Abstract
The natural killer (NK) type of lymphoproliferative disease of granular lymphocytes (LDGL) is associated with the expansion of CD3-, CD16+, and/or CD56+ lymphocytes. We have examined the repertoire of NK receptors expressed on these cells and delineated the functional activity. We found skewed NK receptor expression on patient NK cells. Reactivity to a single anti-killer cell immunoglobulin-like receptor (anti-KIR) antibody was noted in 7 of 13 patients. LDGL patients variably expressed NKp30, NKp44, and NKp46 RNA. In contrast, CD94 and its inhibitory heterodimerization partner NKG2A were homogenously expressed at high levels on these NK cells. Interestingly, these patients expressed a large number of activating KIR receptors by genotype analysis. Semiquantitative reverse transcriptase-polymerase chain reaction (RT-PCR) demonstrated that lower than normal levels of RNA of the inhibitory KIR was present in some patients in contrast to normal NK cells. Consistent with a high level of activating receptors, we found the NK-LDGL cells have potent cytolytic function in both direct and redirected cytotoxicity assays. These results demonstrate that patients with NK-LDGL have an increased activating-to-inhibitory KIR ratio. This altered ratio might induce inappropriate lysis or cytokine production and impact the disease pathogenesis. (Blood. 2004;103:3431-3439)
Introduction
The natural killer (NK) type of lymphoproliferative disease of granular lymphocytes (LDGL) is characterized by the expansion of NK cells that express CD56 and/or CD16 but do not express CD3 or the T-cell receptor (TCR) complex or rearrange TCR genes.1-4 Most cases of LDGL in the United States and Europe follow a chronic clinical course.1,5 Aggressive cases of NK leukemia are more commonly seen in the Far East and are associated with a rapidly progressive clinical course with a median survival of only months. Epstein Barr virus (EBV) has been reported as an etiologic agent of aggressive NK leukemia/lymphoma.6,7
The etiology of LDGL is not known. However, we have found that sera from 67% of LDGL patients have high titer-specific antibody reactivity directed at a 34-amino acid epitope of the envelope protein of human T-cell lymphotropic virus type I (HTLV-I).8 This finding raises the possibility of serologic cross-reactivity to a cellular or retroviral antigen showing some amino acid homology with the transmembrane glycoprotein of HTLV. However, these patients are not infected with any of the known prototypic retroviruses such as HTLV-I, HTLV-II, primate T-lymphotropic virus (PTLV), HIV, or bovine leukemia virus (BLV).9,10 We hypothesize that chronic viral stimulation by a novel HTLV-related retrovirus with homology in the p21e region may contribute to the pathogenesis of this disease.
Multiple NK-associated receptors have now been identified, which might prove useful in the clinical diagnosis of LDGL as well as provide a better understanding of disease pathogenesis. Normal NK cells express 3 types of receptors: C-type lectin receptors (NKG2), immunoglobulin-like receptors (killer cell immunoglobulin-like receptor; KIR), and natural cytotoxic receptors (NCRs).11-14 There are both activating and inhibitory NKG2 and KIR receptors based on the presence of either an ITAM (immunoreceptor tyrosine-based activation motif) or ITIM (immunoreceptor tyrosine-based inhibitory motif) in the cytoplasmic tail, respectively.15-17 Activating receptors recognize their ligand and mediate cytolysis or cytokine production, whereas inhibitory receptors prevent the killing of normal cells and limit inflammatory cytokine production.18,19 The receptors of the three NKG2 family members, NKG2C, NKG2E, and NKG2D, possess activating properties.20-23 NKG2C and NKG2E receptors as well as the inhibitory NKG2A receptor bind the adaptor protein CD94.24,25 The major function of inhibitory KIR and inhibitory NKG2 receptors is the recognition of self-human leukocyte antigen (self-HLA) alleles, thereby limiting the lysis of autologous cells.26,27 NK cells from healthy individuals express between 2 and 8 KIRs per cell and usually express multiple inhibitory KIRs that recognize autologous HLA ligands.28-30 NCR receptors include NKp30, NKp44, and NKp46, which are exclusively found on resting and interleukin 2 (IL-2)-stimulated NK cells but not on T cells.31-33 Activating NK receptors form oligomeric complexes that associate with the transmembrane adaptor proteins, death-associated protein 12 (DAP12), DAP10, CD3ζ, or CD3ζ/FcϵRγ.21,34-39 Through the activating receptors, NK cells mediate target lysis by perforin and granzyme B release.13
Here, we examined the surface expression of several KIRs, CD94, and NKG2 receptors on NK cells from LDGL patients. We found that skewed NK receptor expression occurs on these cells. We also performed DNA- and RNA-based typing studies for the KIR receptors. The RNA expression for NKG2A, NKG2C, NKG2D, and NKG2E and NCRp44, NCRp46, and NCRp30 was also assessed. Lytic assays were performed to address the functional significance of dysregulated NK receptor expression in LDGL.
Patients, materials, and methods
Patients and preparation of peripheral blood
All patients met the clinical criteria of NK-LDGL.40,41 Peripheral blood (∼40 mL of whole blood in heparinized tubes) was obtained from 13 patients with NK-LDGL. All patients had increased total numbers of NK cells. The absolute lymphocyte counts (ALCs) ranged from 1.485 × 109/L to 12.453 × 109/L (mean 6.873 × 109/L; 1485 to 12 453 cells/μL [mean, 6873 cells/μL]) with the percentage of NK cells ranging from 34% to 83%. The average absolute number of NK cells for this patient population was 4.69 × 109/L (4690 cells/μL). In addition to the lymphocytosis, 8 patients had other clinical symptoms such as anemia, neutropenia, splenomegaly, systemic B symptoms, and/or rheumatoid arthritis. One patient had a clinical course characteristic of aggressive NK leukemia (NK-LDGL8) and demonstrated complex clonal cytogenetic abnormalities. No patient was receiving therapy at the time of analysis. Informed consents were signed by all patients to allow the use of their cells for these experiments (institutional review board [IRB] of the University of South Florida). Patient numbers were assigned after accrual into the national LGL leukemia registry initiated at the H. Lee Moffitt Cancer Center and Research Institute.
Normal buffy coats were obtained from the Southwest Florida blood bank. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood of NK-LDGL patients or buffy coats by Ficoll-Hypaque gradient separation, as previously described.42 CD56+ and/or CD16+ cells from buffy coats of healthy individuals were enriched using a negative selection process, RosetteSep, as recommended by the manufacturer (Stem Cell Separation Systems, Vancouver, BC, Canada). We found that 80% to 90% of the cells isolated using this procedure stained positively for CD56 and/or CD16 with fewer than 1% CD3+ T cells and fewer than 5% CD19+ B cells (data not shown).
Surface phenotyping by flow cytometry
Paired healthy and patient NK cells were analyzed by 2-color flow cytometry using CyChrome-conjugated anti-CD56 (clone B159; BD Biosciences, San Diego, CA) or anti-CD16-fluorescein isothiocyanate (FITC)-conjugated antibodies in combination with the following phycoerythrin (PE)-conjugated antibodies: anti-CD158a (KIR2DL1, KIR2DS1), anti-CD158b (KIR2DL2, KIR2DL3, KIR2DS2), anti-NKB1 (KIR3DL1), anti-KARp50 (KIR2DS4), and anti-NKG2A, which were all obtained from BD Biosciences. A purified antibody was used to stain NKG2D (0.5 μg; R&D Systems, Minneapolis, MN) and KIR2DL4 (0.5 μg; provided by Dr Eric Long, National Institute of Allergy and Infectious Diseases [NIAID], Rockville, MD) followed by rat antimouse immunoglobulin (Ig)-conjugated PE secondary antibody. Data acquisition and analysis were carried out on a FACScan flow cytometer (BD Biosciences) using the Cell Quest software program (BD Immunocytometry Systems, San Diego, CA). The FACScan flow cytometer was equipped with a 488-nm laser excitation source. Forward scatter (FSC) and side scatter (SSC) plots were used to identify the lymphocyte population. Measurements for FITC were carried out using the fluorescence 1 (FL1) detector (530/30 nm), PE was detected using the FL2 detector (585/42 nm), and a long pass filter was used to detect CyChrome (FL3, ≥ 640 nm). A total of 1 × 105 total events were collected for each data set, and the percentage of positive cells was determined. We also determined the relative number of receptors per cell based on the value obtained for the mean fluorescence intensity (MFI). Determination of NK receptor expression on CD8+ T cells was carried out in some LDGL patients by 3-color analysis with anti-CD8 or anti-CD3-CyChrome, anti-NK receptor antibodies, and anti-CD16-FITC. In patients whose cells expressed CD56, we used anti-CD56-CyChrome, anti-NK receptor antibodies conjugated to PE, and anti-CD8 or anti-CD3-FITC.
RNA-based PCR typing
Total RNA was prepared from 4 × 107 PBMCs using TRIzol, as directed by the manufacturer (Invitrogen, Carlsbad, CA). To prepare cDNA, the ThermoScript reverse transcriptase-polymerase chain reaction (RT-PCR) System (Invitrogen) was used to prepare a 60-μL reaction containing the following reagents: 15 μg total RNA, 1 mM deoxynucleoside triphosphate (dNTP) mix, 2.5 μM oligo (dT)20, 1 × cDNA synthesis buffer, 5 mM dithiothreitol (DTT), 40 units RNAse inhibitor, and 15 units of ThermoScript RT. The cDNA reaction was incubated in a Mastercycler Gradient Thermal Cycler (Eppendorf, Westbury, NY) using the following parameters: 65°C for 5 minutes (RNA, Oligo (dT)20, and dNTP mix only), 50°C for 60 minutes (after the addition of the remaining reagents), and 85°C for 5 minutes.
Amplification of cDNA was then carried out with 2.5 μL of cDNA for the analysis of NKG and NCR gene expression using primer sequences as previously described.43-46 NKG- and NCR-specific cDNA were amplified in a total volume of 25 μL with the addition of the following reagents: Hot Star Taq master mix from Qiagen (Valencia, CA; 1 × PCR buffer containing 1.5 mM MgCl2, 200 μM each dNTP, 1.25 units HotStar Taq DNA polymerase), 0.5 μM NKG and NCR primers, and 0.1 μM β-actin primers. β-Actin primers were excluded from NKG2C, NKp30, NKp44, and NKp46 samples.
The NKG and β-actin amplicons were amplified in a 96-well PCR plate using the following conditions: HotStar Taq activation for 15 minutes at 95°C; 5 cycles of cDNA denaturation for 60 seconds at 95°C, primer annealing for 60 seconds at 62°C, primer extension for 45 seconds at 72°C; 25 to 30 cycles of cDNA denaturation for 30 seconds at 95°C, primer annealing for 45 seconds at 60°C, primer extension for 45 seconds at 72°C; and primer extension for 5 minutes at 72°C. Identical conditions were used to amplify β-actin for semiquantitative RT-PCR except that the total cycles were reduced to 16.
The NKp30 and NKp46 amplicons were amplified in a 96-well PCR plate using the following conditions: HotStar Taq activation for 15 minutes at 95°C, 30 cycles of cDNA denaturation for 30 seconds at 95°C, primer annealing for 30 seconds at 60°C, primer extension for 30 seconds at 72°C, and primer extension for 7 minutes at 72°C. Identical conditions were used to amplify NKp44 except that the primer annealing temperature was increased to 65°C. Products were analyzed using the entire 25-μL sample, which was electrophoresed through a 3% agarose gel containing ethidium bromide. The Alpha Imager 1220 Documentation and Analysis System (Alpha Innotech, San Leandro, CA) was used for data analysis.
Semiquantitative RNA analysis was also performed on healthy donors and patients. RNA from 3 healthy donors was used as standards for this assay in dilutions of 100%, 20%, 4%, and .8% (Figure 3A), and primers for KIR and β-actin control yielded amplicons that were in a linear range using these dilutions. The gels were analyzed by densitometry and a ratio was obtained to β-actin expression. Ratios of all reactions were then determined relative to healthy donor number 1.
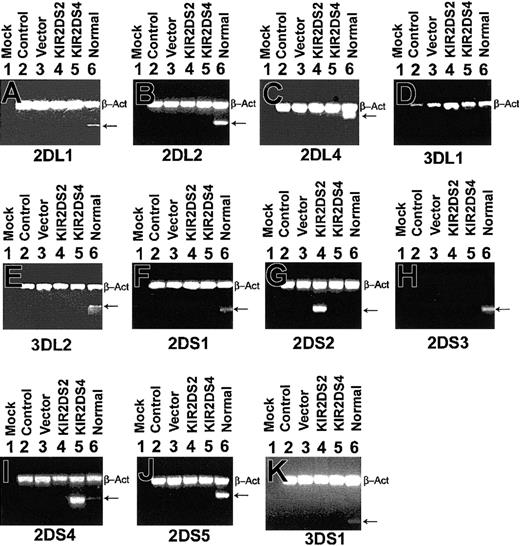
Specific amplification of KIRs in K562. RNA was amplified using primers specific for KIR2DL1, KIR2DL2, KIR2DL4, KIR3DL1, KIR3DL2, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, and KIR3DS1 from K562 cells that were untransfected (Control, lane 2), pcDNA3 vector control transfected (Vector, lane 3), pcDNA3-KIR2DS2 transfected (KIR2DS2, lane 4), and pcDNA3-KIR2DS4 transfected (KIR2DS4, lane 5). Normal 1 was used as a positive control for all KIR primers (lane 6) but was found to be negative for KIR3DL1 (D). β-Actin primers were included to ensure that a similar amount of cDNA was present in each sample (top band, β-Act). Reaction with water was included as a negative control for each set of primers (Mock, lane 1).
Specific amplification of KIRs in K562. RNA was amplified using primers specific for KIR2DL1, KIR2DL2, KIR2DL4, KIR3DL1, KIR3DL2, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, and KIR3DS1 from K562 cells that were untransfected (Control, lane 2), pcDNA3 vector control transfected (Vector, lane 3), pcDNA3-KIR2DS2 transfected (KIR2DS2, lane 4), and pcDNA3-KIR2DS4 transfected (KIR2DS4, lane 5). Normal 1 was used as a positive control for all KIR primers (lane 6) but was found to be negative for KIR3DL1 (D). β-Actin primers were included to ensure that a similar amount of cDNA was present in each sample (top band, β-Act). Reaction with water was included as a negative control for each set of primers (Mock, lane 1).
NK receptor genotyping
Genomic DNA was isolated from PBMCs of patients with NK-LDGL using the Wizard Genomic DNA purification kit (Promega, Madison, WI). DNA amplification was carried out for 100 ng of DNA for KIR genotyping and was amplified in a total volume of 25 μL with the addition of the following reagents: HotStar Taq master mix from Qiagen (1 × PCR buffer containing 1.5 mM MgCl2, 200 μM each dNTP, 1.25 units HotStar Taq DNA polymerase), 0.5 μM KIR primers, except KIR3DL1 and KIR2DS4, which are at 1 μM.43,45,46
Cytotoxicity assay
Cytotoxicity assays were performed as previously described.47 Briefly, a 4-hour 51Cr-release assay was performed using K562 as targets for PBMCs and enriched NK cells from healthy donors or from patients with NK-LDGL at effector-target (E/T) ratios of 50:1, 25:1, 12:1, and 6:1. The percentage of specific 51Cr release was determined by the following equation: ([experimental cpm - spontaneous cpm]/total cpm incorporated) × 100, where cpm indicates count per minute. All experiments were performed in triplicate, and the standard deviation (SD) of all assays was calculated and was typically approximately 5% of the mean or less. Redirected killing assays were performed using the FcγR+ murine mastocytoma (P815) target cell line in 4-hour 51Cr-release assays. Isotype control or purified antibodies to CD158a, CD158b, NKB1, NKG2A, CD94, and NKG2D were added at a concentration of 1 μg/mL in 96-well plates to negative-selected enriched healthy NK cells and NK-LDGLs 30 minutes prior to the addition of labeled P815 cells at a 50:1 E/T ratio. The cells were harvested and analyzed for lysis as described above for determining the percentage of specific 51Cr release.
Results
Skewed NK receptor expression in patients with NK-LDGL
We examined the expression of KIRs on NK cells from 4 healthy donors (CD56+) and NK cells from 13 patients with NK-LDGL (CD56+ and/or CD16+). The KIR antibodies used were as follows: CD158a, CD158b, NKB1, KARp50, and KIR2DL4. KIRs recognized by these antibodies are as follows: KIR2DS1 and KIR2DL1 (anti-CD158a); KIR2DL2, KIR2DL3, and KIR2DS2 (anti-CD158b); KIR3DL1 (anti-NKB1); and KIR2DS4 (anti-KARp50). KIR2DS1, KIR2DS2, and KIR2DS4 are activating receptors and KIR2DL1, KIR2DL2, KIR2DL3, and KIR3DL1 are inhibitory receptors.
NK cells from all 4 healthy donors reacted to all of the KIR antibodies used (Table 1). Dot plot flow cytometry data for one of the healthy donors are shown in Figure 1 (left column). The percentage of KIR-positive cells for the NK cells from this donor ranged from 13% to 47% (double-positive for KIR and CD56). Dot plot flow cytometry results are also shown for 2 exemplary patients, NK-LDGL3 and NK-LDGL6 (Figure 1; middle and right columns, respectively). NK cells from NK-LDGL3 had limited reactivity to each of the KIR antibodies used. Results similar to NK-LDGL3 were observed for NK-LDGL1, NK-LDGL4, and NK-LDGL7 (Table 1). In contrast, NK cells from NK-LDGL6 homogenously reacted to a single KIR antibody (anti-CD158b). Seven NK-LDGL patients displayed this immunophenotype, where more than 50% of total NK cells were homogenously positive for a single KIR antibody (Table 1; *, NK-LDGL2, NK-LDGL5, NK-LDGL6, NK-LDGL8, NK-LDGL9, NK-LDGL10, and NKLDGL12). Of note, a homogenously positive pattern of KIR antibody reactivity (NKB1) was observed in the one patient examined with aggressive NK-LGL leukemia (NK-LDGL8).
KIR immunophenotype of CD56+ or CD16+ NK cells in patients with NK-LDGL
. | CD158a, 2DS1/2DL1, % . | CD158b, 2DS2/2DL2/2DL3, % . | NKB1, 3DL1, % . | KARp50, 2DS4, % . | KIR2DL4, % . | CD94 . | . | NKG2A . | . | NKG2D . | . | % Total CD56+ or CD16+ NK cells† . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibodies . | . | . | . | . | . | % . | MFI . | % . | MFI . | % . | MFI . | . | |||
| NKLDGL1 | 0 | 0.6 | 1.3 | 1.3 | ND | 38 | 392 | 0 | 0 | 66 | 200 | 66 | |||
| NKLDGL2 | 0.4 | 0.8 | 0 | 4 | 21* | 40 | 505 | 42 | 668 | 86 | 204 | 42 | |||
| NKLDGL3 | 0.2 | 3 | 0 | 0 | 1.5 | 45 | 1225 | 46 | 733 | 44 | 35 | 48 | |||
| NKLDGL4 | 0.1 | 0.1 | 0.1 | 0.2 | ND | 74 | 400 | 75 | 32 | ND | ND | 74 | |||
| NKDLGL5 | 76* | 4 | 0 | 0.8 | 19 | 78 | 519 | 78 | 234 | 68 | 135 | 78 | |||
| NKLDGL6 | 0.7 | 80* | 0.3 | 4 | ND | 83 | 1211 | 83 | 161 | 74 | 17 | 83 | |||
| NKLDGL7 | 0.5 | 0.8 | 1.6 | 0.6 | ND | 75 | 304 | 73 | 86 | ND | ND | 75 | |||
| NKLDGL8 | 0.6 | 0.7 | 92* | 0.2 | 4 | 94 | 757 | 3 | 96 | 77 | 20 | 94 | |||
| NKLDGL9 | 0.1 | 3.6 | 36* | 0 | ND | 44 | 198 | 42 | 185 | ND | ND | 45 | |||
| NKLDGL10 | 3 | 0.2 | 0.2 | 41* | ND | 55 | 462 | 58 | 176 | ND | ND | 58 | |||
| NKLDGL11 | 45 | 51 | 26 | 0 | ND | 35 | 124 | 2 | 170 | ND | ND | 60 | |||
| NKLDGL12 | 0.8 | 2.5 | 19 | 0.8 | 81* | 83 | 403 | 83 | 259 | 81 | 249 | 84 | |||
| NKLDGL13 | 8.7 | 12 | 15.6 | 0 | ND | 19 | 140 | 6 | 68 | 1.6 | 22 | 79 | |||
| Average | — | — | — | — | — | 59 ± 23 | 511 ± 356 | 45 ± 33 | 221 ± 226 | 62 ± 28 | 110 ± 97 | 68 ± 16 | |||
| Normal 1 | 1.8 | 2.4 | 0.1 | 0.5 | 1.4 | 5.6 | 200 | 4 | 77 | 1.6 | 28 | 8 | |||
| Normal 2 | 1.4 | 2.8 | 0.6 | 0.4 | 2.2 | 12 | 168 | 5.8 | 88 | 1.5 | 66 | 17 | |||
| Normal 3 | 3.5 | 4 | 0.8 | 0.5 | 3 | 15 | 122 | 9.2 | 70 | 0.3 | 24 | 26 | |||
| Normal 4 | 1.2 | 2.2 | 1.4 | 4.8 | 3.7 | 10 | 170 | 3.7 | 51 | 3 | 25 | 19 | |||
| Average | — | — | — | — | — | 11 ± 4 | 165 ± 32 | 6 ± 3 | 71 ± 16 | 1.6 ± 1 | 36 ± 20 | 18 ± 7 | |||
. | CD158a, 2DS1/2DL1, % . | CD158b, 2DS2/2DL2/2DL3, % . | NKB1, 3DL1, % . | KARp50, 2DS4, % . | KIR2DL4, % . | CD94 . | . | NKG2A . | . | NKG2D . | . | % Total CD56+ or CD16+ NK cells† . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibodies . | . | . | . | . | . | % . | MFI . | % . | MFI . | % . | MFI . | . | |||
| NKLDGL1 | 0 | 0.6 | 1.3 | 1.3 | ND | 38 | 392 | 0 | 0 | 66 | 200 | 66 | |||
| NKLDGL2 | 0.4 | 0.8 | 0 | 4 | 21* | 40 | 505 | 42 | 668 | 86 | 204 | 42 | |||
| NKLDGL3 | 0.2 | 3 | 0 | 0 | 1.5 | 45 | 1225 | 46 | 733 | 44 | 35 | 48 | |||
| NKLDGL4 | 0.1 | 0.1 | 0.1 | 0.2 | ND | 74 | 400 | 75 | 32 | ND | ND | 74 | |||
| NKDLGL5 | 76* | 4 | 0 | 0.8 | 19 | 78 | 519 | 78 | 234 | 68 | 135 | 78 | |||
| NKLDGL6 | 0.7 | 80* | 0.3 | 4 | ND | 83 | 1211 | 83 | 161 | 74 | 17 | 83 | |||
| NKLDGL7 | 0.5 | 0.8 | 1.6 | 0.6 | ND | 75 | 304 | 73 | 86 | ND | ND | 75 | |||
| NKLDGL8 | 0.6 | 0.7 | 92* | 0.2 | 4 | 94 | 757 | 3 | 96 | 77 | 20 | 94 | |||
| NKLDGL9 | 0.1 | 3.6 | 36* | 0 | ND | 44 | 198 | 42 | 185 | ND | ND | 45 | |||
| NKLDGL10 | 3 | 0.2 | 0.2 | 41* | ND | 55 | 462 | 58 | 176 | ND | ND | 58 | |||
| NKLDGL11 | 45 | 51 | 26 | 0 | ND | 35 | 124 | 2 | 170 | ND | ND | 60 | |||
| NKLDGL12 | 0.8 | 2.5 | 19 | 0.8 | 81* | 83 | 403 | 83 | 259 | 81 | 249 | 84 | |||
| NKLDGL13 | 8.7 | 12 | 15.6 | 0 | ND | 19 | 140 | 6 | 68 | 1.6 | 22 | 79 | |||
| Average | — | — | — | — | — | 59 ± 23 | 511 ± 356 | 45 ± 33 | 221 ± 226 | 62 ± 28 | 110 ± 97 | 68 ± 16 | |||
| Normal 1 | 1.8 | 2.4 | 0.1 | 0.5 | 1.4 | 5.6 | 200 | 4 | 77 | 1.6 | 28 | 8 | |||
| Normal 2 | 1.4 | 2.8 | 0.6 | 0.4 | 2.2 | 12 | 168 | 5.8 | 88 | 1.5 | 66 | 17 | |||
| Normal 3 | 3.5 | 4 | 0.8 | 0.5 | 3 | 15 | 122 | 9.2 | 70 | 0.3 | 24 | 26 | |||
| Normal 4 | 1.2 | 2.2 | 1.4 | 4.8 | 3.7 | 10 | 170 | 3.7 | 51 | 3 | 25 | 19 | |||
| Average | — | — | — | — | — | 11 ± 4 | 165 ± 32 | 6 ± 3 | 71 ± 16 | 1.6 ± 1 | 36 ± 20 | 18 ± 7 | |||
KIR expression was determined by flow cytometry on PBMCs. Values indicate the % of CD16+ or CD56+ cells that coexpress KIR. Values shown as % positive = numbers obtained from the top right-hand quadrant of 2-color dot plot flow cytometry graphs.
More than 50% of the NK population has homogenous reactivity to a single KIR antibody.
The number in the top right added to the number in the top left quadrants of dot plot flow cytometry = total number of NK cells.
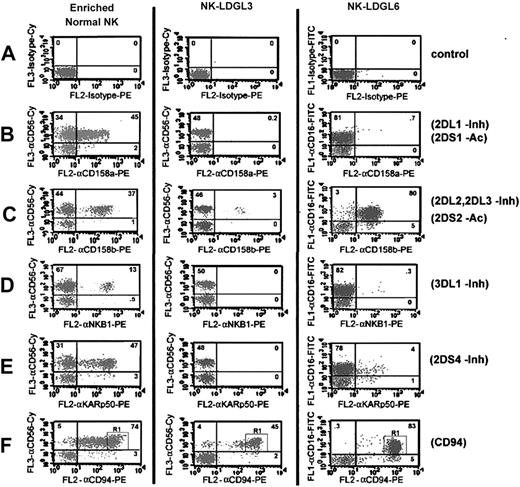
Skewed surface KIR receptor expression in patients with NK-LDGL. Dot plot flow cytometry results for enriched normal NK cells (left column), PBMCs from NK-LDGL3 (middle column), and PBMCs from NK-LDGL6 (right column) after staining with (A) isotype control-CyChrome (y-axis, control for normal NK and NK-LDGL3) and isotype control-FITC (y-axis, control for NK-LDGL6) and/or isotype control-PE (x-axis, control). Two-color analysis is shown for either anti-CD56-CyChrome (NK-LDGL3 and normal NK) or anti-CD16-FITC (NK-LDGL6) with (B) anti-CD158a-PE (KIR2DS1 and KIR2DL1), (C) anti-CD158b-PE (KIR2DL2, KIR2DL3, and KIR2DS2), (D) anti-NKB1-PE (KIR3DL1), (E) anti-KARp50-PE (KIR2DS4), and (F) anti-CD94-PE. The percentage of cells positive in each quadrant is indicated in the top right-hand corner of each dot plot. Adding the numbers in the top left and top right quadrants represents the total number of NK cells (∼48% for NK-LDGL3 and ∼83% for NK-LDGL6). The number in the top right represents the number of NK cells coexpressing the NK receptor. R1 indicates region 1; Inh, inhibitory KIR; and Ac, activating KIR.
Skewed surface KIR receptor expression in patients with NK-LDGL. Dot plot flow cytometry results for enriched normal NK cells (left column), PBMCs from NK-LDGL3 (middle column), and PBMCs from NK-LDGL6 (right column) after staining with (A) isotype control-CyChrome (y-axis, control for normal NK and NK-LDGL3) and isotype control-FITC (y-axis, control for NK-LDGL6) and/or isotype control-PE (x-axis, control). Two-color analysis is shown for either anti-CD56-CyChrome (NK-LDGL3 and normal NK) or anti-CD16-FITC (NK-LDGL6) with (B) anti-CD158a-PE (KIR2DS1 and KIR2DL1), (C) anti-CD158b-PE (KIR2DL2, KIR2DL3, and KIR2DS2), (D) anti-NKB1-PE (KIR3DL1), (E) anti-KARp50-PE (KIR2DS4), and (F) anti-CD94-PE. The percentage of cells positive in each quadrant is indicated in the top right-hand corner of each dot plot. Adding the numbers in the top left and top right quadrants represents the total number of NK cells (∼48% for NK-LDGL3 and ∼83% for NK-LDGL6). The number in the top right represents the number of NK cells coexpressing the NK receptor. R1 indicates region 1; Inh, inhibitory KIR; and Ac, activating KIR.
Using the same samples, we also examined the expression of CD94 and the C lectin-like receptors NKG2A and NKG2D. Inhibitory NKG2A heterodimerizes with CD94; NKG2D does not heterodimerize with CD94 but acts as an activating NK receptor.48 Most of the NK cells from NK-LDGL3 and NK-LDGL6 expressed CD94 with a pattern that was more homogenous for bright receptor expression than was the pattern for normal NK cells (Figure 1). The MFI values for NK-LDGL3, NK-LDGL6, and normal NK cells were 1225, 1211, and 258, respectively. Bright CD94 expression was observed in NK cells from all patients with NK-LDGL (Table 1) and in a subpopulation of normal NK cells as exemplified in Figure 1F (R1).
The pattern of expression for NKG2A and NKG2D paralleled that of CD94 in most patients examined. NKG2A and NKG2D displayed a bright homogenous staining pattern on most of the NK cells from LDGL patients (Table 1). Collectively, our data show that NK cells from patients with NK-LDGL have a more restricted pattern of KIR antibody reactivity than do normal NK cells and generally have bright expression of the C lectin-like receptors and CD94.
KIR genotyping of patients with NK-LDGL
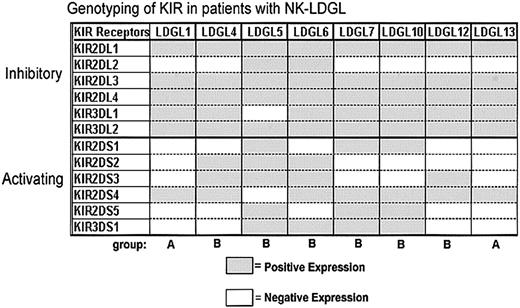
An individual's repertoire of NK receptors is based predominantly on the NK receptor genes he or she inherits.49-51 Therefore, the genotype, the amount of positive RNA expression, and the surface phenotype of KIR receptors are congruent in healthy individuals. In a previously described healthy white population, approximately 20 different phenotypes were observed.43 However, 2 general groups (groups A and B) were defined based on their individual KIR repertoires. Group A KIR haplotypes include many inhibitory KIR genes but only one activating KIR gene (usually KIR2DS4), whereas group B haplotypes consist of multiple activating KIR genes. These 2 groups occur with equal frequency in the healthy population.43
We examined the KIR genotypes of patients with NK-LDGL by PCR analysis, and the data are summarized in Figure 2. Group A and B KIR haplotypes were observed in the NK-LDGL population, but the frequency of group B patterns (multiple activating KIR) in LDGL patients (6 of 8) was greater than in previously described healthy individuals. Additionally, 4 of the 8 NK-LDGL patients expressed the genes for 4 or 5 activating KIRs, a phenomenon seen in only 13% of healthy white individuals in a previous study.43 These results demonstrate that patients with NK-LDGL have genotypes associated with multiple activating KIRs. Large numbers of activating KIRs have been shown previously in patients with NK-LDGL and in other autoimmune diseases, suggesting that this phenotype may contribute to the pathogenesis of the disease.52-54
Genotyping of KIR DNA in patients with NK-LDGL. DNA was amplified for KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR3DL1, and KIR3DL2 (inhibitory) or KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, and KIR3DS1 (activating). The data are summarized for 8 patients with NK-LDGL. Group A haplotype indicates one activating receptor (KIR2DS4); group B haplotype, multiple activating KIR.
Genotyping of KIR DNA in patients with NK-LDGL. DNA was amplified for KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR3DL1, and KIR3DL2 (inhibitory) or KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, and KIR3DS1 (activating). The data are summarized for 8 patients with NK-LDGL. Group A haplotype indicates one activating receptor (KIR2DS4); group B haplotype, multiple activating KIR.
RNA phenotyping
To gain a better understanding of the RNA expression levels of KIRs in patients and healthy individuals, we performed semiquantitative RT-PCR analysis for several KIRs. To confirm the amplification specificity of the RT-PCR assay, we used KIR-negative K562 cells, either untransfected (control), mock-transfected, or transfected with cDNA for KIR2DS2 (CD158j) or KIR2DS4 (pcDNA3-KIR2DS2 and pcDNA3-KIR2DS4). β-Actin primers were included as a positive control in each reaction. The transfection efficiency of the K562 cells was 15% as determined by green fluorescence protein (GFP) expression and analysis by flow cytometry (data not shown). The RNA for KIR2DS2 and KIR2DS4 were present in the appropriate transfected cells (Figure 3G lane 4 and 3I lane 5) but were not detected in control or mock-transfected cells. These data show that the primer pairs used specifically amplify KIR cDNA. We also show expression of several KIR RNA in cells derived from a healthy individual.
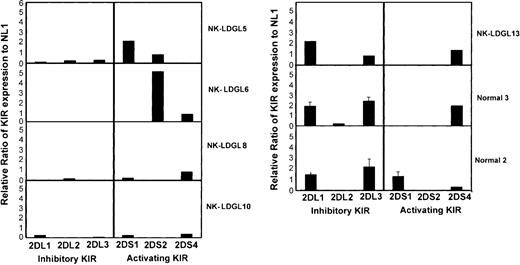
We next analyzed the RNA expression of multiple KIRs in PBMCs from healthy individuals and from patients with NKLDGL. Amplification of KIRs from RNA of 3 healthy donors was used as standards for a semiquantitative RT-PCR assay. Serial dilutions of RNA (100%, 20%, 4%, and .8%) were performed to establish the linear range for each KIR. We found cells from patients with NK-LDGL expressed less RNA for the inhibitory KIRs than did cells from healthy individuals. Additionally, patients with single-antibody reactivity (NK-LDGL5 and NK-LDGL6) preferentially expressed RNA for the activating KIRs. For example, NK-LDGL5 had single-antibody reactivity to CD158a (Table 1), which detects both KIR2DS1 (CD158h, activating) and KIR2DL1 (inhibitory; Figure 4). RNA levels of KIR2DL1 in NK-LDGL5 were greatly reduced compared with those of healthy individuals, while KIR2DS1 RNA levels were greater than normal. NK-LDGL6 had single-antibody reactivity to anti-CD158b, which recognizes 2 inhibitory KIRs (KIR2DL2 and KIR2DL3) and one activating KIR (KIR2DS2, CD158j). RNA for the 2 inhibitory KIRs were less abundant in NK-LDGL6 than in healthy individuals, whereas the RNA for KIR2DS2 (CD158j) was more abundant. NK-LDGL13 had cells that reacted to all of the anti-KIR antibodies. In contrast to other patients, NK-LDGL13 had normal levels of the RNA for KIR2DL1 and KIR2DL3 (inhibitory) and expressed only one RNA for an activating KIR (KIR2DS4 receptor, group A haplotype). Therefore, in addition to the genetic phenotype of these patients, which is consistent with multiple activating KIRs, there was subnormal inhibitory KIR mRNA expression in the NK-LDGL patient samples.
Semiquantitative RT-PCR analysis in patients with NK-LDGL. RT-PCR was performed using primers to amplify the inhibitory receptors KIR2DL1 (2DL1), KIR2DL2 (2DL2), KIR2DL3 (2DL3) and the activating receptors KIR2DS1 (2DS1), KIR2DS2 (2DS2), and KIR2DS4 (2DS4), and β-actin using the conditions described in “Patients, materials, and methods.” Reactions were performed using RNA from 3 healthy individuals and 5 patients with NK-LDGL. The graph represents densitometry results of samples normalized to β-actin and then expressed as a ratio to data obtained from normal 1 that is positive for each reaction. Raw data for normal 1 is shown in Figure 3. Normal 1, normal 2, and normal 3 were run on the same gel with each patient sample and the standard deviation is shown for these samples.
Semiquantitative RT-PCR analysis in patients with NK-LDGL. RT-PCR was performed using primers to amplify the inhibitory receptors KIR2DL1 (2DL1), KIR2DL2 (2DL2), KIR2DL3 (2DL3) and the activating receptors KIR2DS1 (2DS1), KIR2DS2 (2DS2), and KIR2DS4 (2DS4), and β-actin using the conditions described in “Patients, materials, and methods.” Reactions were performed using RNA from 3 healthy individuals and 5 patients with NK-LDGL. The graph represents densitometry results of samples normalized to β-actin and then expressed as a ratio to data obtained from normal 1 that is positive for each reaction. Raw data for normal 1 is shown in Figure 3. Normal 1, normal 2, and normal 3 were run on the same gel with each patient sample and the standard deviation is shown for these samples.
In addition to KIR RNA, we also examined the RNA expression of NKG2 family and NCR (NKp30, NKp44, and NKp46) family members. RNA for NKG2A, NKG2D, and NKG2E/F was positive in all patients examined (9/9). The RNA for the activating NKG2C receptor was weakly positive in normal unactivated PBMCs but was induced by treatment with IL-2 for 3 days. Of the NK-LDGL patients examined (n = 9), 3 were strongly positive, 3 were weakly positive, and 3 were negative for NKG2C RNA. As previously demonstrated, PBMCs from healthy individuals constitutively express the RNA for NKp30 and NKp46 but require IL-2 treatment for the induction of NKp44 RNA.31,32,55,56 Six of 9 LDGL patients expressed NKp30 RNA in PBMCs, and 4 of 9 patients expressed NKp46. None of the patients expressed the NKp44 activation receptor. Therefore, the NCRs are displayed less frequently in the NK-LDGL patients than in healthy individuals, as determined by RT-PCR analysis, whereas NKG2C was more frequently expressed.
Expression of KIR on T cells from patients with LDGL
CD8+ T cells, especially when under antigenic stimulation, up-regulate KIR receptor expression.20,57-59 The expression of NKG2C in NK-LDGL patients suggests possible in vivo activation. Therefore, we examined the expression of KIRs on CD3+ and CD8+/CD56- T cells from 5 patients: NK-LDGL5, NK-LDGL6, NKLDGL7, NK-LDGL10, and NK-LDGL12. NK-LDGL patients have less T cells than do healthy individuals. T-cell populations from NK-LDGL12 (multiple KIR-reactive NK cells) had a few cells that reacted to each antibody, but the total number of T cells was only 15% (data not shown). NK-LDGL7 had very few NK cells or T cells that reacted with any KIR antibody (data not shown). NK-LDGL5, NK-LDGL6, and NK-LDGL10 had the same single-antibody reactivity in T cells as they did in NK cells (exemplary data shown for NK-LDGL5; Figure 5).
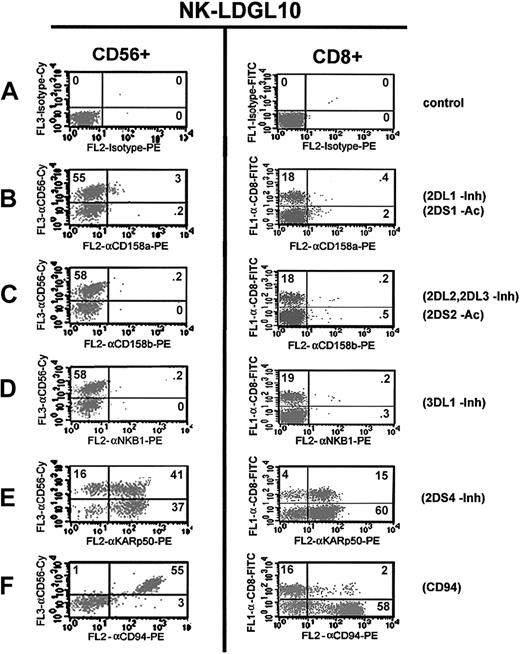
Similar KIR surface reactivity on NK and T cells of patients with NK-LDGL. Flow cytometry dot plots of 3-color flow cytometry staining of an NK-LDGL patient (NK-LDGL10). (A) Isotype control-CyChrome (y-axis, left column) and isotype control-FITC (y-axis, right column) and isotype control-PE (x-axis) were used to set the markers. Anti-CD56-CyChrome was used to identify the KIR expression on NK cells and anti-CD8-FITC was used to identify CD8+ T cells. CD56+ cells were determined to be CD8- by examining the expression of CD56-Cychrome versus CD8-FITC. KIRs were identified using the following antibodies: (B) anti-CD158a-PE, (C) anti-CD158b-PE, (D) anti-NKB1-PE, (E) anti-KARp50-PE, and (F) anti-CD94-PE. The percentages of cells positive in each quadrant are indicated in the top right-hand corner of each dot plot. Adding the numbers in the top left and top right quadrants represents the total number of NK cells or T cells (left and right columns, respectively).
Similar KIR surface reactivity on NK and T cells of patients with NK-LDGL. Flow cytometry dot plots of 3-color flow cytometry staining of an NK-LDGL patient (NK-LDGL10). (A) Isotype control-CyChrome (y-axis, left column) and isotype control-FITC (y-axis, right column) and isotype control-PE (x-axis) were used to set the markers. Anti-CD56-CyChrome was used to identify the KIR expression on NK cells and anti-CD8-FITC was used to identify CD8+ T cells. CD56+ cells were determined to be CD8- by examining the expression of CD56-Cychrome versus CD8-FITC. KIRs were identified using the following antibodies: (B) anti-CD158a-PE, (C) anti-CD158b-PE, (D) anti-NKB1-PE, (E) anti-KARp50-PE, and (F) anti-CD94-PE. The percentages of cells positive in each quadrant are indicated in the top right-hand corner of each dot plot. Adding the numbers in the top left and top right quadrants represents the total number of NK cells or T cells (left and right columns, respectively).
No difference in antibody reactivity was noted between the NK and T cells of most patients. T cells stained more heterogeneously with the anti-CD94 antibody and in a lower percentage than the NK cells from these patients, which expressed a homogenous CD94bright staining pattern (exemplary data are shown for NK-LDGL10; Figure 5F). The percent of T cells and NK cells that express CD94 in these 5 patients with NK-LDGL and 4 healthy donors is shown in Table 2. We conclude that the pattern of KIR expression is similar in both NK and T cells of patients with NK-LDGL. However, T cells display lower CD94 expression than the NK cells in these patients.
Summary of CD94 on T and NK cells of NK-LDGL patients
Anti-CD94 . | T cells % CD94+ (no. total T cells) . | NK cells % CD94+ (no. total NK cells) . |
|---|---|---|
| NKLGL5 | 23 (24) | 78 (78) |
| NKLGL6 | 5 (10) | 83 (85) |
| NKLGL7 | 10 (30) | 75 (75) |
| NKLGL10 | 2 (18) | 55 (58) |
| NKLGL12 | 2 (15) | 83 (84) |
| NL1 | 6 (62) | 15 (25) |
| NL2 | 4 (74) | 10 (13) |
| NL3 | 5 (68) | 6 (14) |
| NL4 | 5 (72) | 9 (18) |
Anti-CD94 . | T cells % CD94+ (no. total T cells) . | NK cells % CD94+ (no. total NK cells) . |
|---|---|---|
| NKLGL5 | 23 (24) | 78 (78) |
| NKLGL6 | 5 (10) | 83 (85) |
| NKLGL7 | 10 (30) | 75 (75) |
| NKLGL10 | 2 (18) | 55 (58) |
| NKLGL12 | 2 (15) | 83 (84) |
| NL1 | 6 (62) | 15 (25) |
| NL2 | 4 (74) | 10 (13) |
| NL3 | 5 (68) | 6 (14) |
| NL4 | 5 (72) | 9 (18) |
Percent of T cells and NK cells that stained positive for CD94 (% CD94+). Values taken from the top right-hand quadrant of 3-color dot plot flow cytometry graphs. NK cells were identified as CD8-.
Target lysis by patients with NK-LDGL
To determine whether the cells from patients with NK-LDGL have NK lytic function, we performed standard Cr-release assays using K562 as targets.60,61 We found that PBMCs from all patients with NK-LDGL effectively killed K562 tumor targets (Figure 6). The amount of killing was comparable to that of enriched normal NK cells and was much greater than that of normal PBMCs in all but one patient (NK-LDGL3).
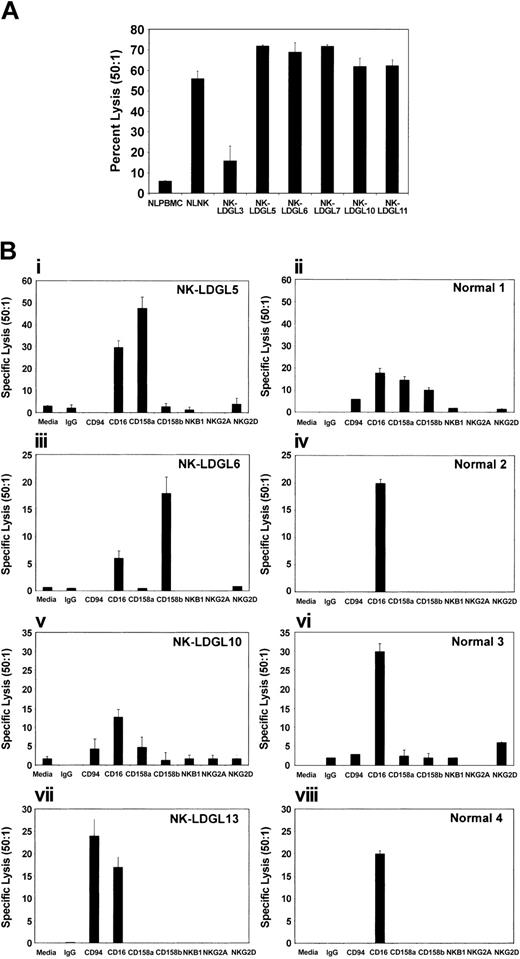
Target lysis by patients with NK-LDGL. (A) Direct killing of K562 cells as targets. To determine whether the cells from patients with NK-LDGL have NK lytic function, we performed standard 51Cr-release assays. The graphic representation of the percent specific lysis at a 50:1 effector-to-target ratio from normal PBMCs (NLPBMC), enriched NK cells (NLNK), and PBMCs from patients with NK-LDGL. (B) Redirected cytolysis using the FcR+ P815 tumor cell line as a target. PBMCs from patients with NK-LDGL and enriched NK cells from healthy individuals were compared using medium (Media), IgG (control antibody), anti-CD94 (coreceptor for some activating and inhibitory NKG2), anti-CD16, anti-CD158a (KIR2DS1, KIR2DS2), anti-CD158b (KIR2DL2, KIR2DL3, and KIR2DS2), anti-NKB1 (KIR3DL1), anti-NKG2A(inhibitory NKG2), and anti-NKG2D (activating NKG2) antibodies. The graphs represent the average of triplicate samples; standard deviation is indicated by the error bars.
Target lysis by patients with NK-LDGL. (A) Direct killing of K562 cells as targets. To determine whether the cells from patients with NK-LDGL have NK lytic function, we performed standard 51Cr-release assays. The graphic representation of the percent specific lysis at a 50:1 effector-to-target ratio from normal PBMCs (NLPBMC), enriched NK cells (NLNK), and PBMCs from patients with NK-LDGL. (B) Redirected cytolysis using the FcR+ P815 tumor cell line as a target. PBMCs from patients with NK-LDGL and enriched NK cells from healthy individuals were compared using medium (Media), IgG (control antibody), anti-CD94 (coreceptor for some activating and inhibitory NKG2), anti-CD16, anti-CD158a (KIR2DS1, KIR2DS2), anti-CD158b (KIR2DL2, KIR2DL3, and KIR2DS2), anti-NKB1 (KIR3DL1), anti-NKG2A(inhibitory NKG2), and anti-NKG2D (activating NKG2) antibodies. The graphs represent the average of triplicate samples; standard deviation is indicated by the error bars.
To determine whether the activating KIR receptors expressed by these patients induced tumor cell lysis, we performed redirected cytolysis assays using purified anti-KIR and anti-NKG2 antibodies to induce the killing of the P815 (FcγR positive) tumor cell line (Figure 6B). Cells from all patients and healthy individuals expressed CD16 and cross-linking of this receptor resulted in cytolysis of the P815 tumor cell line (Figure 6B).
We found that patients with single-antibody reactivity to anti-CD158a or anti-CD158b antibodies (NK-LDGL5 and NK-LDGL6, respectively) were capable of P815 lysis only after antibody cross-linking with these antibodies, and little or no cytotoxicity was induced by other NK receptor antibodies (Figure 6Bi-iii). NK-LDGL13 displayed the highest level of lysis in response to anti-CD94 cross-linking but little killing with anti-CD158a or anti-CD158b. PBMCs from NK-LDGL13 expressed higher levels of NKG2C RNA than unactivated normal cells, suggesting that the redirected killing was mediated by activation of NKG2C/CD94 (Figure 6Bvii). Although capable of inducing cytolysis with anti-CD16 cross-linking, very little killing was observed by cells from NK-LDGL10 using anti-KIR, anti-NKG2, or anti-CD94 antibodies. In contrast to the high NKG2D expression on patient NK cells, NKG2D did not significantly induce P815 cytolysis after cross-linking. NKG2D cross-linking of NKL cells was used as a positive control and resulted in 41% lysis at a 50:1 E/T ratio (data not shown). These results further support the idea that patient NK cells from NK-LDGL patients express predominantly activating NK receptors.
Discussion
Expansion of CD56+ and/or CD16+ NK cell populations in LDGL has been attributed to an undefined antigen that enhances cell survival in the absence of active proliferation.,62-64 We hypothesize that this expansion occurs in response to an unknown, perhaps viral, antigen. Although LDGL patients are not infected with any of the known retroviruses, sera from most patients (67%) react with the p21env protein of HTLV-I.8,63 This antigen also cross-reacts with HTLV-II, PTLV, and BLV, suggesting that these patients might be exposed to a novel retrovirus containing a protein with homology to p21e or to an endogenous protein that mimics the p21e antigen.8
We found that the KIR receptors expressed on NK cells from patients with LDGL have a skewed repertoire in contrast to those of healthy individuals. Most NK cells from 7 patients had homogenous reactivity to a single KIR receptor antibody (NK-LDGL2, NK-LDGL5, NK-LDGL6, NK-LDGL8, NK-LDGL9, NK-LDGL10, and NK-LDGL12; ≥ 50% of total NK cells). The remaining KIRs in each of these patients were largely negative or were expressed on only a small fraction of NK cells. In contrast, healthy individuals had some NK cells that reacted to all anti-KIR antibodies. KIR2DL4, which is reportedly expressed at the RNA level in every NK cell clone derived from healthy humans, was not expressed on most of the NK cells from 3 patients of 5 examined.43
Results of KIR DNA typing studies revealed that NK-LDGL patients possess large numbers of activating receptors. Healthy individuals can be divided into 2 KIR haplotypes based on inhibitory and activating KIR receptor expression (groups A and B). The most common haplotype in white individuals is group A. The group A haplotype consists predominantly of inhibitory KIR receptors with only one activating receptor (KIR2DS4).43 Only 2 patients with NK-LDGL (NK-LDGL1 and NK-LDGL13) displayed this phenotype of 8 patients examined. Instead, most patients with NK-LDGL displayed multiple activating KIRs consistent with a group B haplotype.
The major function of inhibitory KIR and inhibitory NKG2 receptors is the recognition of self-HLA alleles, thereby limiting the recognition of autologous cells.26-30 Individual NK cells usually express multiple KIRs, and ligation by HLA class I molecules overrides activating receptor signals.65 By genotype analysis, inhibitory KIRs were expressed on NK cells from NK-LDGL patients with a normal distribution pattern. Because the balance between inhibitory and activating signals is crucial for protecting against inappropriate autologous recognition, we further analyzed the inhibitory and activating receptors by semiquantitative RTPCR. We found that inhibitory KIR expression was reduced on cells from several patients relative to healthy individuals. Furthermore, the RNA expression level of activating receptors was equal to or greater than that of healthy individuals. Consistent with a high level of activating receptors, we found that NK-LDGL cells have potent cytolytic function in both direct and redirected cytotoxicity assays. Therefore, the semiquantitative RT-PCR analysis and the flow cytometry data suggest that the overrepresentation of activating to inhibitory RNA could be due to the expansion of a dominant population that expresses an activating KIR.
It has always been difficult to assess clonality in LDGL, as there are limited methods available.5,66,67 A previous study from our group using X-linked polymorphic gene analysis did not show clonality for patient NK cells, suggesting a reactive process.67 An attractive hypothesis for the unusual pattern of KIR expression in the patients with LDGL is that a unique antigen, perhaps viral or retroviral in origin, drives the survival of NK cells with excess amounts of activating and subnormal amounts of inhibitory KIRs. Patients with NK-LDGL have KIR genotypes with normal numbers of inhibitory but excess numbers of activating KIRs. Data from our RT-PCR analysis suggest that an NK clone(s) with low inhibitory KIR expression may be selectively amplified. It is also of interest that the small population of residual T cells in these patients has the same skewed KIR repertoire as does NK cells, suggesting a similar regulatory process. These data suggest that both NK cells and T cells from LDGL patients may be responding to a postulated inciting antigen.
Others have established that the molecular mechanism for the clonally expressed KIR repertoire in NK cell lines and NK clones derived from healthy donors is through hypermethylation of a small CpG (cytosine-phosphate-guanosine) island surrounding the transcriptional start site of each KIR gene.49 Transcriptional regulation is thought to restrict the number of KIRs that are expressed on an individual NK cell.28 We found evidence of reduced numbers of KIR transcripts for inhibitory KIRs from 4 patients. We will further explore the mechanism of KIR regulation in future experiments.
Results of our studies suggest an important role of the CD94 adaptor protein and the NKG family proteins. Both CD94bright and NKG2Abright receptors were homogeneously expressed on patient NK cells. The lack of self-recognizing inhibitory KIR has been associated with a compensatory induction of NKG2A-inhibitory receptors.28,68,69 Additionally, uniform bright expression of CD94 on mouse NK cells was associated with increased survival.70 Unlike normal NK cells, NCRs were variably expressed and underrepresented in the patients' NK repertoires.16,71
Our data suggest that expansion of NK cells with skewed NK receptor expression in patients with the NK type of LDGL may be related to disease pathogenesis. Biochemical studies of the exact mechanism leading to the restricted repertoire and the biologic consequences of reduced inhibitory KIR expression will greatly enhance our understanding of NK cell biology.
Prepublished online as Blood First Edition Paper, January 15, 2004; DOI 10.1182/blood-2003-02-0400.
Supported by Veterans Administration Merit Review grants, American Heart Association grant 0256422B, and by National Cancer Institute grants CA90633, CA83146, CA83947, and CA98472.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank the Flow Cytometry Core Facility at H. Lee Moffitt Cancer Center & Research Institute for their assistance with flow cytometry acquisition and analysis. We also thank Mr Larry Kuba of the Molecular Imaging Core Facility and Nancy Olashaw, PhD at H. Lee Moffitt Cancer Center & Research Institute for assistance in the preparation of this manuscript. We thank Ms TyLee McKeown for assistance with blood preparation.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal