Abstract
A missense mutation, FV-Ile359Thr (FV Liverpool), associated with thrombosis has recently been described. This mutation creates an additional potential N-linked glycosylation site (Asn-X-Ser/Thr) in factor V (FV) at Asn357 that could interfere with secretion and/or protein interactions. To investigate the molecular pathology of FV-Ile359Thr, the mutation was created by site-directed mutagenesis and expressed together with other mutations that could help explain the phenotype (FV-Arg306Gln/Ile359Thr/Arg679Gln, FV-Ile359Thr/Arg506Gln/Arg679Gln, and FV-Asn357Gln/Ile359Thr). The FV-Ile359Thr was secreted normally and had full procoagulant activity. Western blot analysis showed that FV-Ile359Thr migrated more slowly, while the FV-Asn357Gln/Ile359Thr was indistinguishable from FV-wild type (FV-WT), indicating that FV-Ile359Thr was expressed with an additional carbohydrate chain. Activated protein C (APC)-mediated inactivation in an FVa degradation assay showed that the Ile359Thr mutation significantly reduced the cleavage at Arg306 both in the presence and absence of protein S, whereas the cleavage at Arg506 was unaffected. When tested in an FVIIIa degradation assay, the FV-Ile359Thr variant exhibited equally low APC cofactor activity as FV Leiden (FVArg506Gln). In conclusion, the Ile359Thr mutation appears to affect anticoagulation by 2 mechanisms, impeding the APC-mediated down-regulation of the FVa molecule and additionally being a poor APC cofactor for the down-regulation of FVIIIa. These findings explain the association of the FV-Ile359Thr mutation with thrombosis. (Blood. 2004;103:3381-3387)
Introduction
Factor V (FV) is a 330-kDa large glycoprotein circulating in plasma at a concentration of 7 μg/mL.1 FV has a mosaic-like structure; its domain organization A1-A2-B-A3-C1-C2 is homologous to that of factor VIII (FVIII).2,3 After activation through limited proteolysis by either thrombin or factor Xa (FXa), the B domain dissociates from FVa.4,5 FVa, composed of the noncovalently associated heavy and light chain, serves as an essential cofactor to FXa in the activation of prothrombin to thrombin. The procoagulant activity of FVa is down-regulated by activated protein C (APC), which, together with its cofactor protein S, inactivates FVa through proteolytic cleavages at Arg306, Arg506, and Arg679.6 In addition to its procoagulant activity, FV has anticoagulant properties, because it has been shown to function as a cofactor to APC, in synergy with protein S, in the down-regulation of FVIIIa.7,8 Consequently, FV has dual functions in the coagulation system and appears to be important for the natural balance between the procoagulant and anticoagulant systems. The biologic fate of the FV molecule is determined by limited proteolysis. After cleavage by thrombin, FVa serves as an essential cofactor in the procoagulant pathways. Conversely, intact FV has anticoagulant properties, which are enhanced after proteolysis by APC at Arg506.8,9
Molecular abnormalities of FV are associated with either hemorrhagic or thrombotic manifestations. Few natural mutations in FV have been reported. Most of the reported mutations have been shown to result in quantitative FV deficiency, resulting in abnormal bleeding.10,11 Less than a decade ago it was found that many patients with venous thrombosis were resistant to APC, and in 90% of the cases the APC resistance was caused by a missense mutation in FV replacing Arg506 with Gln.12,13 As a consequence, this FV variant, designated FV Leiden, is resistant to APC cleavage at Arg506. This impairs not only the down-regulation of the procoagulant activity of FVa but also the anticoagulant cofactor activity of FV, because the 506 cleavage is needed for full anticoagulant activity to occur.8,9 Two other mutations have been reported that eliminate the APC cleavage site at Arg306 site, Arg306Thr and Arg306Gly.14,15 However, these mutations have not been shown to significantly enhance the risk of thrombosis.16-18
We recently reported the identification and characterization of a family with severe early-onset thrombosis.19 Analysis of the FV gene showed that the thrombotic individuals had a complex genotype that included 2 novel point mutations, 529G>T and 1250T>C (cDNA sequence, first described by Jenny et al20 ), resulting in FV-Glu119Stop and FV-Ile359Thr substitutions inherited on different alleles. Individuals in the kindred with either FV-Glu119Stop or FV-Ile359Thr substitutions alone were asymptomatic. We suggested that the FV-Ile359Thr substitution confers prothrombotic risk and APC resistance but that this is only clinically manifest when coinherited with the FV-Glu119Stop allele. The Ile359Thr substitution creates a new consensus sequence for N-linked glycosylation within the FV heavy chain, and we speculated that this abnormal glycosylation might disrupt APC-mediated proteolysis of the variant FV and FVa. To investigate the molecular pathology of FV-Ile359Thr, the recombinant variant together with other mutations that could help explain the phenotype of the disorder were expressed and functionally characterized.
Materials and methods
Materials
BlnI was purchased from Boerhinger Mannheim (Mannheim, Germany) and Bsu36I from New England Biolabs (Beverly, MA). T4 DNA ligase was from Boehringer Mannheim. Oligonucleotides were from DNA Technologies (Aarhus, Denmark). Double-stranded DNA sequencing kit was from Perkin Elmer (Shelton, CT). BioTrace polyvinylidene fluoride (PVDF) membrane was from Pall (Ann Arbor, MI). Chromogenic substrates S-2366, S-2222, and S-2238 and Coatest APC-resistance kits were kindly provided by Chromogenix (Milan, Italy). Human FXa, human protein S, bovine FX, bovine FIXa (beta), and Pefabloc were from Kordia (Leiden, Netherlands). α-Thrombin was from Haematologics (Essex Junction, VT). Human FVIII (Octonative) was from Pharmacia (Uppsala, Sweden) and hirudin from Sigma (St Louis, MO). Human FV was purified from plasma as previously described21 with minor modifications.22 Monoclonal antibody HV-1 against human FV light chain, ovalbumin, and bovine serum albumin were purchased from Sigma. Monoclonal antibody AHV-5146 against the heavy chain was from Haematologics. Monoclonal antibody MK 54 against protein S, raised by our group, has been previously described.23 Cell culture media (Optimem Glutamax) were from Invitrogen (Carlsbad, CA). Egg extracts of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) and brain extracts of phosphatidylserine (PS) were purchased from Avanti Polar Lipids (Alabaster, AL). Vectastain Elite ABC kit was from Vector Laboratories (Burlingame, CA).
Phospholipid vesicle preparation
The phospholipid stocks were dissolved in chloroform-methanol (9:1 vol/vol), and the concentrations were determined by phosphate analysis.24 Mixtures of the lipids were prepared in chloroform-methanol (9:1 vol/vol) and kept at -20°C. Aliquots were drawn from the stocks and dried in a glass tube under a mild flow of nitrogen. The phospholipids were suspended in 25 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 150 mM NaCl (pH 7.5), vigorously vortexed for 2 minutes, and then sonicated for 10 minutes at room temperature at amplitude 3, using an XL 2020 sonicator (Misonix, Farmingdale, NY). Fresh phospholipid vesicles were prepared every day.
Site-directed mutagenesis
Mutations were introduced into the expression vector pMT2 containing the full-length cDNA of human FV using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). For each mutant, 2 complementary oligonucleotides were used containing the appropriate mutations. The FV-Ile359Thr and the FV-Asn357Gln/Ile359Thr variants were constructed using wild-type (WT) FV cDNA as template. The sense primer for the Ile359Thr variant was 5′-CTCAAACCAAACTGGAAAACATTATAAGAAAGTTATGTACACACAG-3′, and the sense primer for the Asn357Gln/Ile359Thr was 5′-GGTCTCAGCATTTGGATAATTTCTCACAGCAAACTGGAAAAC-3′ (underlined nucleotides causing the mutations). To ascertain that no other mutation had occurred during the amplification, the 1.5-kilobase (kb) fragments containing the mutations were isolated after cleavage with restriction enzymes BlnI and Bsu36I. These fragments were inserted into the WT template cleaved with the same enzymes. The sequences of the fragments were confirmed by DNA sequencing. The 3 FV variants, FV-Arg506Gln, Arg306Gln/Arg679Gln, and FV-Arg506Gln/Arg679Gln, have been previously described.9,18 The FV-Arg306Gln/Ile359Thr/Arg679Gln variant was constructed using the FV-Arg306Gln/Arg679Gln construct as template. After polymerase chain reaction (PCR), the DNA fragment encoding the Ile359Thr mutation was isolated by BlnI and Bsu36I digestion. The 1.5-kb fragment was used to replace the corresponding fragment in the Arg306Gln/Arg679Gln construct. The replaced fragment encoding the FV-Arg306Gln and the Ile359Thr mutations was subjected to DNA sequence analysis. In addition, the Arg679Gln mutation was also confirmed by DNA sequence analysis. To construct FV-Ile359Thr/Arg506Gln/Arg679Gln, Ile359Thr was combined with Arg506Gln/Arg679Gln by cleaving both FV-Ile359Thr and FV-Arg506Gln/Arg679Gln vector with BlnI and Bsu36I and then ligating the Ile359Thr fragment into the cleaved FV-Arg506Gln/Arg679Gln vector. The resulting construct was then sequenced at the 3 mutated sites to confirm the sequence.
Expression and quantification of recombinant factor V variants
To obtain recombinant protein, COS1 cells were transiently transfected using the diethyl aminoethyl (DEAE)-dextran transfection method, as described25 with the following modifications: To the mixture of DEAE-dextran and DNA, 0.1 mM chloroquine was added. The cells were then incubated with this mixture for 4 hours, before dimethyl sulfoxide (DMSO) shock was performed as reported. The proteins were harvested in serum-free medium (Optimem, Invitrogen) and concentrated in Vivaspin with a molecular weight cutoff of 100 000 (Vivascience, Hannover, Germany). Aliquots were frozen at -80°C. The concentrations of recombinant proteins were determined with both enzyme-linked immunosorbent assay (ELISA) and prothrombinase (PTase) assay, the assays being performed as previously described.9,26
Western blot analysis of recombinant protein
FV was activated with 0.05 mU/mL (0.5 nM) thrombin for 30 minutes at 37°C and then incubated with 0.25 nM APC and 100 nM protein S in the presence of 25 μM phospholipids (PS/PE/PC wt/wt/wt 20:20:60). Reduced samples were separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to PVDF membranes, and detected using AHV-5146, a monoclonal antibody reacting with an epitope in the 306-506 fragment of heavy chain of human FVa. To develop the Western blots, Vectastain Elite ABC kit was used according to the manufacturer's instructions.
Determination of apparent Kd of FXa for FVa using the PTase assay
The formation of membrane-bound FXa-FVa complexes was measured by determining the rates of prothrombin activation in the presence of phospholipid vesicles at increasing concentrations of FXa and a fixed concentration of FVa as previously described.26 In brief, the activated FV variants were diluted to 50 pM, and increasing concentrations of FXa (0.1-50 000 pM) and phospholipids vesicles (50 μM 10:90 PS/PC) were added, and the mixtures were incubated for 4 minutes at 37°C. Thrombin generation was started by the addition of preheated (to 37°C) prothrombin (0.5 μM) and allowed to continue for 1 minute before the reaction was stopped by dilution in ethylenediaminetetraacetic acid (EDTA)-containing buffer and the amount of formed thrombin being measured by synthetic substrate analysis. The apparent dissociation constant (Kd) for the thrombin-activated FV variants was obtained from plots of the rate of thrombin generation as a function of the FXa concentration.
Inactivation of FVa by activated protein C and kinetic analysis
After thrombin activation of the FV variants, hirudin (final concentration, 1 U/mL) was added in some of the experiments to stop the activation reaction (the figure legends indicate when hirudin was used). Time courses of APC-mediated FVa inactivation were followed by determining the loss of FVa cofactor activity as a function of time.18,27 FVa variants at a concentration of 0.8 nM were incubated with APC (final concentration, 0.025 nM or 0.05 nM) in the absence or presence of 100 nM protein S and 25 μM phospholipid vesicles (PS/PE/PC wt/wt/wt 20:20:60). The remaining FVa cofactor activity was determined as previously described.18 To calculate apparent second-order rates for FVa cleavage, the inactivation curves obtained in the FVa inactivation assay were fitted to an equation as previously reported27 with the following modification due to the fact that only one cleavage occurs in these FV variants. For calculation of Arg506 cleavage, the time curves obtained for FVa-Arg306Gln/Ile359Thr/Arg679Gln and FVa-Arg306Gln/Arg679Gln were fitted to the following equation:
Vat represents the FVa activity at time t, Va0 is FVa activity at time 0, B is the remaining FVa activity after Arg506 cleavage, and k506 is the rate constant of cleavage at Arg506.
For calculation of Arg306 cleavage, the following equation was used:
Vat is the cofactor activity determined at time t, Va0 is the cofactor activity determined before APC is added, and k306 is the rate constant of cleavage at position Arg306. This equation was fitted to the time curve of degradation of FVa-Ile359Thr/Arg506Gln/Arg679Gln and FVa-Arg506Gln/Arg679Gln.
The use of this equation requires that the inactivation curves are independent of FVa concentration and that the rates are linear for varying APC concentration; thus, control experiments were performed, and the inactivation curves were found to fulfill these criteria.
APC resistance testing of plasma containing recombinant FV variants
APC sensitivity ratios were measured using Coatest APC-resistance kit (Chromogenix), as described previously.18 Recombinant FV variants were added to FV-depleted plasma (Biopool, Ventura, CA) to a final concentration of 1.2 nM. The FV-deficient plasma was supplemented with 160 nM human protein S because we observed that some FV-deficient plasmas were low in protein S activity. The samples were then run according to the manufacturer's instructions. The APC sensitivity ratios were also tested with low concentrations of protein S (20 nM). Prior to theAPC resistance test, the FV-deficient plasma was depleted of protein S by subjecting the plasma to a Hi-trap column (Pharmacia), coupled with MK 54, a monoclonal antibody, against human protein S. The protein S concentration of the FV-deficient plasma treated in this way was undetectable. Finally, plasma-purified protein S was added to a final concentration of 20 nM.
APC cofactor activity of recombinant FV variants
The APC cofactor activities of the recombinant FV variants were measured in the FVIIIa degradation assay as described after some modifications.18 For the FV titration, APC (final concentration, 5 nM) and protein S (final concentration, 5 nM) were mixed with increasing concentrations of FV (0.5-1.3 nM) in total volumes of 45 μL. A reaction mix of FVIIIa-FIXa complex and phospholipids was then incubated with the APC-protein S-FV mixture for 2.5 minutes before FXa generation was started by addition of FX, as described.18
Results
Expression and procoagulant activity of the FV-Ile359Thr variant
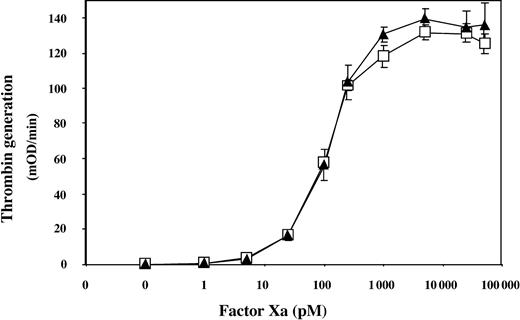
The FV-Ile359Thr construct was generated together with 3 other constructs, FV-Arg306Gln/Ile359Thr/Arg679Gln, FV-Ile359Thr/Arg506Gln/Arg679Gln, and FV-Asn357Gln/Ile359Thr, which were expressed together with FV-WT, FV-Arg506Gln, FV-Arg306Gln/Arg679Gln, and FV-Arg506Gln/Arg679Gln in COS1 cells. The concentrations of all FV variants were determined using both ELISA and PTase assay, and the expression levels were found to be similar (data not shown). To elucidate if FV-Ile359Thr expressed full procoagulant activity, the ability of the thrombin-activated FVa-Ile359Thr variant to support prothrombin activation was investigated at increasing concentrations of FXa (Figure 1). Recombinant FVa-Ile359Thr appeared to interact with FXa in an identical manner to recombinant FVa-WT, with an apparent Kd of 0.2 nM, indicating that FVa-Ile359Thr expressed full procoagulant activity.
Assessing the procoagulant activity of the FVa-Ile359Thr by FXa titration in the PTase assay. The recombinant FV variants (1.25 nM) were incubated with thrombin for 10 minutes at 37°C. After dilution (50 pM final concentration) the FV variants were incubated for 4 minutes with FXa (1-48 000 pM) and 50 μM phospholipids (PS/PC 10:90) at 37°C. Thrombin generation was started by the addition of prothrombin (0.5 μM). After 1 minute, the reactions were stopped by dilution with ice-cold EDTA buffer. The generated thrombin was determined with the chromogenic substrate S-2238. FVa-Ile359Thr, ▴; FVa-WT, □. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
Assessing the procoagulant activity of the FVa-Ile359Thr by FXa titration in the PTase assay. The recombinant FV variants (1.25 nM) were incubated with thrombin for 10 minutes at 37°C. After dilution (50 pM final concentration) the FV variants were incubated for 4 minutes with FXa (1-48 000 pM) and 50 μM phospholipids (PS/PC 10:90) at 37°C. Thrombin generation was started by the addition of prothrombin (0.5 μM). After 1 minute, the reactions were stopped by dilution with ice-cold EDTA buffer. The generated thrombin was determined with the chromogenic substrate S-2238. FVa-Ile359Thr, ▴; FVa-WT, □. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
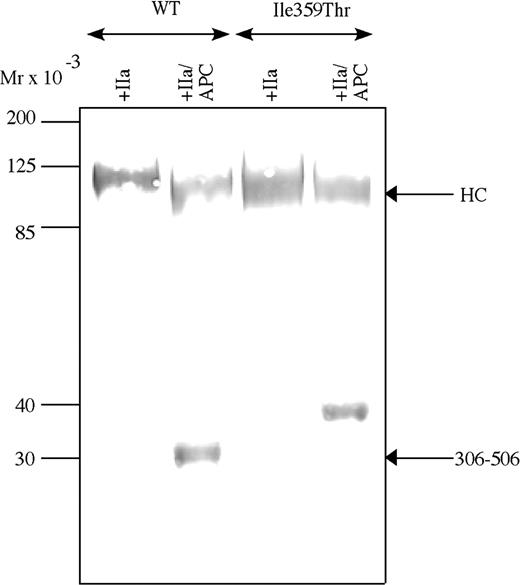
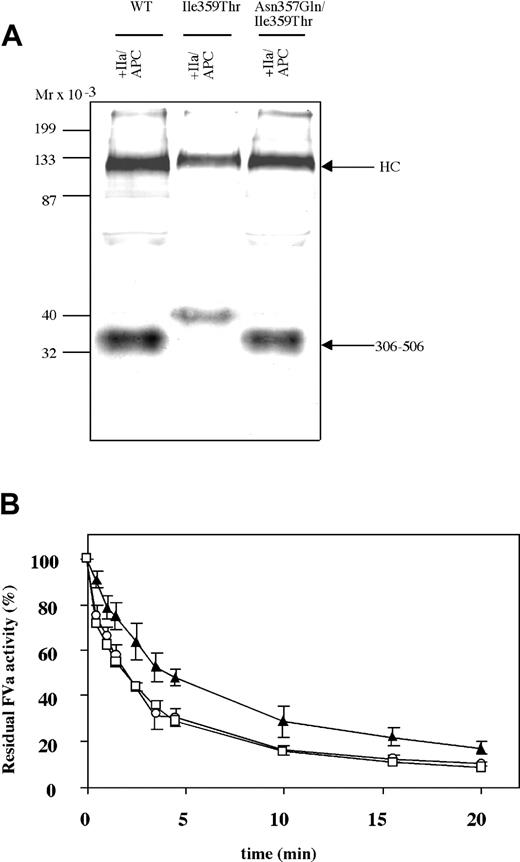
To investigate the thrombin pattern and APC cleavage pattern of FV-Ile359Thr and to elucidate if the point mutation resulted in an extra N-linked glycosylation, the FV-Ile359Thr was analyzed by Western blotting in parallel with FV-WT (Figure 2) using a monoclonal antibody, AHV-5146, reacting with an epitope in the 306-506 fragment of the heavy chain of FVa. The antibody recognized bands of approximately 105 kDa in activated FVa-Ile359Thr and FVa-WT. APC cleavage of FVa-WT yielded a 30-kDa band recognized by the antibody, corresponding to the fragment generated by cleavages at Arg306 and Arg506. In contrast, APC cleavage of the FV-Ile359Thr resulted in a fragment of elevated molecular weight, approximately 35 kDa. The altered mobility of the 306-506 fragment of FV-Ile359Thr is consistent with glycosylation at Asn357 as a consequence of the introduction of a novel N-linked glycosylation site by the Ile359Thr mutation.
Western blot analysis. Western blot analysis of the APC-cleaved FVa-Ile359Thr FV at a concentration of 2 μg/mL incubated with 0.05 U/mL thrombin for 30 minutes at 37°C and then incubated with 0.25 nM APC and 100 nM protein S in the presence of 25 μM phospholipids (PS/PE/PC 20:20:60). Both FVa and APC-cleaved FVa were analyzed by Western blot (12% SDS-PAGE) under reducing conditions. The FVa was detected using a monoclonal against the heavy chain (AHV-5146), and Vectastain Elite ABC kit was used to develop the Western blots. Mr indicates molecular range.
Western blot analysis. Western blot analysis of the APC-cleaved FVa-Ile359Thr FV at a concentration of 2 μg/mL incubated with 0.05 U/mL thrombin for 30 minutes at 37°C and then incubated with 0.25 nM APC and 100 nM protein S in the presence of 25 μM phospholipids (PS/PE/PC 20:20:60). Both FVa and APC-cleaved FVa were analyzed by Western blot (12% SDS-PAGE) under reducing conditions. The FVa was detected using a monoclonal against the heavy chain (AHV-5146), and Vectastain Elite ABC kit was used to develop the Western blots. Mr indicates molecular range.
APC-mediated cleavage of the 506-507 bond of FVa-Ile359Thr
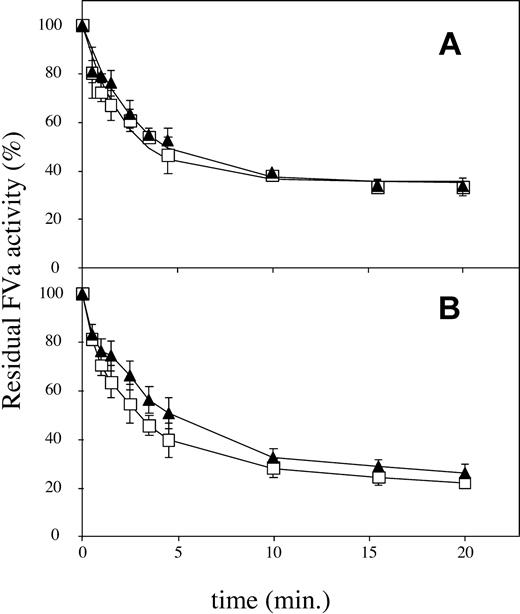
Inactivation of FVa-WT by APC is known to be a biphasic reaction, due to the rapid cleavage at Arg506, yielding a partially active intermediate, followed by the slower cleavage at Arg306 that results in complete loss of FXa cofactor activity.6,27 To investigate if the Ile359Thr mutation had a selective influence on the cleavage at Arg506, the FVa-Arg306Gln/Ile359Thr/Arg679Gln and the FVa-Arg306Gln/Arg679Gln variants were inactivated in parallel over time (Figure 3A). These 2 variants can only be cleaved at Arg506, because the other 2 cleavage sites are eliminated by mutagenesis. No significant differences in the inactivation rates between the FVa-Arg306Gln/Ile359Thr/Arg679Gln and FVa-Arg306Gln/Arg679Gln were detected. This agreed well with results obtained when the FVa-Ile359Thr variant was inactivated in parallel with FVa-WT in the presence of low amounts of APC. At low concentration of APC, only minor cleavage at Arg306 will occur, and thus the loss of activity primarily corresponds to the cleavage at Arg506. The inactivation of FVa-Ile359Thr did not significantly differ from the inactivation of FVa-WT under these conditions (Figure 3B).
APC cleavage at Arg506. Recombinant FV variants (0.8 nM) were incubated with thrombin (0.5 U/mL) for 10 minutes at 37°C, and hirudin (final concentration, 1 U/mL) was then added to inhibit the thrombin. APC was subsequently added together with 10:20:70 PS/PE/PC phospholipids (final concentration, 25 μM). Subsamples were withdrawn at different time points, and FVa activity was measured in the PTase assay in the presence of 1 nM FXa, 0.5 μM prothrombin, and 50 μM 10:90 PS/PE phospholipids. The values were normalized to the value of FVa activity at time zero for each reaction. (A) FVa-Arg306Gln/Arg679Gln, □; FVa-Arg306Gln/Ile359Thr/Arg679Gln, ▴. Final APC concentration was 0.025 nM. (B) FVa-WT, □; FVa-Ile359Thr, ▴. Final APC concentration was 0.033 nM. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
APC cleavage at Arg506. Recombinant FV variants (0.8 nM) were incubated with thrombin (0.5 U/mL) for 10 minutes at 37°C, and hirudin (final concentration, 1 U/mL) was then added to inhibit the thrombin. APC was subsequently added together with 10:20:70 PS/PE/PC phospholipids (final concentration, 25 μM). Subsamples were withdrawn at different time points, and FVa activity was measured in the PTase assay in the presence of 1 nM FXa, 0.5 μM prothrombin, and 50 μM 10:90 PS/PE phospholipids. The values were normalized to the value of FVa activity at time zero for each reaction. (A) FVa-Arg306Gln/Arg679Gln, □; FVa-Arg306Gln/Ile359Thr/Arg679Gln, ▴. Final APC concentration was 0.025 nM. (B) FVa-WT, □; FVa-Ile359Thr, ▴. Final APC concentration was 0.033 nM. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
APC-mediated cleavage of the 306-307 bond of FVa-Ile359Thr
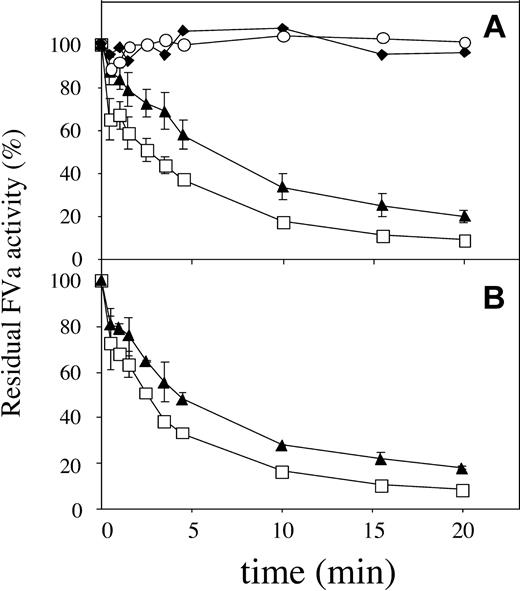
The influence of the FV-Ile359Thr mutation on the Arg306 cleavage was analyzed by comparing the inactivation of FV variants that could only be cleaved at the Arg306 site (FV-Ile359Thr/Arg506Gln/679Gln and FV-Arg506Gln/679Gln). The inactivation reactions were performed at 0.05 nM APC in the presence of protein S and at 1.3 nM APC in the absence of protein S. FVa-Ile359Thr/Arg506Gln/Arg679Gln was inactivated significantly more slowly than FVa-Arg506Gln/Arg679Gln both in the absence and in the presence of protein S (Figure 4A). Using these experimental data, apparent second-order rate constants were calculated for the Arg306 cleavage. In the absence of protein S, the rate constants were 1.4 × 106 M-1s-1 and 3.5 × 106 M-1s-1 for FVa-Ile359Thr/Arg506Gln/Arg679Gln and FVa-Arg506Gln/Arg679Gln, respectively. In the presence of protein S, the rate constants were 4.9 × 107 M-1s-1 and 9.2 × 107 M-1s-1 for FVa-Ile359Thr/Arg506Gln/Arg679Gln and FVa-Arg506Gln/Arg679Gln, respectively. These results suggest that the Ile359Thr mutation resulted in a 2-fold specific reduction of the rate of Arg306 cleavage.
APC cleavage of Arg306. Recombinant FV variants (0.8 nM) were activated with thrombin as described in Figure 3. APC and 10:20:70 PS/PE/PC phospholipids (final concentration, 25 μM) were then added with or without protein S. (A) In the absence of protein S. Final APC concentration was 1.3 nM. FVa-Arg506Gln/Arg679Gln, □; FVa-Ile359Thr/Arg506Gln/Arg679Gln, ▴; controls without APC, FVa-Arg506Gln/Arg679Gln, ○; FVa-Ile359Thr/Arg506Gln/Arg679Gln,  . (B) APC degradation in the presence of protein S (final concentration, 100 nM). Final concentration of APC was 0.05 nM. FVa-Arg506Gln/Arg679Gln, □; FVa-Ile359Thr/Arg506Gln/Arg679Gln, ▴.
. (B) APC degradation in the presence of protein S (final concentration, 100 nM). Final concentration of APC was 0.05 nM. FVa-Arg506Gln/Arg679Gln, □; FVa-Ile359Thr/Arg506Gln/Arg679Gln, ▴.
APC cleavage of Arg306. Recombinant FV variants (0.8 nM) were activated with thrombin as described in Figure 3. APC and 10:20:70 PS/PE/PC phospholipids (final concentration, 25 μM) were then added with or without protein S. (A) In the absence of protein S. Final APC concentration was 1.3 nM. FVa-Arg506Gln/Arg679Gln, □; FVa-Ile359Thr/Arg506Gln/Arg679Gln, ▴; controls without APC, FVa-Arg506Gln/Arg679Gln, ○; FVa-Ile359Thr/Arg506Gln/Arg679Gln,  . (B) APC degradation in the presence of protein S (final concentration, 100 nM). Final concentration of APC was 0.05 nM. FVa-Arg506Gln/Arg679Gln, □; FVa-Ile359Thr/Arg506Gln/Arg679Gln, ▴.
. (B) APC degradation in the presence of protein S (final concentration, 100 nM). Final concentration of APC was 0.05 nM. FVa-Arg506Gln/Arg679Gln, □; FVa-Ile359Thr/Arg506Gln/Arg679Gln, ▴.
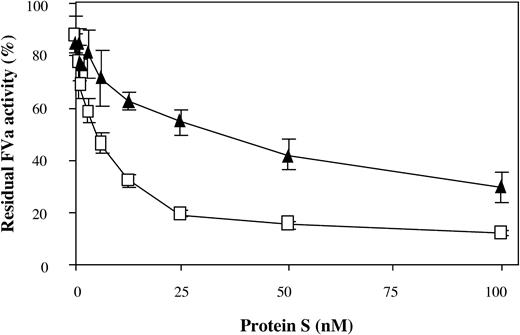
The experiments investigating the rate of Arg306 cleavage in the presence of protein S were performed at physiologic concentrations of protein S. To elucidate whether the mutant FVa variants responded differently than FVa-WT, the protein S concentrations were varied in the APC-mediated inactivation (Figure 5). The FVa variant carrying the Ile359Thr mutation (FVa-Ile359Thr/Arg506Gln/Arg679Gln) was found to require higher levels of protein S for efficient inactivation than FVa without the Ile359Thr mutation, indicating that the attached carbohydrate could possibly interfere with the protein S function.
The effect of increasing protein S concentrations on the APC cleavage at Arg306. Recombinant FV variants (0.8 nM) were activated with thrombin. APC (0.05 nM) was added to the reaction mixture containing protein S (0 to 100 nM) and 10:20:70 wt/wt/wt PS/PE/PC phospholipids (25 μM). The inactivation reactions were stopped after 10 minutes by dilution, and activity was measured as described in the legend to Figure 3. The values were normalized to the value of FVa activity at time zero for each reaction. FVa-Arg506Gln/Arg679Gln, □; FVa-Ile359Thr/Arg506Gln/Arg679Gln, ▴. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
The effect of increasing protein S concentrations on the APC cleavage at Arg306. Recombinant FV variants (0.8 nM) were activated with thrombin. APC (0.05 nM) was added to the reaction mixture containing protein S (0 to 100 nM) and 10:20:70 wt/wt/wt PS/PE/PC phospholipids (25 μM). The inactivation reactions were stopped after 10 minutes by dilution, and activity was measured as described in the legend to Figure 3. The values were normalized to the value of FVa activity at time zero for each reaction. FVa-Arg506Gln/Arg679Gln, □; FVa-Ile359Thr/Arg506Gln/Arg679Gln, ▴. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
Restoring the APC-mediated FVa inactivation by elimination of the glycosylation site
An FV-Ile359Thr variant with the glycosylation site eliminated at 357, FV-Asn357Gln/Ile359Thr, was generated to elucidate if the attenuated APC degradation was specific to the additional glycosylation or if it was caused by the Ile359Thr substitution per se. Western blot analysis following APC cleavage showed the migration of the 306-506 fragment of FVa-Asn357Gln/Ile359Thr and FVa-WT to be indistinguishable (Figure 6A). This observation further supports the interpretation that the elevated apparent molecular weight of the APC fragment 306-506 of the FV-Ile359Thr was as a result of additional glycosylation at Asn357. The FVa-Asn357Gln/Ile359Thr was also tested in the inactivation assay to assess if the APC degradation of FVa was restored when glycosylation at Asn357 was prevented by the Asn357Gln substitution while in the context of the Ile359Thr mutation (Figure 6B). The results indicated that FVa-Asn357Gln/Ile359Thr was inactivated as efficiently as FVa-WT. Furthermore, the APC-mediated inactivation of the FVa-Asn357Gln/Ile359Thr was significantly increased as compared with that of the FVa-Ile359Thr. Taken together, these results suggest that the impaired inactivation of FVa-Ile359Thr is due to additional glycosylation at Asn357 that occurs as a result of the Ile359Thr substitution.
APC-mediated inactivation of FVa-Asn357Gln/Ile359Thr. (A) The recombinant FV variants (2 μg/mL) were incubated with thrombin (0.05 U/mL) for 30 minutes at 37°C and then incubated with 0.25 nM APC and 100 nM protein S in the presence of 25 μM phospholipids (PS/PE/PC 20:20:60). Both FVa and APC-cleaved FVa were analyzed by Western blot (12% SDS-PAGE) under reducing conditions. The FVa was detected using a monoclonal against the heavy chain (AHV-5146), and Vectastain Elite ABC kit was used for detection. (B) The FV variants (0.8 nM) were incubated with thrombin (0.5 U/mL) for 10 minutes at 37°C. APC was subsequently added (final concentration, 0.050 nM) together with protein S (final concentration, 100 nM) and 10:20:70 PS/PE/PC phospholipids (final concentration, 25 μM). Sub-samples were withdrawn at different time points, and activity was measured as in Figure 3. The values were normalized to the value of FVa activity at time zero for each reaction. FVa-WT, □; FVa-Ile359Thr, ▴; FVa-Asn357Gln/Ile359Thr, ○. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
APC-mediated inactivation of FVa-Asn357Gln/Ile359Thr. (A) The recombinant FV variants (2 μg/mL) were incubated with thrombin (0.05 U/mL) for 30 minutes at 37°C and then incubated with 0.25 nM APC and 100 nM protein S in the presence of 25 μM phospholipids (PS/PE/PC 20:20:60). Both FVa and APC-cleaved FVa were analyzed by Western blot (12% SDS-PAGE) under reducing conditions. The FVa was detected using a monoclonal against the heavy chain (AHV-5146), and Vectastain Elite ABC kit was used for detection. (B) The FV variants (0.8 nM) were incubated with thrombin (0.5 U/mL) for 10 minutes at 37°C. APC was subsequently added (final concentration, 0.050 nM) together with protein S (final concentration, 100 nM) and 10:20:70 PS/PE/PC phospholipids (final concentration, 25 μM). Sub-samples were withdrawn at different time points, and activity was measured as in Figure 3. The values were normalized to the value of FVa activity at time zero for each reaction. FVa-WT, □; FVa-Ile359Thr, ▴; FVa-Asn357Gln/Ile359Thr, ○. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
APC resistance testing of the FV-Ile359Thr mutant
To investigate the effect of the Ile359Thr mutation in the APC resistance test, FV-Ile359Thr, FV-WT, and FV-Arg506Gln were added to FV-deficient plasma, which was then subjected to the APC resistance test. In the test, the ratio between the clotting times in the presence and absence of APC was calculated. The ratio obtained in the presence of FV-WT was 2.1, whereas the ratio of FVArg506Gln was 1.3. The FV-Ile359Thr yielded a ratio of 1.9. This is in accordance with the finding that the plasma from the patients with the Ile359Thr mutation yielded an APC ratio intermediate of heterozygous FV Leiden plasma and normal plasma.19 Because the inhibitory effect of the Ile359Thr mutation on the FVa inactivation was more prominent at low concentrations of protein S, the APC sensitivity ratio was also examined at lower concentrations of protein S. At 20 nM protein S, the APC ratios were 1.8, 1.7, and 1.2 for FV-WT, FV-Ile359Thr, and FV-Arg506Gln, respectively, indicating that the concentration of protein S did not significantly affect the APC resistance ratios.
APC cofactor activity of FV-Ile359Thr
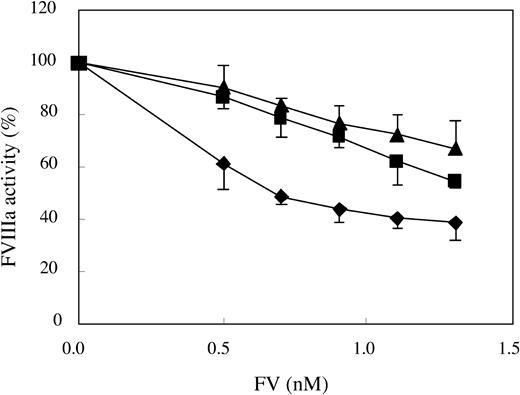
The APC cofactor activity of FV-Ile359Thr was tested in an FVIIIa degradation assay that specifically measures the APC cofactor function of FV. In this assay, a preformed tenase complex (ie, the FVIIIa-FIXa complex assembled on the surface of a phospholipid membrane) was incubated with APC, protein S, and the different FV variants. After the APC-mediated degradation of FVIIIa, the residual ability of the tenase complex to generate FXa was determined. In agreement with earlier reports, FV-WT worked efficiently as APC cofactor, while FV-Arg506Gln exhibited a poor APC cofactor activity.9 Interestingly, the APC cofactor activity of FV-Ile359Thr was equally as low as FV-Arg506Gln in the FV titration (Figure 7). These results suggest that the APC cofactor activity of FV-Ile359Thr is impaired.
Assessing the APC cofactor activity in an FVIIIa degradation assay. Preformed tenase complexes containing phospholipid-bound FIXa and FVIIIa were added to increasing amounts of FV (final concentration, 0.5-1.3 nM) mixed with APC (final concentration, 5 nM) and protein S (final concentration, 5 nM), and the FVIIIa degradation was followed. FV-WT,  ; FV-Ile359Thr, ▪; FV-Arg506Gln, ▴. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
; FV-Ile359Thr, ▪; FV-Arg506Gln, ▴. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
Assessing the APC cofactor activity in an FVIIIa degradation assay. Preformed tenase complexes containing phospholipid-bound FIXa and FVIIIa were added to increasing amounts of FV (final concentration, 0.5-1.3 nM) mixed with APC (final concentration, 5 nM) and protein S (final concentration, 5 nM), and the FVIIIa degradation was followed. FV-WT,  ; FV-Ile359Thr, ▪; FV-Arg506Gln, ▴. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
; FV-Ile359Thr, ▪; FV-Arg506Gln, ▴. Each data point represents the mean of 3 independent experiments performed in duplicate. Error bars represent ± SD.
Discussion
APC resistance, caused by the FV-Arg506Gln mutation, is one of the most common hereditary thrombotic disorders in the Western population. In this report we have characterized the molecular mechanisms by which a missense mutation in FV, Ile359Thr, caused thrombosis. The mutation was first found in 2 brothers with recurring thrombotic episodes. The affected individuals were pseudohomozygous for the Ile359Thr mutation, with a null allele FV-Glu119Stop in addition to the Ile359Thr allele. The mutation Ile359Thr creates an additional potential glycosylation site (Asn-X-Ser/Thr) in FV at position Arg357. To investigate the association of the Ile359Thr mutation with thrombosis, the mutation was introduced into the WT FV cDNA by site-directed mutagenesis together with other FV variants (FV-Arg306Gln/Ile359Thr/Arg679Gln, FV-Ile359Thr/Arg506Gln/Arg679Gln, and FV-Asn357Gln/Ile359Thr). Recombinant proteins were expressed using a well-characterized eukaryotic system,25 and the recombinant FV variants were functionally characterized.
The FV-Ile359Thr appeared to be expressed with an additional glycan as judged by the electrophoretic mobility of FVa-Ile359Thr before and after APC digestion, because the apparent molecular weight of the 306-506 fragment of the FVa-Ile359Thr was increased compared with that of FVa-WT. Thus, the potential glycosylation site at Asn357 created by the Ile359Thr substitution appeared to result in an additional glycosylation. FV-Asn357Gln/Ile359Thr, with the sequence for N-linked glycosylation eliminated, migrated with a mobility indistinguishable from FV-WT, further suggesting that an additional carbohydrate causes the elevated molecular weight in FV-Ile359Thr. The additional carbohydrate chain did not affect the synthesis and secretion of FV-Ile359Thr. This is in agreement with a previous study by us in which we probed FVa with N-linked glycosylations to find regions important for protein binding.28
FV-Ile359Thr had procoagulant activity indistinguishable from FV-WT. However, analysis of the plasma of the original propositus had indicated that FVa degradation was significantly reduced when compared with normal plasma.19 One explanation for the reduced FVa degradation could be a selective reduction of the cleavage at either Arg306 or Arg506; therefore, FV-Ile359Thr variants with 2 of the APC cleavages eliminated were designed to enable selective examination of the effect of the Ile359Thr substitution on each cleavage site in isolation. The results indicated that the Ile359Thr mutation affected inactivation of FVa at Arg306. However, we were not able to detect any significant impairment on the cleavage at Arg506. The rate of the Arg306 cleavage was significantly reduced by the Ile359Thr mutation both in the presence and in the absence of protein S. The inhibitory effect of the Ile359Thr mutation on the Arg306 cleavage was more prominent at low concentrations of protein S. Despite this objective evidence that the Ile359Thr mutation reduces the rate of inactivation of FVa by APC in a purified assay, when examined in the plasma APC resistance (APCR) assay, the APC resistance ratio was only marginally reduced. The plasma APCR assay used in this study has been optimized to demonstrate resistance to APC by the FV-Arg506Gln substitution and may not have sufficient sensitivity to clearly demonstrate APCR due to FV-Ile359Thr.
Little is known about how APC and protein S bind to FV. Based on a peptide inhibition study, it was suggested that protein S could interact with residues 493-506.29 Moreover, residues 1865-1874 in the light chain of FVa have been suggested to be important for APC binding, because peptides encompassing this region were found to inhibit FVa inactivation.30 However, these residues are buried inside the protein core in the 3-dimensional model of FVa, suggesting this region may not have any physiological role.31 Residue 357 is located rather distant from the cleavages at Arg306 and Arg506, which argues against this residue being directly involved in protein S and APC binding. On the other hand, the observed effects appear to be specific to the additional glycosylation, because the nonglycosylated variant, FV-Asn357Gln/Ile359Thr, did not have impaired functional interactions with APC/protein S. This suggests that the Ile359Thr substitution itself does not induce a conformational change but, rather, the large bulky carbohydrate side chains interfere with binding sites located relatively distant from residue Asn357.
FV has been reported to be a cofactor to APC, acting in synergy with protein S, in the inactivation of FVIIIa.7,8 When the APC cofactor activity of FV-Ile359Thr was tested, it was found to be an equally poor APC cofactor as FV-Arg506Gln. This suggests that the APC cofactor activity of FV-Ile359Thr is impaired. The APC-mediated cleavage of Arg506 has been reported to enhance the cofactor activity of FV8,9 ; however, it is not known why the Arg506 cleavage enhances the anticoagulant activity. It has been hypothesized that proteolysis at Arg506 exposes binding sites for APC, protein S, or even FVIIIa.9 The rate of cleavage ofArg506 in FVa-Ile359Thr is normal, and the mechanism by which the APC cofactor activity of the FV Ile359Thr variant is decreased is at present unknown.
Modulation of FV activity appears to be crucially important for the regulation of blood coagulation. After proteolytic processing, FV has the potential to express both procoagulant and anticoagulant activity. The FV molecule is thus important for maintaining the balance between the procoagulant and anticoagulant systems.32,33 This is illustrated by the fact that the Arg506Gln substitution is a well-documented risk factor for thrombosis. Similarly to the Arg506Gln mutation, the Ile359Thr mutation appears to impair both the regulation of FVa and FVIIIa activity. The hypercoagulable state associated with the Ile359Thr mutation emphasizes the importance of balancing both the procoagulant and anticoagulant activities of FVa.
In summary, we have functionally characterized a novel mutation associated with thrombosis. The FV-Ile359Thr is expressed with an additional carbohydrate chain at Asn357 that attenuates down-regulation of FVa. In addition, the anticoagulant APC cofactor function of the FV-Ile359Thr molecule was found to be impaired. As a consequence, FV-Ile359Thr appears to affect anticoagulation by 2 mechanisms: it is a poorer APC substrate impeding the regulation of the FVa molecule, and also the APC cofactor activity is reduced, which results in defective FVIIIa degradation. These findings explain the association of the Ile359Thr mutation with thrombosis.
Supported by Swedish Medical Research Council grant 07143, a Senior Investigator's Award from the Foundation for Strategical Research, the Albert Påhlsson Trust, and the University Hospital, Malmö.
M.S. and E.A.N. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Prepublished online as Blood First Edition Paper, December 24, 2003; DOI 10.1182/blood-2003-06-2092.
We warmly thank Mrs Ing-Marie Persson for expert technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal