with AmpliWax PCR Gem 100 (Perkin Elmer Applied Biosystems). Primers were 5′-AGTGAGATGAACCAAGCATCAACG-3′ (sense), 5′-CTGCACAGTGTATGTTTTAAAGCC-3′ (antisense) for MEL1/1S; 5′-TGTGACGAGTGTGACGAACT-3′ (sense), 5′-GTTTTCACATTCGAAGCGTT-3′ (antisense) for MEL1S; 5′-CCCTGTCTACATTCCTGAAG-3′ (sense), 5′-CCACGTCCGTCAGTATTTGC-3′ (antisense) for MEL1; 5′-CGCCACCTTCAGCAACTTCGGCAT-3′ (sense), 5′-ATCTCCACCTCCTGCAGCGG-3′ (antisense) for CACNA1H; and 5′-CAGTACCTGAGGCTCAACGACAAC-3′ (sense), 5′-ACCTGAGCCTTCTGAGTCACCAGT-3′ (antisense) for Nogo receptor in T-Gradient Thermocycler (Biometra, Gottingen, Germany). The PCRs were performed under the following conditions: MEL1/1S: 2 minutes at 94°C, for 36 cycles for 30 seconds at 94°C, 30 seconds at 56°C, 30 seconds at 72°C; MEL1 and MEL1S: 2 minutes at 94°C, for 40 cycles for 30 seconds at 94°C, 30 seconds at 58°C, 30 seconds at 72°C; CACNA1H: 2 minutes at 94°C, for 40 cycles for 30 seconds at 94°C, 30 seconds at 60°C, 30 seconds at 72°C; and Nogo receptor: 3 minutes at 94°C, for 40 cycles for 1 minute at 94°C, 40 seconds at 57°C, and 50 seconds at 72°C.
Western blot analysis
Cells were lysed in a buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Triton X, 10% glycerol, 0.5 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM Na3VO4, 10 μg/mL aprotinin, and 10 μg/mL leupeptin. After 30 minutes on ice, lysates were cleared by centrifugation at 12 500g (15 000 rpm) for 15 minutes at 4°C. Proteins (20 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by electrotransfer to a polyvinylidene difluoride membrane (ATTO, Tokyo, Japan). The blots were blocked in blocking buffer for 1 hour at room temperature and incubated in 3000-fold diluted rabbit serum anti-MEL1 DNA binding domain 2 for 1 hour at room temperature. After 5 washes in 0.5% Tween-phosphate-buffered saline (PBS), the blots were incubated in 4000-fold diluted antirabbit immunoglobulin-biotinylated specific whole antibody (from donkey) (Amersham Biosciences) for 30 minutes at room temperature. After 5 washes in 0.5% Tween-PBS, the blots were incubated in 4000-fold diluted streptavidin-biotinylated horseradish peroxidase complex (Amersham Biosciences) for 30 minutes at room temperature. After 2 washes in Tween-PBS, bound antibodies were detected using the Western blotting Chemiluminescence Luminol Reagent (Santa Cruz Biotechnology, Santa Cruz, CA). The blots were then exposed on Hyperfilm ECL film (Amersham Biosciences).
Transfection and assay for cell proliferation
For the construction of vectors expressing MEL1 and MEL1S genes, MEL1 and MEL1S cDNA were subcloned into pME18Sneo (K. Maruyama and A. Miyajima, unpublished data, 2000), which was modified from pCEV4.33 The mouse interleukin 2 (IL-2)-dependent T-cell line, CTLL-2, was maintained in RPMI 1640 supplemented with 10% fetal calf serum (FCS), 50 μM 2-mercaptoethanol, and 50 units/mL of human recombinant IL-2. Expression vectors and control vector were transfected into CTLL-2 cells by electroporation, and those cells were selected by G418 selection (0.75 mg/mL). Three transfectants, CTLL-2/pME18Sneo, CTLL-2/MEL1, and CTLL-2/MEL1S, were established. The expression of MEL1 mRNA in cell clones was investigated by RT-PCR. For cell proliferation studies, MEL-expressing clones (CTLL-2/MEL1 and CTLL-2/MEL1S) and the control clone (CTLL-2/pME18Sneo) were plated at a density of 4 × 103 cells/well in 96-well microtiter plates. Cells were treated with increasing concentrations of transforming growth factor β (TGF-β; R&D Systems, Minneapolis, MN) for 96 hours and assayed for cell growth by the methyl thiazolyl tetrazolium (MTT) assay. Each experiment was performed 3 times, and typical results are shown.
Rapid amplification of cDNA 5′ ends (5′-RACE)
The 5′ terminus of MEL1S mRNA was determined by the SMART (Switching mechanism at the 5′ end of RNA transcript) RACE (rapid amplification of complementary DNA ends) cDNA Amplification Kit (BD Biosciences Clontech) according to the protocol provided by the manufacturer. The cDNA was synthesized by reverse transcription (PowerScript Reverse Transcriptase; BD Biosciences Clontech) of 1 μg of ATL-55T total RNA using 5′CDS primer (modified oligo [dT] primer) and SMARTIIA oligonucleotide. The first-strand cDNA was used directly in 5′-RACE PCR. For nested PCR amplifications, primers specific for the MEL1 gene (5′-CTCGGTGTGCACATGAAGAATACCGCG-3′ and 5′-AGGTCGAACCTGACCTTCTCAAGCAGT-3′) were used. The PCR products were subcloned into a TA cloning vector (pGEM-T Easy Vector; Promega) and sequenced.
Results
Identification of differentially hypomethylated CpG islands in ATL by MCA/RDA
To identify the differentially hypomethylated DNA regions in ATL, we used the MCA/RDA method. Genomic DNA extracted from peripheral blood mononuclear cells (PBMCs) of a carrier was used as a tester and that from acute ATL cells was used as a driver. After 2 rounds of RDA, the PCR products were cloned into plasmids and sequenced, resulting in identification of 16 DNA fragments. Thereafter, we checked that isolated DNA specifically hybridized to the tester amplicon by Southern blot analysis, in which MCA products were used as substrates as described in “Materials and methods.” Specific hybridization to the tester amplicon implied that isolated DNAs were hypomethylated in ATL cells compared with PBMCs from a carrier. Finally, we identified 3 differentially hypomethylated DNA fragments in ATL cells as shown in Table 1. The MEL1 (MDS1/EVI1-like gene 1) gene, which was mapped to human chromosome 1p36, was transcriptionally activated in acute myeloid leukemia or myelodysplastic syndrome with t(1;3)(p36;q21).34 Another gene was CACNA1H (α1H T type Ca2+ channel), which is mapped to human chromosome 16p13.3, and was normally transcribed in kidney and heart.35 The Nogo receptor gene is associated with inhibition of neurites by interaction with its ligand, and its expression was observed in neurons.36
Hypomethylated DNA clones isolated by MCA/RDA in ATL cells
Clone no. . | Blast homology . | Size, bp . | Identities, % . | GenBank accession no. . |
|---|---|---|---|---|
| 3 | CACNA1H gene (intron 1) | 640 | 99 | AE006466 |
| 6 | MEL1 complete cds (exon 9) | 428 | 100 | AB078876 |
| 12 | Upstream of homo sapiens similar to Nogo receptor, reticulon 4 receptor (LOC96184), mRNA | 508 | 100 | XM_015620 |
Clone no. . | Blast homology . | Size, bp . | Identities, % . | GenBank accession no. . |
|---|---|---|---|---|
| 3 | CACNA1H gene (intron 1) | 640 | 99 | AE006466 |
| 6 | MEL1 complete cds (exon 9) | 428 | 100 | AB078876 |
| 12 | Upstream of homo sapiens similar to Nogo receptor, reticulon 4 receptor (LOC96184), mRNA | 508 | 100 | XM_015620 |
CACNA1H indicates voltage-dependent T-type Ca2+ channel; and MEL1, MDS1/EVI-like gene 1.
DNA methylation of MEL1, CACNA1H, and Nogo receptor genes
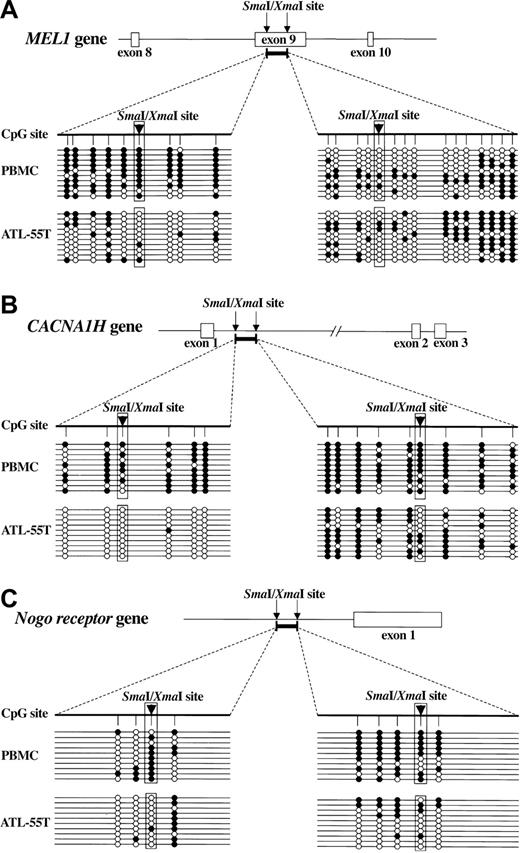
To reveal the detailed DNA methylation status, we studied DNA methylation of MEL1, CACNA1H, and Nogo receptor genes by sequencing using sodium bisulfite-treated genomic DNAs. In the case of MEL1, a DNA fragment within exon 9 was isolated by MCA/RDA. Although the 5′ region of this DNA fragment was hypermethylated in normal PBMCs, the number of methylated CpG sites decreased in an ATL cell line, ATL-55T (Figure 1A), confirming that the MCA/RDA method isolated the differentially hypomethylated DNA regions. On the other hand, DNA methylation in the 3′ region showed no difference between normal PBMCs and ATL-55T. In CACNA1H, CpG sites in both the 5′ and 3′ regions were hypomethylated in ATL-55T cells compared with PBMCs (Figure 1B). In addition, the Nogo receptor gene was also undermethylated in ATL-55T (Figure 1C). Thus, these data confirmed that CpG sites in the isolated DNA regions were hypomethylated in ATL cells when compared with control PBMCs.
Methylation status of DNA fragments isolated by MCA/RDA. Genomic DNA was treated by sodium bisulfite and amplified by primers specific for DNA regions identified by MCA/RDA. Then, PCR products were subcloned into plasmid DNA, and the sequences were determined in 10 clones of each gene: (A) MEL1, (B) CACNA1H, and (C) Nogo receptor gene. The schemas show the structures of isolated DNA regions (bold bars), and arrowheads indicate the SmaI/XmaI sites. The methylation status of each CpG site is shown (•, methylated CpG; ○, unmethylated CpG).
Methylation status of DNA fragments isolated by MCA/RDA. Genomic DNA was treated by sodium bisulfite and amplified by primers specific for DNA regions identified by MCA/RDA. Then, PCR products were subcloned into plasmid DNA, and the sequences were determined in 10 clones of each gene: (A) MEL1, (B) CACNA1H, and (C) Nogo receptor gene. The schemas show the structures of isolated DNA regions (bold bars), and arrowheads indicate the SmaI/XmaI sites. The methylation status of each CpG site is shown (•, methylated CpG; ○, unmethylated CpG).
Expression of MEL1, CACNA1H, and Nogo receptor mRNAs in ATL cell lines
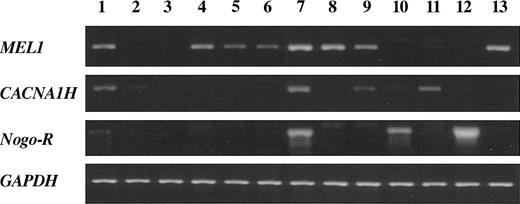
Since hypomethylation of the MEL1, CACNA1H, and Nogo receptor genes might be associated with their aberrant expression in ATL cells, we analyzed the transcription of these genes by RT-PCR. As shown in Figure 2, transcripts of the MEL1 gene were detected in all HTLV-I-associated cell lines and another T-cell line (Kit225), although its expression was not detectable in PBMCs and activated T cells. Transcripts of the CACNA1H and Nogo receptor genes were also detected in some HTLV-I-associated T-cell lines or non-HTLV-I-associated T-cell lines. Thus, these results showed that the MEL1, CACNA1H, and Nogo receptor genes were aberrantly transcribed in ATL cells, suggesting a linkage between DNA hypomethylation and the aberrant transcription. Among them, MEL1 gene was most frequently transcribed in ATL cells.
Expression of genes isolated by MCA/RDA in various cell lines. Expression of the MEL1, CACNA1H, and Nogo receptor genes, as identified by MCA/RDA, was analyzed by RT-PCR. Transcripts of the GAPDH gene were used as a control. Lane 1, 293 cells; lane 2, PBMCs; lane 3, activated T cells; lane 4, MT-4; lane 5, ATL-35T; lane 6, ATL-43T; lane 7, ATL-55T; lane 8, ED; lane 9, ATL-2; lane 10, Hut78; lane 11, Jurkat; lane 12, SupT1; and lane 13, Kit225.
Expression of genes isolated by MCA/RDA in various cell lines. Expression of the MEL1, CACNA1H, and Nogo receptor genes, as identified by MCA/RDA, was analyzed by RT-PCR. Transcripts of the GAPDH gene were used as a control. Lane 1, 293 cells; lane 2, PBMCs; lane 3, activated T cells; lane 4, MT-4; lane 5, ATL-35T; lane 6, ATL-43T; lane 7, ATL-55T; lane 8, ED; lane 9, ATL-2; lane 10, Hut78; lane 11, Jurkat; lane 12, SupT1; and lane 13, Kit225.
Expression of MEL1S gene product in ATL cell lines
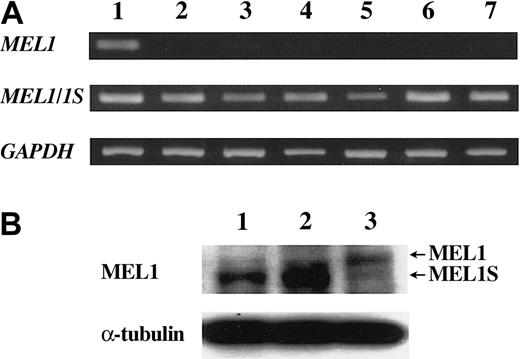
MEL1 is a member of the ecotropic viral integration site 1 (EVI1) gene family34 and is also a PR (positive regulatory domain I binding factor 1 and retinoblastoma-interacting zinc finger protein) domain member (PRDM16).37 Two forms of MEL1 gene transcripts were identified, one encoded the full-length MEL1 protein with a PR domain and the other was the short form lacking PR domain (designated as MEL1S). MEL1S, but not MEL1, gene was expressed in t(1;3)(p36;q21)-positive myeloid leukemia cells by translocation, suggesting that MEL1S lacking the PR domain was associated with oncogenesis. To investigate which type of MEL1 gene was expressed in ATL cells we analyzed MEL1 and MEL1S gene expression by RT-PCR. As shown in Figure 3A, the 293 cell line expressed both types of MEL1 gene transcripts; however, all ATL cell lines (MT-4, ATL-35T, ATL-43T, ATL-55T, ED, and ATL-2) predominantly expressed MEL1S type transcript. Moreover, we detected MEL1S protein in ATL-55T and ED, whereas the 293 cell line produced both MEL1 and 1S proteins by Western blot analysis (Figure 3B). Predominant expression of the MEL1S gene was also observed in Kit225 (data not shown). Taken together, these results showed that ATL cells produced MEL1S but not MEL1 protein.
Short form products of the MEL1 gene were expressed in ATL cell lines. (A) Transcription of MEL1 and MEL1S genes. The transcription of MEL1 and MEL1S genes were studied with the specific primers in ATL cell lines and control cell line as follows: lane 1, 293 cells; lane 2, MT-4; lane 3, ATL-35T; lane 4, ATL-43T; lane 5, ATL-55T; lane 6, ED; and lane 7, ATL-2. RT-PCR that could detect both MEL1 and MEL1S gene transcripts revealed the transcription (330 bp) in all cell lines, whereas RT-PCR with primers specific for MEL1 gene detected the transcript (197 bp) only in 293 cells. (B) Western blot analysis of MEL1 and MEL1S. Cell lysates were analyzed by Western blot using antibodies against the MEL1/1S protein. Lane 1, ATL-55T; lane 2, ED; and lane 3, 293 cells.
Short form products of the MEL1 gene were expressed in ATL cell lines. (A) Transcription of MEL1 and MEL1S genes. The transcription of MEL1 and MEL1S genes were studied with the specific primers in ATL cell lines and control cell line as follows: lane 1, 293 cells; lane 2, MT-4; lane 3, ATL-35T; lane 4, ATL-43T; lane 5, ATL-55T; lane 6, ED; and lane 7, ATL-2. RT-PCR that could detect both MEL1 and MEL1S gene transcripts revealed the transcription (330 bp) in all cell lines, whereas RT-PCR with primers specific for MEL1 gene detected the transcript (197 bp) only in 293 cells. (B) Western blot analysis of MEL1 and MEL1S. Cell lysates were analyzed by Western blot using antibodies against the MEL1/1S protein. Lane 1, ATL-55T; lane 2, ED; and lane 3, 293 cells.
Transcriptional initiation sites of the MEL1S gene have been determined in leukemic cells with t(1;3)(p36;q21), which showed that they existed in exon 2.30 In acute myeloid leukemia cells with t(1;3)(p36;q21), translocation occurred in the promoter region or intron 1 of the MEL1 gene and the Ribophorin I gene in 3q21, indicating that ectopic expression of MEL1S gene was driven by the Ribophorin I gene.34,38 However, transcriptional initiation sites of the MEL1S gene in ATL cells remain unknown. Therefore, we determined the initiation sites of the MEL1S gene by 5′-RACE (Figure 4A) in ATL-55T. The 2 major PCR products observed in Figure 4A were cloned into plasmid DNA, and their sequences were determined. All 8 clones containing the upper band had a 633-bp sequence upstream from exon 6 (Figure 4B, number 1). On the other hand, among the 8 clones that contained the lower band, 6 contained a 410-bp sequence upstream from exon 6 (Figure 4B, number 2), and the remaining 2 clones started from 477-bp upstream of exon 4 and a putative exon (154 bp) spliced to exon 5 (Figure 4B, number 3). All transcripts lacked the PR domain, which was encoded by exons 2, 3, 4, and 5. Thus, 5′-RACE identified 3 different initiation sites of MEL1S gene transcription (Figure 4B). In other cell lines, ED and ATL-43T, the majority of transcripts of MEL1S initiated from the putative exon upstream exon 4 (Figure 4B, number 3). Therefore, this transcriptional initiation site is predominant for MEL1S gene transcription in ATL cells. We next determined the methylation status of DNA regions surrounding these initiation sites (Figure 4C). As observed in the coding region of the MEL1 gene (Figure 1A), these regions surrounding the transcriptional initiation sites were also hypomethylated in ATL cells compared with normal PBMCs, suggesting that aberrant expression of the MEL1S gene was associated with hypomethylation in the putative promoter region of the MEL1S gene. In contrast to the MEL1S gene, the promoter region of MEL1 was not methylated in PBMCs but was hypermethylated in ATL-55T (Figure 4C), showing that in contrast to the MEL1S gene, transcription of MEL1 was silenced in ATL cells by DNA hypermethylation.
Identification of transcriptional initiation sites of the MEL1S gene, and DNA methylation. (A) The 5′-RACE was performed with cDNA synthesized using mRNA from an ATL cell line, ATL-55T. Primers in exon 6 were used to amplify cDNA. Two major bands were detected. (B) Schema of transcriptional initiation sites of the MEL1S gene in ATL cells. PCR products of 5′-RACE were cloned into plasmid DNA and then their sequences were determined. The majority of MEL1S transcripts initiated from upstream of exon 6 (633 bp [1] and 410 bp [2] upstream from exon 6 as shown in Figure 4B). Transcription started 477 bp upstream of the exon 4, which contained putative exon (▦; 154 bp [3]). From these data, 2 major initiation sites were identified as shown in 1 and 2. (C) The methylation status surrounding the transcriptional initiation sites of MEL1S gene and promoter region of MEL1 gene. Genomic DNA was treated by sodium bisulfite and amplified by specific primers. The methylation status was determined by sequencing of these PCR products. The schema shows the structure of the MEL1/1S gene and transcriptional initiation sites of the MEL1S gene are shown by arrows (1, 2, and as described for panel B). Methylation of CpG sites surrounding the transcriptional initiation sites of MEL1S is shown (•, methylated; ○, unmethylated).
Identification of transcriptional initiation sites of the MEL1S gene, and DNA methylation. (A) The 5′-RACE was performed with cDNA synthesized using mRNA from an ATL cell line, ATL-55T. Primers in exon 6 were used to amplify cDNA. Two major bands were detected. (B) Schema of transcriptional initiation sites of the MEL1S gene in ATL cells. PCR products of 5′-RACE were cloned into plasmid DNA and then their sequences were determined. The majority of MEL1S transcripts initiated from upstream of exon 6 (633 bp [1] and 410 bp [2] upstream from exon 6 as shown in Figure 4B). Transcription started 477 bp upstream of the exon 4, which contained putative exon (▦; 154 bp [3]). From these data, 2 major initiation sites were identified as shown in 1 and 2. (C) The methylation status surrounding the transcriptional initiation sites of MEL1S gene and promoter region of MEL1 gene. Genomic DNA was treated by sodium bisulfite and amplified by specific primers. The methylation status was determined by sequencing of these PCR products. The schema shows the structure of the MEL1/1S gene and transcriptional initiation sites of the MEL1S gene are shown by arrows (1, 2, and as described for panel B). Methylation of CpG sites surrounding the transcriptional initiation sites of MEL1S is shown (•, methylated; ○, unmethylated).
MEL expression conferred resistance against TGF-β
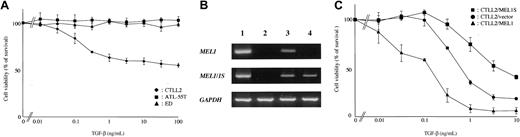
The EVI1 gene is an alternative splicing form of the MDS1/EV1 gene; MDS1/EVI1 retains PR domain whereas EV1 lacks it. EVI1 has been reported to repress the growth suppression mediated by TGF-β against a mouse IL-3-dependent myeloid cell line, 32Dcl3, whereas MDS1/EVI1 enhanced TGF-β signaling and strengthened its growth-inhibitory effect.39 Structural similarity between MEL1S and EVI1 suggested that MEL1S might be associated with the growth inhibitory effect of TGF-β. ATL cell lines expressing MEL1S were resistant to TGF-β, whereas mouse T-cell line, CTLL-2, was sensitive to it (Figure 5A). To determine whether MEL could influence the antiproliferative effects of TGF-β, we established CTLL-2 cell lines stably expressing MEL1 and MEL1S by transfection of the respective expression vectors. RT-PCR confirmed their expressions (Figure 5B). TGF-β suppressed the growth of a CTLL-2 cell line in a dose-dependent manner. Although CTLL-2/MEL1 cells showed increased susceptibility to TGF-β, CTLL-2/MEL1S cells exhibited resistance against TGF-β (Figure 5C). These results showed that MEL1 protein with the PR domain and MEL1S protein without the PR domain had differential influences on susceptibility to TGF-β responsiveness and that MEL1S protein lacking the PR domain rendered ATL cells resistant to the inhibitory effect of TGF-β as well as EVI1.
Sensitivity to TGF-β in ATL cell lines and CTLL-2 cell lines expressing MEL1 or MEL1S gene. (A) Resistance of ATL cell lines to TGF-β. ATL cell lines, ATL-55T and ED, and mouse T-cell line, CTLL-2, were treated with the indicated concentrations of TGF-β for 72 hours. Proliferation of each cell was examined by MTT assay. The results are shown as percentages of the values obtained from control TGF-β-free culture. The experiment was performed at least 3 times. The results are the mean ± SD of 3 experiments. (B) Expression of MEL1 and MEL1S gene in CTLL-2 transfected with vectors expressing MEL1 or MEL1S genes. CTLL-2 was transfected with vectors expressing MEL1 or MEL1S genes, and their expressions were studied by RT-PCR as described in Figure 3. Lane 1 shows 293 cells (control); lane 2, CTLL-2/empty vector; lane 3, CTLL-2/MEL1; and lane 4, CTLL-2/MEL1S. (C) MEL1S expression confers resistance against TGF-β in CTLL-2. MEL1-expressing (▴, MEL1; ▪, MEL1S) and unmodified (•) CTLL-2 were treated with the indicated concentrations of TGF-β for 96 hours. The results are shown as percentages of the values obtained from control TGF-β-free culture. The results are the mean ± SD of 3 experiments.
Sensitivity to TGF-β in ATL cell lines and CTLL-2 cell lines expressing MEL1 or MEL1S gene. (A) Resistance of ATL cell lines to TGF-β. ATL cell lines, ATL-55T and ED, and mouse T-cell line, CTLL-2, were treated with the indicated concentrations of TGF-β for 72 hours. Proliferation of each cell was examined by MTT assay. The results are shown as percentages of the values obtained from control TGF-β-free culture. The experiment was performed at least 3 times. The results are the mean ± SD of 3 experiments. (B) Expression of MEL1 and MEL1S gene in CTLL-2 transfected with vectors expressing MEL1 or MEL1S genes. CTLL-2 was transfected with vectors expressing MEL1 or MEL1S genes, and their expressions were studied by RT-PCR as described in Figure 3. Lane 1 shows 293 cells (control); lane 2, CTLL-2/empty vector; lane 3, CTLL-2/MEL1; and lane 4, CTLL-2/MEL1S. (C) MEL1S expression confers resistance against TGF-β in CTLL-2. MEL1-expressing (▴, MEL1; ▪, MEL1S) and unmodified (•) CTLL-2 were treated with the indicated concentrations of TGF-β for 96 hours. The results are shown as percentages of the values obtained from control TGF-β-free culture. The results are the mean ± SD of 3 experiments.
Discussion
Hypomethylation has been demonstrated to be associated with genetic instability and aberrant gene expression.14 Most hypomethylated genes have been identified by analyses of known genes, such as oncogenes29 and genes associated with drug resistance.28 However, several different methods have been applied for isolation of aberrantly methylated DNA regions, which include restriction landmark genomic screening (RLGS)40 and methylation-sensitive-representational difference analysis (MS-RDA),41 in addition to MCA/RDA.31 For isolation of hypomethylated DNA regions, the RLGS method revealed that the peri-centromeric region of human acrochromosomes was hypomethylated in hepatitis B virus (HBV)-integrated hepatocellular carcinoma (HCC) cells.42 It has also been shown that a 1.4-kb repetitive sequence was hypomethylated in sperm and HCC cells.43 MS-RDA could also detect hypomethylated DNA regions in mouse HCC, which included the long interspersed nuclear element 1 (LINE-1) and α-enolase genes.41 In addition to these methods, MCA/RDA also identified hypomethylated DNA regions as shown in the present study. Among the 3 hypomethylated genes identified in the present study, the CACNA1H and Nogo receptor genes were not as frequently expressed in ATL cells, although their expression was associated with hypomethylation. On the other hand, the MEL1S gene was aberrantly expressed in ATL cells and also exhibited hypomethylation in ATL cells compared with PBMCs. Thus, the MCA/RDA method could isolate hypomethylated DNA regions in addition to hypermethylated DNA regions, as reported previously.31 Although repetitive sequences are hypomethylated in cancer cells, MCA/RDA could not isolate these sequences in this study. Since MCA/RDA detected differentially methylated SmaI/XmaI sites, the rarity of such sites in the repetitive sequences might account for the inability of MCA/RDA to isolate the repetitive sequences. DNA methylation of promoter regions influences transcription in general, and the DNA regions isolated by MCA/RDA method were not the promoter regions of the MEL1S and CACNA1H genes. However, hypomethylation in the nonpromoter DNA regions was linked with aberrant expression of these genes. Indeed, the putative promoter region of the MEL1S gene was also hypomethylated in ATL cells as shown in our study, indicating that hypomethylation of nonpromoter regions was associated with aberrant transcription and hypomethylation of promoter regions.
The MEL1 gene, which was mapped to human chromosome 1p36, was originally isolated as the gene that was transcriptionally activated by t(1;3)(p36;q21) in acute myeloid leukemia.34 This gene encodes 2 types of transcripts, MEL1 and MEL1S. The MEL1 gene that retains the PR domain is highly homologous to MDS1/EVI1, whereas the MEL1S gene is an alternatively spliced transcript of MEL1 gene, which lacks the PR domain as observed in EVI1. In 293 cells, both MEL1 and MEL1S genes were transcribed, although only MEL1S gene transcripts were detected in AML cells with t(1;3)(p36;q21). Another PR domain-containing gene, RIZ (the retinoblastoma protein-interacting zinc finger gene) was identified,44 which also encodes 2 types of transcripts, a PR-positive and PR-negative form. In the MDS1/EVI1, MEL1, and RIZ genes, transcripts lacking the PR domain have all been associated with oncogenesis as observed in the present study. The EVI1 gene that lacked the PR domain was transcribed in leukemic cells by retroviral integration45 or translocation.46,47 The expression of RIZ1 (PR domain positive) was suppressed by DNA hypermethylation,48 whereas expression of RIZ2 (PR domain negative) gene was intact in colorectal cancer49 and breast cancer cells,50 resulting in predominant expression of the PR domain-negative form. Moreover, inactivation of the RIZ1 gene allowed tumor formation in vivo.51 Taken together, these findings indicate that the PR domain had a tumor-suppressive character, whereas transcripts without the PR domain were oncogenic. In normal tissues, transcripts both with and without the PR domain were observed.50,52 In MEL1 gene, the pattern of DNA methylation of MEL1 and MEL1S gene promoter regions is contrast between ATL cells and PBMCs. Since both genes are not transcribed in PBMCs and activated T cells, lack of transcriptional factors necessary for MEL1 and MEL1S genes transcription or the presence of silencer is considered the cause of an absence of transcription, since the MEL1 promoter is hypomethylated regardless of lack of MEL1 gene expression. In the fetal kidney, both MEL1 and MEL1S genes are transcribed, suggesting that common transcriptional machinery is involved in their transcriptions. On the other hand, DNA hypermethylation in the promoter region of MEL1 gene associated with DNA hypomethylation in that of MEL1S gene alters the expression pattern of MEL1/MEL1S gene in ATL cells, as observed in RIZ1/RIZ2 genes.
Predominant expression of the PR domain-negative form by retroviral insertion, translocation, hypermethylation, and hypomethylation (this study) was linked to oncogenesis. Increased expression of the MEL1S gene with suppressed expression of the PR domain-positive form is considered to facilitate proliferation of leukemic cells. One mechanism by which PR domain-negative forms might be associated with oncogenesis is that the PR domain-negative form of both EVI1 and MEL1S genes inhibits the tumor-suppressive effect of TGF-β. However, the findings that both EVI1 and MEL1S could inhibit the differentiation of myeloid cells30 suggested another mechanism in the oncogenesis of ATL.
The PR domain is composed of about 130 amino acid residues and shares significant sequence identity to the SET domain. This domain is present in suppressor of variegation 3-9 homolog 1 (SUV39H1),53 SET1 (yeast),54 and human trithorax,55 which are contained in chromosomal proteins that function in modulating gene expression. Recent studies have identified the suppressor of variegation 3-9 family as being H3 lysine 9-specific histone methyltransferase.53 These proteins, which play an important role in the establishment and maintenance of heterochromatin, mediate their methylation activities via the SET domain. The finding that the shared residues between PR and SET are contained in the most conserved residues in each domain indicates that they might share a common function.56 The tumor-suppressive effects shared by all PR domain-containing genes suggested that they influenced the function of PR domain-negative forms, such as MEL1S and EVI1, by acting on chromatin structures. The mechanism(s) by which PR domain-negative forms, such as MEL1S and EVI1, are associated with oncogenesis remains to be elucidated.
The EVI1 gene (PR domain negative) is an alterative splicing form of MDS1/EV1 gene (PR domain positive). It has been reported that EVI1 represses TGF-β signaling through inhibition of Smad3,57 however, MDS1/EVI1 enhanced TGF-β signaling and strengthened its growth-inhibitory effect.39 Consequently, the inappropriate expression of the PR-negative oncogenic product, EVI1, in hematopoietic cells has been implicated in the oncogenesis of acute myeloid leukemia. In this study, we also reported that MEL1S, lacking the PR domain, conferred resistance against TGF-β, whereas MEL1 increased susceptibility to TGF-β. In HTLV-I-infected cells, the expression of Tax conferred resistance against TGF-β, as reported previously.58-60 However, Tax protein could not be produced by genetic changes of the HTLV-I provirus genome in most fresh ATL cells, suggesting an alternative mechanism for resistance against TGF-β.61,62 Although the ATL cell lines ATL-55T, 43T, and ED could not produce Tax protein due to mutation or deletion of the tax gene and hypermethylation of the 5′-LTR,63 they secreted TGF-β and were resistant to it (Figure 5A). The MEL1S gene was transcribed in Tax-negative ATL cells, such as ATL-43T, ED, and ATL-55T, suggesting that MEL1S rendered ATL cells resistant to TGF-β in the absence of Tax.
The CACNA1H (α1H T type Ca2+ channel) gene was also identified as a hypomethylated gene in ATL cells, although the frequency of aberrant expression was not as high (2 of 6 ATL cell lines). It has been reported that antagonists of T-type Ca2+ channels (CACNA1H) inhibited cell proliferation,64 suggesting that its aberrant expression is associated with cell proliferation.
In conclusion, we have demonstrated in the present study that MCA/RDA is a powerful method for identifying hypomethylated and aberrantly transcribed genes in cancer cells. Identification of aberrantly expressed genes associated with hypomethylation should help elucidate mechanisms of oncogenesis.
Prepublished online as Blood First Edition Paper, December 4, 2003; DOI 10.1182/blood-2003-07-2482.
Supported by a Grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Ei-ichi Kodama and Yoshihiro Koya for valuable suggestions. The authors also thank Dr F. G. Issa (word-medex.com.au) for the careful reading and editing of the manuscript.

![Figure 4. Identification of transcriptional initiation sites of the MEL1S gene, and DNA methylation. (A) The 5′-RACE was performed with cDNA synthesized using mRNA from an ATL cell line, ATL-55T. Primers in exon 6 were used to amplify cDNA. Two major bands were detected. (B) Schema of transcriptional initiation sites of the MEL1S gene in ATL cells. PCR products of 5′-RACE were cloned into plasmid DNA and then their sequences were determined. The majority of MEL1S transcripts initiated from upstream of exon 6 (633 bp [1] and 410 bp [2] upstream from exon 6 as shown in Figure 4B). Transcription started 477 bp upstream of the exon 4, which contained putative exon (▦; 154 bp [3]). From these data, 2 major initiation sites were identified as shown in 1 and 2. (C) The methylation status surrounding the transcriptional initiation sites of MEL1S gene and promoter region of MEL1 gene. Genomic DNA was treated by sodium bisulfite and amplified by specific primers. The methylation status was determined by sequencing of these PCR products. The schema shows the structure of the MEL1/1S gene and transcriptional initiation sites of the MEL1S gene are shown by arrows (1, 2, and as described for panel B). Methylation of CpG sites surrounding the transcriptional initiation sites of MEL1S is shown (•, methylated; ○, unmethylated).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/7/10.1182_blood-2003-07-2482/6/m_zh80070459060004.jpeg?Expires=1769825637&Signature=jj8OU623WVQZ-T3iedZnEeCmmHQXadAXoX2klIMMX~8yTqQq8YLujv9--lXGMV9d6G0y~Od9AhNhFX5o43cj6H1EDrlD37GgNvLLQwM-5-ErHs6itUdgopqRASRDnDCxsxXOwp2zyDl9XjwuGs2gX7WBC2ddrZaaMUcYQ2UQsqKnzYwv686GgA0pTZUCKEctOsIvV4FppoB8TOnT6CSoa-ZJ-nbboipIOuCowZDFAzkfrCci9uhR~Ac00cJ16urAhij-cw8KWkrewaksxw~ZN0levJOxFpabxp5zIk8fsoJXR3UOXHAi7dvylx2Akwlwnzh3mvYtSDk90664Y3iLTg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal