Abstract
Comparative expressed sequence hybridization (CESH) to chromosomes is a recently introduced technique that identifies chromosomal regions corresponding to a differential gene expression. This technique is analogous to comparative genomic hybridization (CGH) that detects genomic imbalances. We applied CESH for the study of hairy cell leukemia (HCL), a disorder with a largely unknown expression profile. Twelve HCL cases with spleen involvement were investigated by CESH and CGH. While the latter analysis identified only a few nonrecurrent genomic imbalances, CESH showed a consistent expression profile in all HCL cases. In addition, pairing normal spleen with normal lymph node, a “spleen signature” was established by CESH. This signature most likely reflects the expression profile of spleen-specific components, such as the sinusoidal lining cells from the red pulp and the marginal zone B cells from the white pulp. Imprint of the spleen signature was found in the HCL expression profile, suggesting that HCL may originate from a particular B-cell subset present in these splenic components. Besides pairing HCL with normal lymph node and spleen, we identified an “HCL signature” comprising several chromosome regions with altered expression. The most significantly underexpressed regions include 3p24, 3p21, 3q13.3-q22, 4p16, 11q23, 14q22-q24, 15q21-q22, 15q24-q25, and 17q22-q24; and 13q31 and Xq13.3-q21 were the most significantly overexpressed. These regions possibly harbor genes related to the biology and the pathogenesis of HCL. Their identification warrants further molecular investigations.
Introduction
Hairy cell leukemia (HCL) is an indolent chronic lymphoproliferative disorder accounting for 2% of all lymphoid leukemias.1 This disorder predominantly affects middle-aged male adults (median age, 55 years). The disease is characterized by the presence of a small number of circulating B cells with a villous morphology, splenomegaly, bone marrow involvement, and pancytopenias. HCL cells express pan–B-cell markers (CD19, CD20, CD22, and CD79a) and more typically express CD11c, CD25, CD103, and FMC7. Moreover, most HCL cases are characterized by cytoplasmic expression of tartrate-resistant acid phosphatase (TRAP). HCL is highly sensitive to unconventional agents, including interferon-α and nucleosides, and shows high incidence of complete remission.2
So far, the origin of hairy cells remains unknown. One of the most tempting candidates for the normal counterpart of HCL is a marginal zone B cell.3,4 Neither genetic nor molecular events associated with the development of HCL are well understood. Scarce cytogenetic data confirmed by comparative genomic hybridization (CGH) studies show that the most HCL cases have a normal (or at least balanced) karyotype.5-7 Among a few recurrent chromosomal changes identified in HCL cases, the most frequent are abnormalities of chromosome 5 (trisomy 5, 5q13 aberrations) and chromosome 14 (add(14)(q32) and del(14)(q)).8,9 Overexpression of cyclin D1, found in about 50% to 75% of HCL cases, did not appear to be associated with a genomic rearrangement of 11q13/BCL1.10
To get insight into the molecular mechanism(s) underlying the pathogenesis of this poorly investigated leukemia, we applied a recently developed molecular cytogenetic technique termed comparative expressed sequence hybridization (CESH) for the study of 12 HCL cases. This technique gives a genome-wide view of relative expression patterns within tissues according to chromosomal location in a way similar to that of CGH.11 Until now, more than 100 cases of solid tumors have been successfully analyzed using CESH.12 The established profiles allowed the authors to distinguish particular groups and subgroups of tumors and to identify tumors with differential biologic behaviors.
We present here the expression profile of HCL, the first hematologic malignancy analyzed using CESH.
Materials and methods
Samples
Two cell lines, IMR-32 and DEL, and Epstein-Barr virus (EBV)–transformed normal B lymphocytes were used to validate the CESH protocol. IMR-32 (CCL 127; American Type Culture Collection [ATCC], Manassas, VA) is a neuroblastoma cell line with amplification and overexpression of the NMYC (2p24) and MEIS1 (2p15) genes,13 and DEL is an anaplastic lymphoma kinase (ALK)–positive anaplastic lymphoma cell line carrying a cryptic t(2;5)(p23;q35).14
Twelve HCL cases documented by snap-frozen tissue blocks taken from splenectomy specimens were selected. On review, all spleen biopsies showed a massive involvement of the red pulp by HCL with almost no or minimal residual white pulp. These cases were diagnosed in our department between 1990 and 2003 according to the Revised European-American Lymphoma (REAL) classification.15 Eleven of the patients underwent splenectomy at the time of diagnosis, and in one patient splenectomy was performed during the course of the disease.
In addition, snap-frozen biopsies of 5 lymph nodes, 5 spleens, and 1 muscle sample, all without obvious morphologic anomalies, were used as reference and/or test material in the CESH experiments.
This study was approved by the institutional ethics commission of the K.U. Leuven. Informed consent was provided according to the Declaration of Helsinki.
Comparative genomic hybridization
CGH studies performed on DNA from the IMR-32 and DEL cell lines and HCL biopsies using standard reference DNA followed a previously described protocol.16 The image analysis was performed using either a Leica DMRB epifluorescence microscope (Leica, Wetzlar, Germany) equipped with a cooled black-and-white CCD camera (Photometrics, Tucson, AZ) run by Quips software (Vysis, Downers Grove, IL) (initial validation studies) or using a Zeiss Axioplan 2 fluorescence microscope (Zeiss, Zaventem, Belgium) equipped with a cooled charge-coupled device camera COHU 4910 (Diagnostic Instruments, Detroit, MI) and controlled by Cytovision software version 2.81 (Applied Imaging International, Newcastle upon Tyne, United Kingdom) (HCL studies). Ten good-quality metaphases were analyzed in each experiment.
RNA preparation and reverse transcription
Total RNA was extracted from the cell lines and from eight 20-μm sections of each frozen tissue sample by using Trizol reagent (Life Technologies, Merelbeke, Belgium) following the manufacturer's instructions. The concentration of RNA was measured spectrophotometrically. Twenty-five micrograms of RNA was treated with 2 units of RNase-free DNase (Promega, Leiden, The Netherlands) during 45 minutes and further purified using phenol-chloroform. RNA was reverse transcribed using random hexamers and Superscript II (Life Technologies).
Complementary DNA labeling
Briefly, cDNAs reverse transcribed from 10 ng total RNA from the test and reference samples were amplified and differentially labeled with, respectively, SpectrumGreen-dUTP and SpectrumRed-dUTP (Vysis, Ottignies, Belgium) during 2 rounds of degenerate oligonucleotide-primed polymerase chain reaction (PCR) using the degenerate primer UN-1.17 Labeled probes were purified using Qiaquick PCR purification columns (Westburg, Leusden, The Netherlands) according to the manufacturer's instructions. Products were checked by agarose gel electrophoresis (1%) and showed a size of 300 to 1500 base pairs.
Comparative expressed sequence hybridization
CESH followed a previously described protocol.11 The CGH hybridization kit (Vysis, Ottignies, Belgium) was used to prepare hybridization mixtures containing labeled cDNA probes. To reduce background noise, target slides were additionally pretreated with RNase (200 μL RNase [0.1 μg/μL] per slide) at 37° C for 30 minutes. Hybridization was carried out in a humid chamber at 37° C for 72 hours.
Ten good-quality metaphases, counterstained with DAPI (4,6 diamidino-2-phenylindole), were analyzed in each experiment. CESH analysis was performed using the same system and CGH analysis software as described for CGH. As thresholds for determination of a relative overexpression or underexpression, we used dynamic standard reference intervals (SRIs)18,19 basing on systematic variations seen in normal samples. The SRIs were created in our system by the analysis of 100 metaphases of self-self hybridizations after CESH pairing cDNA from the same normal lymph node. The 90%, 95%, and 99% confidence limits (CL), applied to record regions with a differential expression, are recognized statistical indications of how accurately the slide profile reflects the real abnormalities in the test DNA/cDNA sample.
Results
Validation experiments
For initial validation of CESH analysis we used cDNA from the IMR-32 and DEL cell lines with a respective amplification/overexpression of NMYC (2p24) and MEIS1 (2p15) and a rearrangement/overexpression of ALK (2p23), paired with cDNA from EBV-transformed B lymphocytes. Both cell lines were also analyzed by CGH. CESH experiments showed numerous differentially expressed chromosomal regions, including hybridization peaks at 2p24-p25 and 2p15 in IMR-32 and at 2p23-p24 in DEL (data not shown). Several of these regions, including 2p23-p24 in DEL, showed balanced signals by CGH. The reliability of CESH was further demonstrated in experiments comparing normal tissues, namely, lymph node versus muscle and lymph node versus lymph node. While no differential expression was found in the latter CESH experiment, comparison of lymph node with muscle showed 7 chromosomal regions with a relative overexpression (6pter-p22, 6p21.2, 14q32, 16p12, 17q12-q22, 22q11.2, and 22q13) and 2 regions with a relative underexpression (3q26.1 and Xp11.4) identified using the 99% CL (data not shown). Overexpression of 6p, possibly reflecting the differential expression of genes in the MHC cluster, was previously also observed by Lu et al11 in CESH experiments pairing lymphocytes with fibroblasts and muscle cells. Of particular interest is our finding of overexpression of the 14q32 region in the experiment pairing normal lymph node and muscle cells and underexpression of the same region in the CESH experiment comparing cDNA from IMR-32 and EBV-transformed B lymphocytes (Figure 1). This region corresponds with a localization of the immunoglobulin heavy chain gene (IGH) cluster specifically expressed in B lymphocytes and not expressed in neuroblastoma and muscle cells.
CESH validation studies. Expression profile of chromosome 14 in the CESH experiments pairing cDNA from the IMR-32 cell line and EBV-transformed B lymphocytes (A) and cDNA from normal lymph node and muscle cells (B), respectively. Note underexpression of the 14q32 region corresponding to the IGH localization in panel A and overexpression of the same region in panel B.
CESH validation studies. Expression profile of chromosome 14 in the CESH experiments pairing cDNA from the IMR-32 cell line and EBV-transformed B lymphocytes (A) and cDNA from normal lymph node and muscle cells (B), respectively. Note underexpression of the 14q32 region corresponding to the IGH localization in panel A and overexpression of the same region in panel B.
CGH analysis of HCL
Eleven of 12 HCL cases were successfully subjected to CGH analysis. No genomic imbalances were found in 5 cases. In the remaining 6 cases nonrecurrent imbalances including gain of 1p32-p36 (case 2), loss of 8q21.3, gain of 10p12 (case 5), loss of 11q14-q23 (case 6), gain of 6p22-p24, 7q32, 10p12, 16q22, 17q22 (case 7), gain of 14q23-q24 (case 9), and gain of 5q13-q31 (case 10) were detected.
CESH profiling of HCL
CESH analysis of 12 HCL cases was performed in 2 sets of experiments pairing cDNA from HCL with cDNA from either normal lymph nodes or normal spleens as reference. In addition, the relative CESH profile of normal spleen versus normal lymph node was determined. CESH results are illustrated and summarized in Figure 2; only those regions that were underexpressed (↓) or overexpressed (↑) in at least 3 cases (25%) are represented. Like in CGH, all centromeric regions and chromosome Y, as well as critical hybridization regions identified in 6 random color-flip experiments (data not shown), were excluded from the final evaluation. To recognize regions with a potential significance but a moderate level of differential expression, we included in the final analysis all differentially expressed regions detected at the 99%, 95%, and 90% CL.
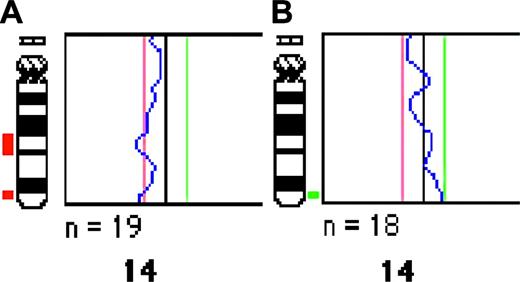
CESH profiling of hairy cell leukemia. (A) A representative CESH image from the experiment pairing cDNA from HCL (spleen) with cDNA from normal lymph nodes. Original magnification, ×100 (Plan Neofluor lens 100×/1.30 Oil [Zeiss]). (B) The average CESH profile of 10 analyzed metaphase cells from the same experiment showing the red-green fluorescence intensity ratio along the chromosomes (pink lines) using the 95% CL (yellow lines). Dynamic standard reference intervals (SRIs) offered by the high-resolution Cytovision CGH analysis software are shown as black lines. A ratio outside SRI is indicative of a relative underexpression/overexpression, indicated as red and green bars, respectively. (C) Schematic summary of CESH results obtained in all 3 sets of experiments showing chromosomal regions with a relative underexpression (left side) and overexpression (right side). Color bars represent differentially expressed regions identified in the HCL versus lymph node (green), HCL versus normal spleen (red), and normal spleen versus normal lymph node (blue) experiments. Each bar marks an averaged region with differential expression recorded in at least 25% (3 of 12) of analyzed cases. Dotted, regular, and thick bars point to regions with altered expression detected using the 90%, 95%, and 99% CL, respectively. The shown level of differential expression (90%, 95%, and 99% CL) is representative for at least 25% of informative cases.
CESH profiling of hairy cell leukemia. (A) A representative CESH image from the experiment pairing cDNA from HCL (spleen) with cDNA from normal lymph nodes. Original magnification, ×100 (Plan Neofluor lens 100×/1.30 Oil [Zeiss]). (B) The average CESH profile of 10 analyzed metaphase cells from the same experiment showing the red-green fluorescence intensity ratio along the chromosomes (pink lines) using the 95% CL (yellow lines). Dynamic standard reference intervals (SRIs) offered by the high-resolution Cytovision CGH analysis software are shown as black lines. A ratio outside SRI is indicative of a relative underexpression/overexpression, indicated as red and green bars, respectively. (C) Schematic summary of CESH results obtained in all 3 sets of experiments showing chromosomal regions with a relative underexpression (left side) and overexpression (right side). Color bars represent differentially expressed regions identified in the HCL versus lymph node (green), HCL versus normal spleen (red), and normal spleen versus normal lymph node (blue) experiments. Each bar marks an averaged region with differential expression recorded in at least 25% (3 of 12) of analyzed cases. Dotted, regular, and thick bars point to regions with altered expression detected using the 90%, 95%, and 99% CL, respectively. The shown level of differential expression (90%, 95%, and 99% CL) is representative for at least 25% of informative cases.
In general, the CESH patterns were highly consistent. Regions showing differential expression were detected with a mean recurrence of 62.5% (range, 8.3%-100%). There were more underexpressed than overexpressed regions detected. The highest fluorescence intensity we ever noted in all individual experiments was detected at the 99.9% CL. Chromosomal regions showing such a high differential expression in more than 2 cases were localized at 12q21 and 13q21 (↑) and 12q23-q24.1, 14q31-q32, 15q24, and 17q22-q23 (↓).
CESH profiling of HCL versus normal lymph node (HCL/nLN) using the 90% CL led to the detection of 78 differentially expressed chromosomal regions (47↓ and 31↑) of which only 19 (15↓ and 4↑) were still identified using the 99% CL (green lines in Figure 2C). All chromosomes were affected, and the number of differentially expressed regions per chromosome ranged from 1 to 7. The highest number of involved regions (7) was found on chromosome 3.
CESH profiling of HCL versus normal spleen (HCL/nS) using the 90% CL resulted in the identification of 80 differentially expressed chromosomal regions (50↓ and 30↑), including 5 (3↓ and 2↑) detected at the 99% CL (red lines in Figure 2C). The pattern of chromosome involvement was analogous to that seen in the previous set of experiments.
CESH profiling of normal spleen versus normal lymph node (nS/nLN) using the 90% CL resulted in detection of 37 differentially expressed chromosomal regions (24↓ and 13↑) of which only 3 (1p35-p34↓, 1p34-p32↓, 1q22-q25↓) were identified using the 99% CL (blue lines in Figure 2C). The affected regions were found on all but 5 chromosomes (15, 17, 19, 21, and 22). The highest number (3 to 6) of regions with altered expression was detected on chromosomes 1, 2, 4, and 6.
Compilation of the CESH results from the HCL/nLN, HCL/nS, and nS/nLN experiments led to the identification of regions consistently affected in all 3 sets of experiments and regions detected exclusively in one or both HCL pairing experiments (Figure 2C). The first ones (31 regions) comprise part of the relative “spleen signature” identified in the nS/nLN experiment, composed of 37 differentially expressed chromosomal regions. Interestingly, 4 of the remaining 6 regions (1p31↑, 6q15-q21↓, 8p21-p22↑, and 10p12-p13↑) showed an opposite expression change in the HCL/nLN and/or HCL/nS pairing experiments.
Forty-nine differentially expressed chromosomal regions, identified exclusively in the HCL/nLN and HCL/nS experiments, were considered as the relative “HCL signature” (Table 1 and Figure 2C). Two of them, 13q31 and Xq13.3-q21, were significantly overexpressed, and 9 regions, including 3p24, 3p21, 3q13.3-q22, 4p16, 11q23, 14q22-q24, 15q21-q22, 15q24-q25, and 17q22-q24, were significantly underexpressed (at least 95% CL). The differential expression of the remaining 38 regions was less prominent (95% CL or below), although most of them were recurrently recorded in both HCL experiments.
Chromosomal regions with differential expression composing the HCL signature and relevant genes mapped to these regions
Chromosomal regions with differential expression* . | Some relevant genes involved in the biology of hematopoietic cells and oncogenesis† . |
|---|---|
| 1p31 ↓ /1q31 ↑ | —/RGS18, RGS1, RGS13, RGS2, CD45 |
| 2q22-q24 ↑ /2q24-q32.1 ↓ | ZFHX1B/HOXD |
| 3p24 ↓ /3p21-p23 ↓ /3p21 ↓ /3p14-p21 ↓ /3q13.3-q22 ↓ /3q26 ↓ | —/CCR4, PDCD6IP, ITGA9, CTNNB1/MST1R, RASSF1A, HYAL1-3/—/RYK, EPHB1/TRAIL, WIG1 |
| 4p16 ↓ /4q22-q24 ↑ /4q26-q27 ↑ | FGFR3, RGS12/TMSL4, NFKB1,PGD2‡/IL2, IL21,FGF2‡ |
| 5p14-p15.1 ↑ /5q14-q15 ↑ /5q23-q31 ↑ | —/—/PPIC‡, IL3,GM-CSF, IRF1, IL5, IL4, IL13, CDC25C, EGR1 |
| 6p21.2-p21.3 ↓ /6q14 ↑ /6q15-21 ↑ /6q25-q27 ↓ | DDR1, TNFA, LTA, LTB, MHC2, DAXX, BAK, ETV7, CDKN1A, PIM1/—/MAP3K7, EPHA7, FYN/ST8 |
| 7p14-p15 ↓ /7q31 ↑ | IL6, HOXA/MET |
| 8p21-p22 ↓ /8q13-q21.3 ↑ /8q22 ↓ /8q23 ↑ | TNFRSF10A-D/—/—/— |
| 9p21 ↑ /9q21-q22 ↑ | CDKN2A/B, TEK/ANXA1, NTRK2, SYK, ROR2, PTCH,PHF2‡ |
| 10p12-p13 ↓ /10q22 ↓ /10q24-q25 ↓ | —/—/PTEN, FAS, HOX11 |
| 11q23 ↓ | NCAM1, IL10RA, MLL,CXCR5‡ |
| 12p13 ↓ /12p11.2-p12 ↑ | CCND2, ETV6, p27/CAZ3, LRMP, KRAS1 |
| 13q31 ↑ | SPRY2‡, Y918‡ |
| 14q22-q24 ↓ | CDKN3, RGS6, TGFB3 |
| 15q21-q22 ↓ /15q24-q25 ↓ | B2M/CSK, KIP2, NTRK3 |
| 17p11.2-p13.1 ↓ /17q12-q21 ↓ /17q22-q24 ↓ | TP53, TNK1/ERBB2, CSF3, RARA, CDC6, TOP2A, CCR7, STAT3, STAT5, NME1/2/RGS9, PRKAR1A |
| 18q12-q21.1 ↑ | MAPK4, MADH4 |
| 19p13.2 ↑ /19q13.2-q13.3 ↓ /19q13.2-q13.3 ↑ | INSR, TYK2, CDC37, ILF3, ACP5 (TRAP), JUNB/AXL, TGFB1, KIAA1883, BAX, NSPL1‡, RRAS‡ |
| 20p12-p13 ↓ | PCNA |
| 21q21-q22 ↓ | IL10RB, AML1 |
| 22q12-q13.1 ↓ | NF2, OSM, LIF |
| Xq13.3-q21, ↑ /Xq25q26 ↑ | LAMR1, BTK/CD40L |
Chromosomal regions with differential expression* . | Some relevant genes involved in the biology of hematopoietic cells and oncogenesis† . |
|---|---|
| 1p31 ↓ /1q31 ↑ | —/RGS18, RGS1, RGS13, RGS2, CD45 |
| 2q22-q24 ↑ /2q24-q32.1 ↓ | ZFHX1B/HOXD |
| 3p24 ↓ /3p21-p23 ↓ /3p21 ↓ /3p14-p21 ↓ /3q13.3-q22 ↓ /3q26 ↓ | —/CCR4, PDCD6IP, ITGA9, CTNNB1/MST1R, RASSF1A, HYAL1-3/—/RYK, EPHB1/TRAIL, WIG1 |
| 4p16 ↓ /4q22-q24 ↑ /4q26-q27 ↑ | FGFR3, RGS12/TMSL4, NFKB1,PGD2‡/IL2, IL21,FGF2‡ |
| 5p14-p15.1 ↑ /5q14-q15 ↑ /5q23-q31 ↑ | —/—/PPIC‡, IL3,GM-CSF, IRF1, IL5, IL4, IL13, CDC25C, EGR1 |
| 6p21.2-p21.3 ↓ /6q14 ↑ /6q15-21 ↑ /6q25-q27 ↓ | DDR1, TNFA, LTA, LTB, MHC2, DAXX, BAK, ETV7, CDKN1A, PIM1/—/MAP3K7, EPHA7, FYN/ST8 |
| 7p14-p15 ↓ /7q31 ↑ | IL6, HOXA/MET |
| 8p21-p22 ↓ /8q13-q21.3 ↑ /8q22 ↓ /8q23 ↑ | TNFRSF10A-D/—/—/— |
| 9p21 ↑ /9q21-q22 ↑ | CDKN2A/B, TEK/ANXA1, NTRK2, SYK, ROR2, PTCH,PHF2‡ |
| 10p12-p13 ↓ /10q22 ↓ /10q24-q25 ↓ | —/—/PTEN, FAS, HOX11 |
| 11q23 ↓ | NCAM1, IL10RA, MLL,CXCR5‡ |
| 12p13 ↓ /12p11.2-p12 ↑ | CCND2, ETV6, p27/CAZ3, LRMP, KRAS1 |
| 13q31 ↑ | SPRY2‡, Y918‡ |
| 14q22-q24 ↓ | CDKN3, RGS6, TGFB3 |
| 15q21-q22 ↓ /15q24-q25 ↓ | B2M/CSK, KIP2, NTRK3 |
| 17p11.2-p13.1 ↓ /17q12-q21 ↓ /17q22-q24 ↓ | TP53, TNK1/ERBB2, CSF3, RARA, CDC6, TOP2A, CCR7, STAT3, STAT5, NME1/2/RGS9, PRKAR1A |
| 18q12-q21.1 ↑ | MAPK4, MADH4 |
| 19p13.2 ↑ /19q13.2-q13.3 ↓ /19q13.2-q13.3 ↑ | INSR, TYK2, CDC37, ILF3, ACP5 (TRAP), JUNB/AXL, TGFB1, KIAA1883, BAX, NSPL1‡, RRAS‡ |
| 20p12-p13 ↓ | PCNA |
| 21q21-q22 ↓ | IL10RB, AML1 |
| 22q12-q13.1 ↓ | NF2, OSM, LIF |
| Xq13.3-q21, ↑ /Xq25q26 ↑ | LAMR1, BTK/CD40L |
Regions with differential expression found in only one HCL pairing experiment are italicized; bold are regions found to be differentially expressed in at least 50% of analyzed cases.
Genes were selected from the human genome map (http://www.ensembl.org/); — indicates no relevant genes were identified in this region; bold are genes coding proteins with known underexpression/overexpression in HCL.
Genes found to be up- and down-regulated in HCL by Basso et al.20
Discussion
Genetic changes associated with malignant transformation and tumor progression critically affect the expression of key genes. Therefore, the recently introduced molecular approaches including microarray-based technologies that directly evaluate the level of gene expression have shown to be potential tools in cancer research.21,22 The global expression profiling has been successfully used for the molecular classification of leukemias, lymphomas, and solid tumors; identification of genetic markers of clinical behavior of tumors; and predictive oncogenic pathways.23-28 Recently, an alternative molecular cytogenetic approach to expression profiling termed CESH has been developed.11 This technique gives a genome-wide view of relative expression patterns within tissues according to chromosomal location. Although the resolution of CESH is significantly lower than that of cDNA microarray techniques, it has been shown that the potential of CESH in tumor classification and even in predicting their clinical behavior may be comparable to that of microarray approaches.12 Importantly, no prior knowledge of genes or cloning is required for CESH. Moreover, this low-cost analysis using a minimal amount of tissue/RNA can be performed using standard fluorescence in situ hybridization (FISH) equipment.
Having such a potential, we validated and applied CESH for profiling of HCL, a leukemia with a largely unknown gene expression pattern. Twelve HCL cases with massive spleen involvement were successfully subjected to CESH analysis. In addition, we established the CGH pattern of genomic imbalances in 11 of these cases. Interestingly, all of them showed a strikingly uniform expression profile that contrasted with their either balanced or heterogeneously unbalanced genomic profiles. These observations are in line with the previously reported CESH and CGH findings in rhabdomyosarcoma.11 Notably, such a consistent expression pattern of HCL remains in agreement with its uniform morphologic, phenotypic, and clinical features that allowed us to recognize HCL as a distinct lymphoma entity.1,15 The HCL profiles obtained in 2 sets of experiments, pairing HCL with normal lymph nodes and normal spleens, were further compared with the relative profile of normal spleen paired with normal lymph node. Such a compilation allowed us to discriminate between the relative HCL signature and the relative spleen signature. The latter was significantly imprinted not only in the HCL/nLN but also in the HCL/nS experiment, possibly due to its unbalanced enhancement by massively infiltrating HCL cells. The identified relative spleen signature composed by 37 differentially expressed regions seems to reflect the expression profile of splenic components not present or underrepresented in the lymph node, including the red pulp with its sinusoidal lining cells as well as components of the marginal zone (MZ). Regarding a well-documented B-cell origin of HCL, one might assume that HCL originates from a particular B-cell subset residing in the spleen but being absent or underrepresented in the lymph node. Of these, MZ B cells are the best candidates. The hypothesis of MZ origin of HCL was already postulated by our and other groups in the early years.3,4 Moreover, this concept has been supported by significant morphologic and immunophenotypic similarities between HCL and MZ cells. It must be stressed, however, that unlike HCL, MZ cells do not express CD11c, TRAP, and PCA-1 but do express CD21.29
Besides the spleen signature, manifested in all 12 analyzed HCL cases, we were able to identify the HCL signature composed of 11 regions with a significantly altered expression (at least 95% CL) and 38 regions with a less prominently (95% CL or below) changed expression. These regions possibly harbor differentially expressed genes associated with the biology and/or the pathogenesis of HCL. The most significant regions are 3p21↓, 3p24↓, 3q13.3-q22↓, 4p16↓, 11q23↓, 13q31↑, 14q22-q24↓, 15q21-q22↓, 15q24-q25↓, 17q22-q24↓, and Xq21↑. Regarding that aberrations of chromosome 5, including trisomy 5, are the most frequent karyotypic abnormalities in HCL,8,9 our finding of overexpression of 3 chromosome 5 regions (5p14-p15.2, 5q14-q15, and 5q23-q31.1) in all analyzed cases is intriguing. On the other hand, however, only one of our cases showed gain of 5q by CGH. Also, underexpression of the 14q22-q24 region correlates with a genomic loss of this area recurrently observed in HCL9 as well as in other lymphoma types.5,30 Further compilation of the HCL signature found in this study with chromosomal localization of genes encoding proteins with previously documented altered expression in HCL led to the following matches: 4q26-q27↑-FGF2↑,31,32 5q23-q31↑-GM-CSF↑,33 12p13↓-p27↓,34,35 19p13↑-TRAP↑,36 and 19q13.2-q13.3↑-TGFB↑.31 Differential expression of regions containing other genes shown by immunohistochemistry and/or by reverse transcriptase (RT)–PCR to be affected in HCL, including FN1/2q34,37 CXCR3/8p12,38 INFA/9p22,39 IL2RA/10p15,40 LSP1/11p15.5,41 CCND1/11q13,10 CD44/11q15,42 CD11c/16p11.2,43 CD103/17p13,44 iNOS/17q11,45 δCD79b/17q23,46 BCL2/18q21,47 SRC/20q12,48 L2RB/22q12,40 and F-actin/Xp11,49 was not detected by CESH. This might be caused by either an unfavorable localization of these genes in chromosomal regions critical for CESH (eg, near-centromeric [BCL1, CD11c, iNOS, F-actin] and near-telomeric [LSP1, CD44, CD103]), or by their expression by other components of lymph node and spleen, or by a limited sensitivity and resolution of CESH. Besides this, CESH profiling of HCL led to the identification of new chromosomal regions containing genes with a differential expression in this leukemia. Although targeted loci could not be directly identified by this molecular cytogenetic technique, browsing through the human genome map (http://www.ensembl.org/) showed a number of candidate genes mapped in the highlighted regions that are involved in fundamental cellular functions and/or oncogenesis (Table 1). Further investigations are required to identify critical genes involved in the pathogenesis of HCL.
During the editorial processing of this paper, Basso et al20 published their results of gene expression profiling of 14 HCL cases using Affymetrix platform representative of about 12 000 genes. The analyzed HCL material included bone marrow biopsies and peripheral blood and only one spleen sample. Like in our study, HCL showed a very homogenous profile, which is clearly distinct from that of other B-cell NHLs. By comparing the HCL profile with profiles of different normal B-cell subpopulations and corresponding B-cell lymphomas, the authors postulated derivation of HCL from memory B cells. This suggestion can be in line with our findings that support a marginal zone B-cell origin of HCL, because memory B cells constitute one of the B-cell components of MZ. Looking for localization of the genes indicated by Basso et al20 as up- and down-regulated in HCL when compared with normal memory B cells, we found at least 9 matches marked in Table 1. The regions carrying the remaining genes pointed out by the authors were not identified in our CESH experiments. Because these genes mainly control adhesion and response to chemokines, one might assume that at least part of them are related to the homing of HCL cells in the bone marrow and the peripheral blood. Regarding the design of our CESH study (HCL/nLn and HCL/nS) we presumably identified only those regions that were differentially expressed in HCL cells, excluding those commonly expressed in B cells.
In conclusion, our study supports previously published data11,12 showing that CESH is a reliable and potential technique for expression profiling of human malignancies. CESH analysis of HCL cases showed a strikingly consistent expression pattern in all analyzed cases, independent from underlying genomic imbalances. Imprinting of the relative spleen signature in the HCL profiles suggests that HCL may originate from a spleen-related B-cell population, of which marginal zone B cells are the best candidates. In addition, chromosomal regions harboring differentially expressed genes in HCL, possibly related to the biology and the pathogenesis of this disease, have been elucidated. Identification of targeted genes present in these regions warrants further molecular investigations and may indicate new therapeutic endeavors.
Prepublished online as Blood First Edition Paper, March 11, 2004; DOI 10.1182/blood-2004-01-0181.
Supported by grant G.0223.03 from the Fund for Scientific Research (FWO), Flanders, Belgium. V.V. is a fellow of the FWO Flanders (grant G.0362.01).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This text presents research results of the Belgian Programme of Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming. The scientific responsibility is assumed by the authors.

![Figure 2. CESH profiling of hairy cell leukemia. (A) A representative CESH image from the experiment pairing cDNA from HCL (spleen) with cDNA from normal lymph nodes. Original magnification, ×100 (Plan Neofluor lens 100×/1.30 Oil [Zeiss]). (B) The average CESH profile of 10 analyzed metaphase cells from the same experiment showing the red-green fluorescence intensity ratio along the chromosomes (pink lines) using the 95% CL (yellow lines). Dynamic standard reference intervals (SRIs) offered by the high-resolution Cytovision CGH analysis software are shown as black lines. A ratio outside SRI is indicative of a relative underexpression/overexpression, indicated as red and green bars, respectively. (C) Schematic summary of CESH results obtained in all 3 sets of experiments showing chromosomal regions with a relative underexpression (left side) and overexpression (right side). Color bars represent differentially expressed regions identified in the HCL versus lymph node (green), HCL versus normal spleen (red), and normal spleen versus normal lymph node (blue) experiments. Each bar marks an averaged region with differential expression recorded in at least 25% (3 of 12) of analyzed cases. Dotted, regular, and thick bars point to regions with altered expression detected using the 90%, 95%, and 99% CL, respectively. The shown level of differential expression (90%, 95%, and 99% CL) is representative for at least 25% of informative cases.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/1/10.1182_blood-2004-01-0181/6/m_zh80130463640002.jpeg?Expires=1767722374&Signature=xKtoc~VKEYRO1DSLHxw~weUKbEjT87yjzIdyv6XyYMh~kF8052R7AbnMPRvc1Pg0QOtAmKS7~0ktWO2UHuZTov3TjFdE6YCiH4dTfIGjQO43GcEaMCbeMokLiIEBCDDOa7Fk8VF~ZuhJ~XJbPS-8atSTUeIBLk2gRhNHAW-ZV1S4rxejUW2fOn0jk7-2mMs6JW0Kk32zn5avrbVxdHjjmEskNKmBVa2vKoTXOI4Q02L9tRyskwz0d6XqtzLSziOqW0zRdH39fC1sqzDqlIQeAW9KbpWND2TwV9riZY1W4XwIfCMrrLvs~9TQ5Zb4NTPYLe5zT20iuq1tFyb0Kp-eyw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal