Abstract
It is generally recognized that postmitotic neutrophils give rise to polymorphonuclear neutrophils alone. We obtained evidence for a lineage switch of human postmitotic neutrophils into macrophages in culture. When the CD15+CD14- cell population, which predominantly consists of band neutrophils, was cultured with granulocyte macrophage–colony-stimulating factor, tumor necrosis factor-α, interferon-γ, and interleukin-4, and subsequently with macrophage colony-stimulating factor alone, the resultant cells had morphologic, cytochemical, and phenotypic features of macrophages. In contrast to the starting population, they were negative for myeloperoxidase, specific esterase, and lactoferrin, and they up-regulated nonspecific esterase activity and the expression of macrophage colony-stimulating factor receptor, mannose receptor, and HLA-DR. CD15+CD14- cells proceeded to macrophages through the CD15-CD14- cell population. Microarray analysis of gene expression also disclosed the lineage conversion from neutrophils to macrophages. Macrophages derived from CD15+CD14- neutrophils had phagocytic function. Data obtained using 3 different techniques, including Ki-67 staining, bromodeoxyuridine incorporation, and cytoplasmic dye labeling, together with the yield of cells, indicated that the generation of macrophages from CD15+CD14- neutrophils did not result from a contamination of progenitors for macrophages. Our data show that in response to cytokines, postmitotic neutrophils can become macrophages. This may represent another differentiation pathway toward macrophages in human postnatal hematopoiesis. (Blood. 2004;103:2973-2980)
Introduction
One apparent characteristic of the hematopoietic system is that all types of mature cells with distinct functions are continuously replaced by cells derived from hematopoietic stem cells that reside mainly in the bone marrow and have the ability for self-renewal and multilineage differentiation.1,2 In the process of differentiation, hematopoietic stem cells sequentially lose multilineage potential and generate lineage-committed progenitors with limited developmental capacity. Lineage-committed progenitors give rise to precursors that eventually differentiate into mature blood cells. After commitment to specific lineages, hematopoietic progenitor cells are incapable of producing mature cells of other lineages. For example, neutrophilic monopotent progenitors proliferate and differentiate into neutrophils, and progenitors restricted to a macrophage lineage cannot give rise to mature cells other than macrophages.
Several studies have focused on a lineage switch in the hematopoietic differentiation pathway. Boyd and Schrader3 reported that murine pre–B-cell lines differentiate into macrophage-like cells in response to the demethylating drug, 5-azacytidine. B-lineage cells have been also shown to transit into neutrophils or natural killer (NK)/T-lineage cells.4-8 Furthermore, the macrophage cell line differentiates into B-lineage cells.9 Common lymphoid progenitors that exogenously express cytokine receptors or transcriptional factors develop myeloid lineage cells.10-12 In addition to these lineage switches, lineage conversions within myeloid lineages have also been observed in experiments using cell lines and transformed cells. Enforced expression of transcriptional factors such as GATA-1 and PU.1 leads to the lineage switch of myeloid cells to cells with another myeloid phenotype.12-17 These lineage switches include myeloid to megakaryocytic, myeloid to eosinophilic, myeloid to erythroid, and erythroid to myeloid conversions. The ectopic expression of PU.1 and C/EBP also results in the commitment of hematopoietic progenitors to an eosinophilic lineage.18-20 With respect to normal primary cells, it has been reported that murine B-cell precursor cells acquire the potential to differentiate into macrophage-like cells when they are transferred to myeloid culture conditions.21 Normal murine early T progenitor cells also generate macrophages in cultures supplemented with conditioned medium from a thymic stromal cell line or in the presence of cytokines.22 Thus, normal murine lymphoid progenitors may retain the potential to differentiate into macrophages. Reynaud et al23 reported that human cord blood–derived pro-B cells with DJH rearrangements of the immunoglobulin (Ig) locus generate macrophages, NK cells, and T cells. However, it is unknown whether the lineage switch occurs in normal human mature hematopoietic cells.
We conducted research to determine whether normal postmitotic neutrophils are able to differentiate into other types of mature cells. We found that human postmitotic neutrophils switched their differentiation program and acquired the features of macrophages in cultures supplemented with cytokines. These data support the possibility that postmitotic neutrophils commonly thought to be restricted to the neutrophilic differentiation pathway can become macrophages, and we argue that the lineage switch of human mature blood cells may, at least in part, be relevant to normal hematopoietic differentiation.
Materials and methods
Cytokines
Recombinant human macrophage colony-stimulating factor (M-CSF) and interferon-γ (IFN-γ) were purchased from R&D Systems (Minneapolis, MN). Recombinant human granulocyte macrophage–colony-stimulating factor (GM-CSF) was provided by Kirin Brewery (Tokyo, Japan). Recombinant human tumor necrosis factor-α (TNF-α) was a gift from Dainippon Pharmaceutical (Suita, Japan). Recombinant human interleukin-4 (IL-4) was provided by Ono Pharmaceutical (Osaka, Japan). Cytokines were used at the following concentrations: M-CSF, 100 ng/mL; IFN-γ, 25 IU/mL; GM-CSF, 10 ng/mL; TNF-α, 20 ng/mL; and IL-4, 10 ng/mL.
Cell preparation
Peripheral blood was obtained from healthy Japanese donors given subcutaneous injections of granulocyte colony-stimulating factor (G-CSF) to harvest peripheral blood stem cells. Each donor gave written, informed consent. Peripheral blood mononuclear cells (PBMCs) were separated by centrifugation on Ficoll-Hypaque, washed with Ca2+-, Mg2+-free phosphate-buffered saline (PBS), and suspended in PBS with 0.1% bovine serum albumin (BSA) (Sigma Chemical, St Louis, MO). CD15+CD14- cells were separated from PBMCs using CD14 and CD15 immunomagnetic beads (MACS; Miltenyi Biotec, Auburn, CA), according to the manufacturer's instructions. CD14+ and CD8+ cells were also separated from PBMCs using CD14 and CD8 immunomagnetic beads (MACS; Miltenyi Biotec), respectively. Healthy donors gave written, informed consent. The purity of CD15+CD14-, CD14+, or CD8+ cells exceeded 99%.
Culture
Culture medium was RPMI 1640 (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 2 mM l-glutamine, 50 U/mL penicillin, 50 μg/mL streptomycin, and 10% fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT). CD15+CD14- cells (1 × 106/mL) were cultured with designated combinations of cytokines in a 24-well tissue culture plate (Nunc, Roskilde, Denmark) for 18 days. Half of the culture medium was replaced with fresh medium containing cytokines every 3 to 4 days. In some experiments, cells were plated with designated combinations of cytokines; on day 11 of culture, the cells were washed 3 times with PBS and replated in cultures containing M-CSF. CD14+ and CD15+CD14- cells (1 × 106/mL) were cultured in the presence of M-CSF for 7 days. Viable cells were counted using trypan blue dye exclusion methods.
Morphologic cell analysis
Cytospin preparations of freshly isolated and cultured cells were stained with May-Grünwald-Giemsa solution, myeloperoxidase (MPO) staining kit, and double-specific (naphthol AS-D chloroacetate esterase)/nonspecific (α-naphthyl butyrate esterase) esterase staining kit.
Cell cycle analysis
Cell cycle analysis was made using the CycleTEST PLUS DNA Reagent Kit (Becton Dickinson, San Jose, CA) according to the manufacturer's instructions. Nuclear DNA content of freshly isolated CD15+CD14- cells was analyzed on a FACSCalibur flow cytometer (Becton Dickinson) with ModFit software (Verity Software House, Topsham, ME). HPB-NULL cells (American Type Culture Collection, Manassas, VA) were used as a control. HPB-NULL cells were passaged every 24 hours for 3 days before use to minimize differences among experimental conditions.
Flow cytometric analysis
The following murine or rat monoclonal antibodies (mAbs) were used: anti–CD14-phycoerythrin (anti–CD14-PE), anti–CD15-fluorescein isothiocyanate (anti–CD15-FITC), and anti–HLA-DR-PE (Becton Dickinson); anti–MPO-FITC (DAKO, Glostrup, Denmark); anti–lactoferrin-PE (Immunotech, Marseille, France); anti–c-fms/M-CSF receptor (anti–c-fms/M-CSFR) (Oncogene Research, Boston, MA); anti–mannose receptor-PE, anti–Ki-67-FITC, which reacts with a proliferation-associated nuclear antigen, antibromodeoxyuridine (anti-BrdU)–FITC, and anti–rat IgG2b-FITC (Becton Dickinson PharMingen, San Diego, CA). Rat immunoglobulin and FITC- or PE-labeled mouse immunoglobulin served as isotype control: rat IgG2b and mouse IgM-FITC (Becton Dickinson PharMingen); mouse IgG1-FITC, IgG1-PE, and IgG2a-PE (Becton Dickinson); and mouse IgG2b-PE (Coulter, Miami, FL).
For membrane staining, cells were incubated with mAbs for 30 minutes on ice and washed 3 times with PBS. Intracellular molecules were stained, using Cytofix/Cytoperm Kit (PharMingen, San Diego, CA). Cells were fixed for 20 minutes at 4°C with formaldehyde-based fixation medium, washed, resuspended in permeabilization buffer containing sodium azide and saponin, and stained with mAbs for 30 minutes at 4°C. Cells were washed with the permeabilization buffer and PBS. HPB-NULL cells were used as a positive control for Ki-67 staining. Flow cytometric analysis was performed, using a FACSCalibur flow cytometer. CellQuest software (Becton Dickinson) was used for data acquisition and analysis.
BrdU and carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling
For BrdU pulse labeling, freshly isolated and cultured cells (1 × 106/mL) were incubated with 5 μg/mL BrdU (Sigma Chemical) for 40 minutes. The cells were washed with 0.1% BSA PBS, fixed with ice-cold 70% ethanol for 30 minutes at -20°C, and treated with 4 N HCl/0.5% Tween-20 at room temperature for 20 minutes for DNA denaturation. Acid was neutralized with 0.1 M Na2B4O7. After washing with 0.1% BSA PBS, the cells were stained with FITC-conjugated anti-BrdU mAb. BrdU incorporation was analyzed using a FACSCalibur cytometer.24 HPB-NULL cells served as a positive control for BrdU labeling.
Freshly isolated CD15+CD14- cells (1 × 106/mL) were labeled for 10 minutes at 37°C with 0.5 μM CFSE (Lambda, Graz, Austria) PBS, a cytoplasmic dye that is equally diluted between daughter cells, and were washed 3 times with 10% FBS RPMI 1640. CFSE-labeled CD15+CD14- cells were cultured as described above. CD8+ cells (1 × 106/mL) were labeled with CFSE, washed with 10% FBS RPMI 1640, and cultured with T-cell expander beads (CD3/CD28 T-cell expander; DynalBiotech, Oslo, Norway) at a 1:3 bead-to-cell ratio for 5 days. CFSE-labeled freshly isolated and cultured cells were analyzed for fluorescence intensity using a FACSCalibur cytometer.25
Microarray
Microarray assay was carried out as described.26,27 Total RNA extracted from freshly isolated and cultured cells by the acid guanidinium method was used to synthesize double-stranded cDNA. Biotin-labeled cRNA was prepared from each cDNA by ENZO BioArray High-Yield RNA Transcript Labeling kit (Affymetrix, Santa Clara, CA), and hybridized with a GeneChip HGU133A microarray (Affymetrix) harboring more than 22 000 human probe sets. Hybridization, washing, and detection of signals on the arrays were performed using the GeneChip system (Affymetrix) according to the manufacturer's protocols. Fluorescence intensity for each gene was normalized on the basis of the median expression value of the positive control genes (Affymetrix; HGU133Anorm.MSK) in each hybridization. Every microarray analysis was repeated twice. Hierarchical clustering and Welch analysis of variance (ANOVA) of the data set were conducted by using GeneSpring 6.0 software (Silicon Genetics, Redwood, CA).
Phagocytosis
Cultured cells (2 × 105/mL) were incubated with FITC-dextran (40 000 molecular weight; Sigma Chemical) or FITC-latex beads (2 μm; Polysciences, Warrington, PA) for 1 hour at 37°C or 4°C. The uptake of FITC-dextran and FITC-latex beads was halted by the addition of cold 1% FBS PBS. After washing 3 times with 1% FBS PBS, cells were analyzed on a FACS flow cytometer.28,29
Results
CD15+CD14- neutrophils generate macrophages
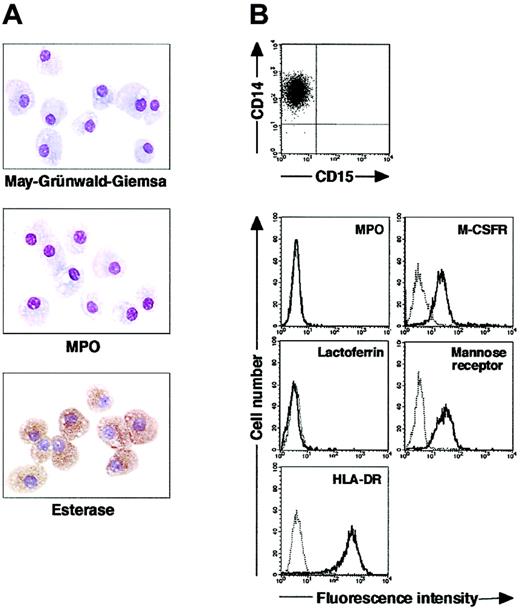
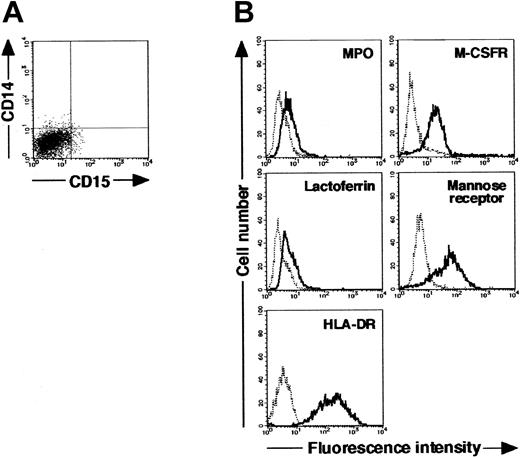
We isolated CD15+CD14- cells from human blood samples. Portions of May-Grünwald-Giemsa–, MPO-, and double-specific/nonspecific esterase–stained preparations are presented in Figure 1A. The CD15+CD14- cell fraction consisted of myelocytes, metamyelocytes, band cells, and segmented cells. Proportions of myelocytes, metamyelocytes, band cells, and segmented cells were 0.6% ± 0.2%, 9.2% ± 2.5%, 87.6% ± 2.5%, and 2.6% ± 0.4%, respectively (n = 5). CD15+CD14- cells were positive for MPO and specific esterase but negative for nonspecific esterase. These staining patterns were found in all portions of cytospin preparations. We analyzed the cell cycle characteristics of CD15+CD14- cells. All CD15+CD14- cells were found in G1 phase of the cell cycle (Figure 1B), supporting the notion that these cells are postmitotic. The expression of MPO, M-CSFR, lactoferrin, mannose receptor, and HLA-DR was assessed by phenotypic analysis (Figure 1C). CD15+CD14- cells were MPO+, M-CSFR-, lactoferrin+, mannose receptor-, and HLA-DR-, a finding compatible with typical features of mature neutrophils. CD14+ cells were also prepared from blood samples. Cultures of CD14+ cells in the presence of M-CSF gave rise to cells with small nuclei and intracytoplasmic vacuoles. These cells showed nonspecific esterase activity but not MPO or specific esterase activity (Figure 2A), and they exhibited the phenotype of CD15- CD14+, MPO-, M-CSFR+, lactoferrin-, mannose receptor+, and HLA-DR+ (Figure 2B), which suggested that the resultant cells were macrophages. CD15+CD14- cells were also cultured in the presence of M-CSF for 7 days. The yield was less than 2% of the starting population. Surviving cells retained the features of neutrophils (data not shown).
Cytochemistry and phenotype of freshly isolated CD15+CD14- cells. (A) Photographs of May-Grünwald-Giemsa–, MPO-, and double-specific/nonspecific esterase–stained cytospin preparations; original magnification, × 400. (B) Nuclear DNA analysis of CD15+CD14- cells was performed using a FACSCalibur flow cytometer. HPB-NULL cells were used as controls. Cell cycle distribution of HPB-NULL cells was as follows: with G1 phase, 42.9%; S phase, 30.5%; and G2/M phase, 26.6%. (C) The expression of CD15/CD14, MPO, M-CSFR, lactoferrin, mannose receptor, and HLA-DR was analyzed using a FACSCalibur flow cytometer. In the histograms, the thick and thin lines show the expression of the indicated molecules and isotype controls, respectively. Representative data from 5 independent experiments are shown.
Cytochemistry and phenotype of freshly isolated CD15+CD14- cells. (A) Photographs of May-Grünwald-Giemsa–, MPO-, and double-specific/nonspecific esterase–stained cytospin preparations; original magnification, × 400. (B) Nuclear DNA analysis of CD15+CD14- cells was performed using a FACSCalibur flow cytometer. HPB-NULL cells were used as controls. Cell cycle distribution of HPB-NULL cells was as follows: with G1 phase, 42.9%; S phase, 30.5%; and G2/M phase, 26.6%. (C) The expression of CD15/CD14, MPO, M-CSFR, lactoferrin, mannose receptor, and HLA-DR was analyzed using a FACSCalibur flow cytometer. In the histograms, the thick and thin lines show the expression of the indicated molecules and isotype controls, respectively. Representative data from 5 independent experiments are shown.
Cytochemistry and phenotype of cells obtained from 7-day culture of freshly isolated CD14+ cells with M-CSF. (A) Photographs of May-Grünwald-Giemsa–, MPO-, and double specific/nonspecific esterase–stained cytospin preparations; original magnification, × 400. (B) The expression of CD15/CD14, MPO, M-CSFR, lactoferrin, mannose receptor, and HLA-DR was analyzed using a FACSCalibur flow cytometer. In the histograms, the thick and thin lines show the expression of the indicated molecules and isotype controls, respectively. Yield of cultured cells was 52.8% ± 9.8% (n = 5). Representative data from 5 independent experiments are shown.
Cytochemistry and phenotype of cells obtained from 7-day culture of freshly isolated CD14+ cells with M-CSF. (A) Photographs of May-Grünwald-Giemsa–, MPO-, and double specific/nonspecific esterase–stained cytospin preparations; original magnification, × 400. (B) The expression of CD15/CD14, MPO, M-CSFR, lactoferrin, mannose receptor, and HLA-DR was analyzed using a FACSCalibur flow cytometer. In the histograms, the thick and thin lines show the expression of the indicated molecules and isotype controls, respectively. Yield of cultured cells was 52.8% ± 9.8% (n = 5). Representative data from 5 independent experiments are shown.
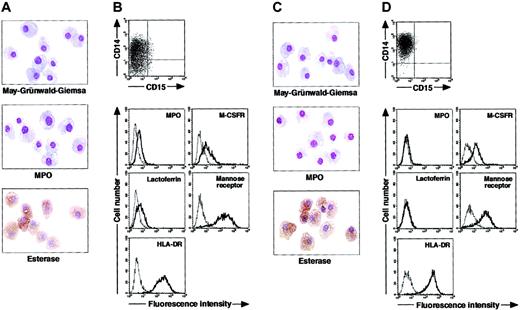
To determine whether postmitotic neutrophils have the potential to alter the lineage, we attempted to drive CD15+CD14- cells to become cells of a monocyte/macrophage lineage, using cultures supplemented with cytokines. The combination of GM-CSF, M-CSF, TNF-α, and IFN-γ was chosen because GM-CSF, M-CSF, TNF-α, and IFN-γ are known to favor the differentiation of monocytes and their progenitors into macrophages.1,30-36 Because M-CSF is a cytokine specific for, and late-acting in, a monocyte/macrophage lineage, we cultured CD15+CD14- cells in the presence of GM-CSF, TNF-α, and IFN-γ for 11 days and subsequently replated the cells in cultures with M-CSF alone. On day 18 after the initiation of culture, cultured cells were harvested and characterized in morphologic, cytochemical, and phenotypic analyses. These cells had macrophage morphology (Figure 3A). Their MPO or specific esterase activity was not detected using light microscopy; however, the nonspecific esterase reaction was positive. CD14 expression was induced in a substantial, although not the entire, population of the resultant cells, but CD15 expression was completely lost (Figure 3B). When compared with freshly isolated CD15+CD14- cells, the resultant cells did not entirely down-regulate MPO and lactoferrin yet they considerably up-regulated M-CSFR, mannose receptor, and HLA-DR. This culture condition yielded 1.8% ± 0.6% (n = 5) of the starting CD15+CD14- cell population. These data suggest that GM-CSF, TNF-α, IFN-γ, and M-CSF allow CD15+CD14- cells to become macrophages.
Cytochemistry and phenotype of cells obtained from culture of freshly isolated CD15+CD14- neutrophils. (A-B) Freshly isolated CD15+CD14- neutrophils were cultured with GM-CSF, TNF-α, and IFN-γ for 11 days, followed by additional 7-day culture with M-CSF alone. (C-D) Freshly isolated CD15+CD14- neutrophils were cultured with GM-CSF, TNF-α, IFN-γ, and IL-4 for 11 days, followed by additional 7-day culture with M-CSF alone. (A,C) Photographs of May-Grünwald-Giemsa-, MPO-, and double specific/nonspecific esterase–stained cytospin preparations; original magnification, × 400. (B,D) The expression of CD15/CD14, MPO, M-CSFR, lactoferrin, mannose receptor, and HLA-DR was analyzed using a FACSCalibur flow cytometer. In the histograms, the thick and thin lines show the expression of the indicated molecules and isotype controls, respectively. Representative data from 5 independent experiments are shown.
Cytochemistry and phenotype of cells obtained from culture of freshly isolated CD15+CD14- neutrophils. (A-B) Freshly isolated CD15+CD14- neutrophils were cultured with GM-CSF, TNF-α, and IFN-γ for 11 days, followed by additional 7-day culture with M-CSF alone. (C-D) Freshly isolated CD15+CD14- neutrophils were cultured with GM-CSF, TNF-α, IFN-γ, and IL-4 for 11 days, followed by additional 7-day culture with M-CSF alone. (A,C) Photographs of May-Grünwald-Giemsa-, MPO-, and double specific/nonspecific esterase–stained cytospin preparations; original magnification, × 400. (B,D) The expression of CD15/CD14, MPO, M-CSFR, lactoferrin, mannose receptor, and HLA-DR was analyzed using a FACSCalibur flow cytometer. In the histograms, the thick and thin lines show the expression of the indicated molecules and isotype controls, respectively. Representative data from 5 independent experiments are shown.
Next, we searched for cytokine(s). The addition of cytokines to cultures would allow cells derived from CD15+CD14- cells to acquire features more typical of macrophages. After testing several cytokines, we found that the appropriate conditions could be satisfied by supplementation with IL-4. When cultured with GM-CSF, TNF-α, IFN-γ, and IL-4 for 11 days and with M-CSF alone for 7 days, CD15+CD14- cells gave rise to cells with morphologic characteristics of macrophages. The resultant cells were negative for MPO and specific esterase activities and positive for nonspecific esterase activity on cytospin preparations (Figure 3C). Phenotypic analysis showed that the resultant cells lacked CD15 and exclusively expressed CD14. MPO and lactoferrin were completely down-regulated, whereas the high levels of M-CSFR, mannose receptor, and HLA-DR expression were retained (Figure 3D), as in the macrophage phenotype induced from CD14+ cells by M-CSF. Strikingly, the yield increased to 15.1% ± 3.6% (n = 5) of the starting CD15+CD14- cell population. These observations unambiguously demonstrated a cytokine-induced lineage switch of postmitotic neutrophils to macrophages. We also cultured CD15+CD14- cells in the presence of GM-CSF, TNF-α, IFN-γ, and IL-4 for 11 days and analyzed their phenotypes using flow cytometry. Cultured cells expressed neither CD15 nor CD14 (Figure 4A). MPO and lactoferrin were detected at low levels, whereas the expression levels of M-CSFR, mannose receptor, and HLA-DR were high (Figure 4B). These data suggest that the expression levels of molecules that characterize postmitotic neutrophils or macrophages are not simultaneously altered during this lineage switch program.
Cytochemistry and phenotype of cells obtained from culture of freshly isolated CD15+CD14- neutrophils with GM-CSF, TNF-α, IFN-γ, and IL-4 for 11 days. (A) Expression pattern of CD15/CD14. (B) The expression of CD15/CD14, MPO, lactoferrin, M-CSFR, mannose receptor, and HLA-DR was analyzed using a FACSCalibur flow cytometer. In the histograms, the thick and thin lines show the expression of the indicated molecules and isotype controls, respectively. Representative data from 5 independent experiments are shown.
Cytochemistry and phenotype of cells obtained from culture of freshly isolated CD15+CD14- neutrophils with GM-CSF, TNF-α, IFN-γ, and IL-4 for 11 days. (A) Expression pattern of CD15/CD14. (B) The expression of CD15/CD14, MPO, lactoferrin, M-CSFR, mannose receptor, and HLA-DR was analyzed using a FACSCalibur flow cytometer. In the histograms, the thick and thin lines show the expression of the indicated molecules and isotype controls, respectively. Representative data from 5 independent experiments are shown.
Gene expression profiles of CD15+CD14- neutrophils and macrophages
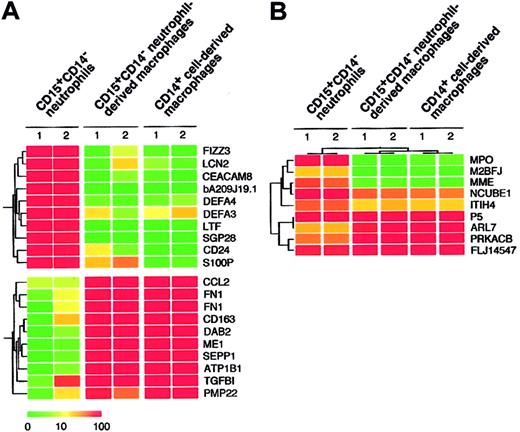
Given that the combination of cytokines consisting of GM-CSF, TNF-α, IFN-γ, IL-4, and M-CSF allowed for the generation of macrophages from CD15+CD14- neutrophils, as determined by morphologic, cytochemical, and phenotypic analyses, we compared the gene expression profiles of freshly isolated CD15+CD14- neutrophils, CD15+CD14- neutrophil–derived macrophages, and CD14+ cell–derived macrophages using high-density oligonucleotide microarrays. Because every microarray was repeated twice, the mean expression intensity was calculated for each gene and was used for the following analysis. Among our expression data set, first searched were the genes expressed abundantly in CD15+CD14- neutrophils but not in CD14+ cell–derived macrophages. Table 1 shows 10 such genes that had an expression level of more than 100 arbitrary units in CD15+CD14- neutrophils and the highest ratio of the expression level between CD15+CD14- neutrophils and CD14+ cell-derived macrophages. Interestingly, all these CD15+CD14- neutrophil-specific genes were also transcriptionally silent in CD15+CD14- neutrophil-derived macrophages, as in CD14+ cell-derived macrophages. A well-known marker for granulocytes, CD24 (GenBank accession number AA761181) was only expressed in CD15+CD14- neutrophils but not in CD15+CD14- neutrophil-derived macrophages or CD14+ cell-derived macrophages. Conversely, we also tried to extract CD14+ cell-derived, macrophage-specific genes by comparing CD14+ cell-derived macrophages and CD15+CD14- neutrophils (Table 2). Again, expression levels of these genes in the CD15+CD14- neutrophil-derived macrophages were highly similar to those in CD14+ cell-derived macrophages. A monocyte/macrophage-specific cell surface antigen, CD163 (Z22969), was abundantly expressed in the CD15+CD14- neutrophil-derived macrophages and the CD14+ cell-derived macrophages, but not in the CD15+CD14- neutrophils. Hierarchical clustering analysis of these lineage-specific genes showed the similarity between gene expression profiles of CD15+CD14- neutrophil-derived macrophages and CD14+ cell-derived macrophages (Figure 5A). Next, to statistically examine this similarity, we directly compared the 4 expression data sets of CD15+CD14- neutrophils (n = 2) and CD14+ cell-derived macrophages (n = 2) and attempted to identify the genes, whose expression was different in the 2 groups (Welch ANOVA, P < .001). The expression profiles of such 9 lineage-dependent genes (Table 3) were then used to measure the similarity between CD14+ cell–derived macrophages and the other 2 groups. As shown in Figure 5B, 2-way clustering analysis37 of 6 data sets (3 groups) clearly indicated that, with regard to gene expression profile, CD15+CD14- neutrophil–derived macrophages were similar to CD14+ cell–derived macrophages, separated from CD15+ CD14- neutrophils.
Genes specifically expressed in CD15+CD14−neutrophils
Gene symbol . | GenBank accession no. . | CD15+CD14−neutrophil/CD14+ cell-derived macrophage ratio . | CD15+CD14−neutrophils . | CD14+cell-derived macrophages . | CD15+CD14−neutrophil-derived macrophages . |
|---|---|---|---|---|---|
| bA209J19.1 | AL390736 | 5412.83 | 1623.85 | 0.30 | 0.60 |
| LTF | NM_002343 | 2795.52 | 3773.95 | 1.35 | 1.50 |
| DEFA3 | NM_004084 | 1142.16 | 15 761.80 | 13.80 | 9.65 |
| S100P | NM_005980 | 1099.67 | 3189.05 | 2.90 | 28.05 |
| SGP28 | NM_006061 | 699.78 | 314.90 | 0.45 | 0.45 |
| CEACAM8 | M33326 | 588.24 | 1323.55 | 2.25 | 3.30 |
| LCN2 | NM_005564 | 550.64 | 2945.90 | 5.35 | 8.90 |
| DEFA4 | NM_001925 | 499.83 | 2849.05 | 5.70 | 3.85 |
| CD24 | BG327863 | 474.81 | 759.70 | 1.60 | 8.75 |
| FIZZ3 | NM_020415 | 452.96 | 1223.00 | 2.70 | 5.90 |
Gene symbol . | GenBank accession no. . | CD15+CD14−neutrophil/CD14+ cell-derived macrophage ratio . | CD15+CD14−neutrophils . | CD14+cell-derived macrophages . | CD15+CD14−neutrophil-derived macrophages . |
|---|---|---|---|---|---|
| bA209J19.1 | AL390736 | 5412.83 | 1623.85 | 0.30 | 0.60 |
| LTF | NM_002343 | 2795.52 | 3773.95 | 1.35 | 1.50 |
| DEFA3 | NM_004084 | 1142.16 | 15 761.80 | 13.80 | 9.65 |
| S100P | NM_005980 | 1099.67 | 3189.05 | 2.90 | 28.05 |
| SGP28 | NM_006061 | 699.78 | 314.90 | 0.45 | 0.45 |
| CEACAM8 | M33326 | 588.24 | 1323.55 | 2.25 | 3.30 |
| LCN2 | NM_005564 | 550.64 | 2945.90 | 5.35 | 8.90 |
| DEFA4 | NM_001925 | 499.83 | 2849.05 | 5.70 | 3.85 |
| CD24 | BG327863 | 474.81 | 759.70 | 1.60 | 8.75 |
| FIZZ3 | NM_020415 | 452.96 | 1223.00 | 2.70 | 5.90 |
Mean expression levels of genes in CD15+CD14− neutrophils, CD14+ cell-derived macrophages, and CD15+CD14− neutrophil-derived macrophages are shown in arbitrary units. The ratio between the first 2 is indicated in the CD15+CD14− neutrophil/CD14+ cell-derived macrophage ratio.
Genes specifically expressed in CD14+cell-derived macrophages
Gene symbol . | GenBank accession no. . | CD14+cell-derived macrophage/CD15+CD14−neutrophil ratio . | CD14+cell-derived macrophages . | CD15+CD14−neutrophils . | CD15+CD14−neutrophil-derived macrophages . |
|---|---|---|---|---|---|
| SEPP1 | NM_005410 | 643.60 | 1769.90 | 2.75 | 1373.45 |
| DAB2 | NM_001343 | 307.52 | 1183.95 | 3.85 | 513.20 |
| CD163 | Z22969 | 256.43 | 2602.75 | 10.15 | 1034.35 |
| ME1 | NM_002395 | 197.02 | 551.65 | 2.80 | 310.10 |
| CCL2 | S69738 | 196.38 | 1708.50 | 8.70 | 351.10 |
| ATP1B1 | BC000006 | 180.35 | 775.50 | 4.30 | 408.85 |
| FN1 | BC005858 | 169.67 | 972.70 | 5.75 | 155.60 |
| FN1 | AK026737 | 162.44 | 1210.15 | 7.45 | 191.65 |
| TGFBI | NM_000358 | 131.99 | 4045.55 | 30.65 | 3030.45 |
| PMP22 | L03203 | 129.14 | 1091.25 | 8.45 | 59.20 |
Gene symbol . | GenBank accession no. . | CD14+cell-derived macrophage/CD15+CD14−neutrophil ratio . | CD14+cell-derived macrophages . | CD15+CD14−neutrophils . | CD15+CD14−neutrophil-derived macrophages . |
|---|---|---|---|---|---|
| SEPP1 | NM_005410 | 643.60 | 1769.90 | 2.75 | 1373.45 |
| DAB2 | NM_001343 | 307.52 | 1183.95 | 3.85 | 513.20 |
| CD163 | Z22969 | 256.43 | 2602.75 | 10.15 | 1034.35 |
| ME1 | NM_002395 | 197.02 | 551.65 | 2.80 | 310.10 |
| CCL2 | S69738 | 196.38 | 1708.50 | 8.70 | 351.10 |
| ATP1B1 | BC000006 | 180.35 | 775.50 | 4.30 | 408.85 |
| FN1 | BC005858 | 169.67 | 972.70 | 5.75 | 155.60 |
| FN1 | AK026737 | 162.44 | 1210.15 | 7.45 | 191.65 |
| TGFBI | NM_000358 | 131.99 | 4045.55 | 30.65 | 3030.45 |
| PMP22 | L03203 | 129.14 | 1091.25 | 8.45 | 59.20 |
Mean expression levels of the genes in CD14+ cell-derived macrophages, CD15+CD14− neutrophils, and CD15+CD14− neutrophil-derived macrophages are shown in arbitrary units. The ratio between the first 2 is indicated in the CD14+ cell-derived macrophage/CD15+CD14− neutrophil ratio.
Expression profiles of neutrophil- and macrophage-specific genes. (A) Hierarchical clustering based on the expression intensities in CD15+CD14- neutrophils, CD15+CD14- neutrophil–derived macrophages, and CD14+ cell–derived macrophages was conducted for 10 genes with a specific expression in CD15+CD14- neutrophils and CD14+ cell–derived macrophages (top panel) or in CD14+ cell–derived macrophages (bottom panel). Each row represents a single gene on the microarray, and each column represents a separate sample. Expression intensity of each gene is shown color coded, according to the scale at the bottom, and the gene symbols are indicated on the right. Expression data of these genes are available on request. (B) The gene tree was constructed using 2-way clustering analysis of the genes that are differentially (Welch ANOVA, P < .001) expressed between CD15+CD14- neutrophils and CD14+ cell–derived macrophages. Each row represents a single gene on the microarray, and each column represents a separate sample. Expression intensity of each gene is shown color coded, according to the scale in panel A.
Expression profiles of neutrophil- and macrophage-specific genes. (A) Hierarchical clustering based on the expression intensities in CD15+CD14- neutrophils, CD15+CD14- neutrophil–derived macrophages, and CD14+ cell–derived macrophages was conducted for 10 genes with a specific expression in CD15+CD14- neutrophils and CD14+ cell–derived macrophages (top panel) or in CD14+ cell–derived macrophages (bottom panel). Each row represents a single gene on the microarray, and each column represents a separate sample. Expression intensity of each gene is shown color coded, according to the scale at the bottom, and the gene symbols are indicated on the right. Expression data of these genes are available on request. (B) The gene tree was constructed using 2-way clustering analysis of the genes that are differentially (Welch ANOVA, P < .001) expressed between CD15+CD14- neutrophils and CD14+ cell–derived macrophages. Each row represents a single gene on the microarray, and each column represents a separate sample. Expression intensity of each gene is shown color coded, according to the scale in panel A.
Genes with statistically different expression between CD15+CD14−neutrophils and CD14+cell-derived macrophages
Gene symbol . | GenBank accession no. . |
|---|---|
| MPO | J02694 |
| H2BFJ | NM_003524 |
| MME | NM_007287 |
| NCUBE1 | AF151039 |
| ITIH4 | D38535 |
| P5 | BC001312 |
| ARL7 | NM_005737 |
| PRKACB | AA130247 |
| FLJ14517 | AV7113053 |
Gene symbol . | GenBank accession no. . |
|---|---|
| MPO | J02694 |
| H2BFJ | NM_003524 |
| MME | NM_007287 |
| NCUBE1 | AF151039 |
| ITIH4 | D38535 |
| P5 | BC001312 |
| ARL7 | NM_005737 |
| PRKACB | AA130247 |
| FLJ14517 | AV7113053 |
Gene symbols and GenBank accession numbers are shown for genes that exhibited significant differences in expression level between the 2 groups (Welch ANOVA, P < .001).
Phagocytic activity of CD15+CD14- neutrophil–derived macrophages
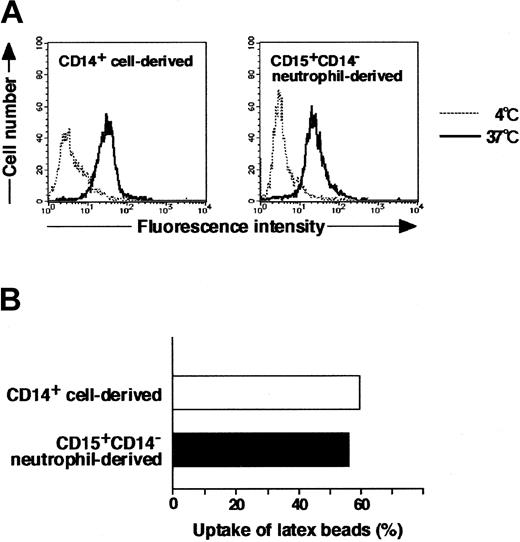
Morphology, cytochemistry, phenotype, and gene expression of cultured cells in the presence of GM-CSF, TNF-α, IFN-γ, IL-4, and M-CSF indicated that CD15+CD14- neutrophils became macrophages. Therefore, we next evaluated the phagocytic activity of these macrophages using FITC-dextran and FITC-latex beads. The potential for CD15+CD14- neutrophil–derived macrophages to incorporate dextran and latex beads was comparable to that of CD14+ cell–derived macrophages (Figure 6).
Phagocytic assay of CD15+CD14- neutrophil–derived macrophages with FITC-dextran and FITC-latex beads. (A) CD15+CD14- neutrophil– and CD14+ cell–derived macrophages were incubated with FITC-dextran for 1 hour at 37°C or 4°C, washed with cold PBS supplemented with 1% FBS, and analyzed using a FACSCalibur flow cytometer. Data are presented using histograms. (B) CD15+CD14- neutrophil– and CD14+ cell–derived macrophages were incubated with FITC-latex beads for 1 hour at 37°C or 4°C, washed with cold PBS supplemented with 1% FBS, and analyzed with a FACSCalibur flow cytometer. Data are expressed as percentage of positive cells. Experiments were repeated 5 times with identical results.
Phagocytic assay of CD15+CD14- neutrophil–derived macrophages with FITC-dextran and FITC-latex beads. (A) CD15+CD14- neutrophil– and CD14+ cell–derived macrophages were incubated with FITC-dextran for 1 hour at 37°C or 4°C, washed with cold PBS supplemented with 1% FBS, and analyzed using a FACSCalibur flow cytometer. Data are presented using histograms. (B) CD15+CD14- neutrophil– and CD14+ cell–derived macrophages were incubated with FITC-latex beads for 1 hour at 37°C or 4°C, washed with cold PBS supplemented with 1% FBS, and analyzed with a FACSCalibur flow cytometer. Data are expressed as percentage of positive cells. Experiments were repeated 5 times with identical results.
Proliferative characteristics during culture
It is possible that macrophages induced from CD15+CD14- neutrophils were derived from a small number of hematopoietic progenitor cells for macrophages that contaminated the CD15+CD14- cell population and consequently proliferated. To exclude this possibility, we analyzed proliferative characteristics of the cultured cells; representative data are presented in Figure 7. In Figure 7A, the yield of cultured cells was 15.1%, on day 18 of culture. Reactivity with Ki-67 and incorporation of BrdU were tested on the indicated days of culture.24,38 Ki-67+ or BrdU+ cells were not evident throughout the culture. Ki-67 expression and BrdU incorporation were observed most and approximately 30%, respectively, of HPB-NULL cells, which served as positive controls. We also used the carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling technique to confirm that CD15+CD14- neutrophils passed through no cell division during culture.25 Analysis of the CFSE labeling pattern in CD8+ T cells, which had elicited several rounds of the cell cycle in response to CD3/CD28 T-cell expander beads, displayed a number of peaks of fluorescence (Figure 7B). However, the CFSE fluorescence remained a single peak in the cells generated by culturing CD15+CD14- neutrophils with GM-CSF, TNF-α, IFN-γ, and IL-4 and subsequently with M-CSF alone. These data indicate that the generation of macrophages from the CD15+CD14- cell population in culture is not associated with cell division.
Analysis of proliferation profile. (A) CD15+CD14- neutrophils were cultured with GM-CSF, TNF-α, IFN-γ, and IL-4 for 11 days, washed, and recultured with M-CSF alone for an additional 7 days. CD15+CD14- neutrophils and the cultured cells were tested for Ki-67 staining and BrdU incorporation. Ki-67+ and BrdU+ cells were counted using a FACSCalibur flow cytometer, and the numbers were expressed as percentages. HPB-NULL cells were used as positive controls. Numbers of cultured cells are also indicated. Representative data from 5 independent experiments are shown. (B) CD15+CD14- neutrophils were cultured with GM-CSF, TNF-α, IFN-γ, and IL-4 for 11 days, washed, and recultured with M-CSF alone for another 7 days. CD15+CD14- neutrophils and the cultured cells were incubated with CFSE. Cell division patterns were analyzed using a FACSCalibur flow cytometer. CD8+ cells that underwent several rounds of the cell cycle in response to CD3/CD28 beads were used as positive controls. Experiments were repeated 5 times with identical results.
Analysis of proliferation profile. (A) CD15+CD14- neutrophils were cultured with GM-CSF, TNF-α, IFN-γ, and IL-4 for 11 days, washed, and recultured with M-CSF alone for an additional 7 days. CD15+CD14- neutrophils and the cultured cells were tested for Ki-67 staining and BrdU incorporation. Ki-67+ and BrdU+ cells were counted using a FACSCalibur flow cytometer, and the numbers were expressed as percentages. HPB-NULL cells were used as positive controls. Numbers of cultured cells are also indicated. Representative data from 5 independent experiments are shown. (B) CD15+CD14- neutrophils were cultured with GM-CSF, TNF-α, IFN-γ, and IL-4 for 11 days, washed, and recultured with M-CSF alone for another 7 days. CD15+CD14- neutrophils and the cultured cells were incubated with CFSE. Cell division patterns were analyzed using a FACSCalibur flow cytometer. CD8+ cells that underwent several rounds of the cell cycle in response to CD3/CD28 beads were used as positive controls. Experiments were repeated 5 times with identical results.
Discussion
Lineage switch of normal primary cells has been noted in murine lymphoid progenitor cells.21,22 Montecino-Rodriguez et al21 observed that a subpopulation of B-cell precursors in the bone marrow gave rise not only to B cells but also to macrophages in cultures supplemented with IL-3, IL-6, c-kit ligand, and GM-CSF. Lee et al22 demonstrated that fetal thymocytes differentiate into macrophages in the presence of M-CSF, IL-6, and IL-7 in vitro. A recent report stated that human B-cell progenitors obtained from cultures of cord blood CD34+CD10-CD19- cells gave rise to macrophages, NK cells, and T cells when exposed to the appropriate culture conditions.23 Our study shows the lineage switch of human primary postnatal cells and further extends the existence of lineage switch to postmitotic cells. This notion is also supported by our observations in the microarray analysis that gene expression profiles of the resultant cells differed from those of the starting CD15+CD14- neutrophils and were similar to those of CD14+ cell–derived macrophages.
When we first cultured CD15+CD14- neutrophils in the presence of GM-CSF, TNF-α, IFN-γ, and M-CSF, the resultant cells displayed morphologic and cytochemical features of macrophages. However, they preserved a low level of MPO activity and lactoferrin expression, as determined by flow cytometry. These data suggest that the resultant cells did not fully exhibit the phenotypic characteristics of macrophages. Our surprise was that the addition of IL-4 to cultures was sufficient for the resultant cells to acquire typical features of macrophages. It is also of note that in the presence of IL-4, the yield of the resultant cells increased to approximately 15%. The phagocytic activity of macrophages generated in IL-4–containing cultures was of a similar magnitude compared with that observed with CD14+ cell–derived macrophages. Previous studies demonstrated the inhibitory activities of IL-4 on the development of monocytes/macrophages from progenitors supported by GM-CSF.39,40 IL-4 has the potential to suppress TNF-α–induced effects on hematopoietic cells.41 IL-4 also down-regulates the expression of 2 distinct receptors for TNF-α, p60, and p80 and induces shedding of these receptors, resulting in blockage of the cellular signaling elicited by TNF-α.42 Moreover, several investigators have shown that IL-4 antagonizes IFN-γ–induced responses in human myeloid progenitor and mature cells.43-45 Therefore, we have no plausible explanation for the mechanism of action of IL-4 on CD15+CD14- neutrophils during their lineage switch to macrophages. Complex networks by multiple cytokines may be involved in the generation of macrophages from CD15+CD14- neutrophils. Interestingly, phenotypic analysis indicated that when CD15+CD14- neutrophils turn their lineage toward macrophages, they lose CD15 expression and acquire CD14. This finding demonstrates that the down-regulation of CD15 occurs before the up-regulation of CD14. The cascade of several different events may lead to the lineage conversion of CD15+CD14- neutrophils to macrophages.
Our concern was whether a rare population of hematopoietic progenitors, contaminating the CD15+CD14- fraction, could proliferate and differentiate into macrophages. If such were the case, the cultured cells would show signs of proliferation at several time points during culture. To address this issue, we used Ki-67 antibody staining, BrdU incorporation, and CFSE labeling. Neither Ki-67+ nor BrdU+ cells were detectable throughout culture, suggesting that cell division did not occur. In addition, if cultured cells had a history of successive cell divisions, we would have expected to observe separate peaks of CFSE fluorescence in the histogram. However, the narrow peak was observed with the resultant cells, as was the peak seen with the starting cell population. On the basis of these data and because the yield was approximately 15%, we assumed it was not possible for the cultured cells to have undergone more than one division throughout culture. Therefore, we propose that the generation of macrophages from CD15+CD14- neutrophils with GM-CSF, TNF-α, IFN-γ, IL-4, and M-CSF was not caused by contamination of progenitor cells for macrophages but was the result of their lineage switch to macrophages. It was also possible that a small number of monocyte/macrophage precursors contaminated the starting CD15+CD14- cell population. The yield in culture of CD15+CD14- cells with M-CSF and neutrophilic features of a marginal number of the surviving cells could conceivably exclude this possibility.
Our observation that postmitotic neutrophils could generate macrophages raises the issue of developmental origin of human macrophages and may represent another developmental pathway from hematopoietic stem cells toward macrophages. However, it is unclear whether such a neutrophil-to-macrophage lineage switch occurs under physiologic conditions. Such a lineage switch may occur under specified conditions, such as inflammation, because GM-CSF, TNF-α, IFN-γ, IL-4, and M-CSF are inflammatory cytokines. Oehler et al46 demonstrate that neutrophil granulocyte-committed cells acquire dendritic cell features in the presence of GM-CSF, IL-4, and TNF-α. Our results indicated that when CD15+CD14- cells were cultured with GM-CSF, TNF-α, IFN-γ, and IL-4, the resultant cells exhibited a partial appearance of macrophages. IFN-γ may play a crucial role in the conversion of neutrophils into the macrophage lineage. In addition, it seems that neutrophils are capable of generating more types of mature cells than is generally recognized. Further studies on the reprogramming of already differentiated cells into other cell types are expected to yield new insights into events related to human hematopoiesis.
Prepublished online as Blood First Edition Paper, December 30, 2003; DOI 10.1182/blood-2003-08-2742.
Supported in part by grants from the Japan Society for the Promotion of Science.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank M. Ohara (Fukuoka) for critical comments and language assistance.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal