Abstract
The early stages of human lymphopoiesis are poorly characterized. Here, we compared the lymphoid potential of a novel umbilical cord blood CD34+CD45RAhiCD7+ hematopoietic progenitor cell (HPC) population with that of CD34+CD45RAhiLin-CD10+ HPCs, previously proposed as candidate common lymphoid progenitors. Limiting-dilution and clonal analysis, fetal thymic organ cultures, and culture onto Notch ligand Delta-like-1-expressing OP9 cells, showed that although CD34+CD45RAhiCD7+ HPCs could generate cells of the 3 lymphoid lineages, their potential was skewed toward the T/natural killer (T/NK) lineages. In contrast, CD34+CD45RAhiLin-CD10+ HPCs predominantly exhibited a B-cell potential. Gene expression profiling with DNA microarrays confirmed that CD34+CD45RAhiCD7+ HPCs selectively expressed T-lymphoid and NK lineage-committed genes while retaining expression of genes affiliated to the granulomonocytic lineage, whereas CD34+CD45RAhiLin-CD10+ HPCs displayed a typical pro-B-cell transcription profile and essentially lacked genes unrelated to the B lineage. In addition, both populations could be generated in vitro from CD34+CD45RAintCD7- and CD34+CD45RAhiLin- HPCs with mixed lymphomyeloid potential, from which they emerged independently with different growth/differentiation factor requirements. These findings indicate that CD34+CD45RAhiCD7+ and CD34+CD45RAhiLin-CD10+ HPCs correspond to multipotent early lymphoid progenitors polarized toward either the T/NK or B lineage, respectively. (Blood. 2004;104: 3918-3926)
Introduction
The immediate progeny of pluripotent hematopoietic stem cells is thought to correspond to common myeloid progenitors (CMPs) and common lymphoid progenitors (CLPs). CMPs are assumed to give rise to granulocytes and macrophages, as well as to the erythroid and megakaryocytic lineages, whereas CLPs are committed to generate either B lymphocytes (BLs) or T lymphocytes (TLs) and natural killer (NK) cells.1,2 Evidence for a primary segregation between CLPs and CMPs stems from in vivo transfer experiments in adult mice, where 2 populations of c-KitloScaloIL-7R+ and FcγRloCD34+ hematopoietic progenitor cells (HPCs) isolated from the postnatal bone marrow (BM) were shown to selectively reconstitute either the lymphoid3 or erythromegakaryocytic and granulomonocytic lineages.4 Such a dichotomous model of hematopoiesis remains however debated because there is also evidence that multilineage precursors coexpress lymphoid as well as myeloerythroid genes5-7 and that populations of early lymphoid progenitors (ELPs) retain some degree of multipotency.8-11 For example, AA4.1+FcγR+ fetal precursors with TL and BL potential retain significant macrophage potential but fail to generate erythroid or granulocytic cells.8 In line with these findings, early GFPloc-kithiSca-1+ BL precursors from RAG1/GFP (recombination activating gene 1/green fluorescent protein) knock-in mice still express TL and macrophage potential when cultured under appropriate conditions.11 Lineage relationships among lymphoid progenitors also remain poorly characterized, because it has been proposed that ELPs subsequently differentiate into bipotent precursors of TLs and BLs12 or further segregate into biased CLP-B or CLP-T populations.13 As to early T-lineage progenitors, although the immediate B220+c-Kit- progeny of murine c-KitloScaloIL-7R+ HPCs has been shown to be present among the CD44+CD25- thymic double negative-1 (DN1) subset, their contribution to postnatal murine thymopoiesis remains controversial because ikaros-deficient mice, which lack this population, have normal numbers of early thymic progenitors.12,14-16 Finally, that TL/NK cell and BL progenitors emerge independently during mouse embryogenesis, in the absence of prototypic CLPs,17-20 strongly suggests that fetal and postnatal lymphopoiesis correspond to largely independent processes, adding further complexity to the picture.21
Only few reports address the ontogenetic relationships of human early lymphoid progenitors which remain poorly characterized.22,23 To date, 2 CD34+ HPC populations with apparently lymphoid-restricted potential have been proposed as candidate CLPs: CD45RAhiLin-CD10+ cells from the BM and CD45RA+CD38-CD7+ cells from the umbilical cord blood (UCB).24,25 Both were identified on the basis of reduced capacity to generate granulomonocytic and erythroid cells, and substantial T-cell, B-cell, and NK cell differentiation potential. However, the lack of comparative analysis of their intrinsic capacity to generate either BLs or TLs/NK cells hampers definitive conclusion, and the hypothesis that these populations actually correspond to progenitors already polarized T/NK or B lineages has to be considered. We have identified in the UCB a novel population of CD34+CD45RAhiCD7+ HPCs with lymphoid potential that comprises bipotent NK and dendritic cell precursors and shares with CD34+CD1a- postnatal thymocytes the capacity to differentiate into Langerhans cells via a transforming growth factor β1 (TGF-β1)-independent pathway.26 Here, UCB CD34+CD45RAhiCD7+ and CD34+CD45RAhiLin-CD10+ HPCs were compared for their lymphoid potentials and gene expression profiles.25,26 We show that, although both populations retain some degree of multipotency, the CD34+CD45RAhiCD7+ HPCs are biased toward the TL/NK cell lineage, whereas the CD34+CD45RAhiLin-CD10+ HPCs correspond to pro-B cells.
Material and methods
Isolation and immunolabeling of UCB and postnatal BM CD34+ cells
Normal UCB (Laboratoire Sender, Hôpital Saint-Vincent de Paul; Service de Gynécologie-Obstétrique, Hôpital Saint-Antoine; Maternité, Hôpital Robert Debré; Paris, France) and BM cells obtained from healthy BM graft donors were both collected according to institutional guidelines and processed in the same manner. Cord blood samples were collected only after informed consent was provided according to the Declaration of Helsinki. After Pancoll (Dutscher, Brumath, France) centrifugation, CD34+ cells, enriched to equal 85% with the midiMACS system (Miltenyi Biotech France, Paris, France), were incubated for 30 minutes at 4°C in phosphate-buffered saline (PBS), 2% fetal calf serum (FCS; Dutscher), with CD34-phycoerythrocyanin 5 (PECy5) or CD34-allophycocyanin (APC) (clone 581), CD7-fluorescein isothiocyanate (FITC) (clone 8H8.1) (all from Beckman Coulter, Villepinte, France), CD45RA-phycoerythrin (PE) (clone HI100; BD Pharmingen, Le Pont de Claix, France), or CD10-PECy5 (clone A1B1; Beckman Coulter) monoclonal antibodies (mAbs) diluted 1:50 final. Cells were sorted based on CD7, CD45RA, and CD10 expression (purity > 93%), using a FACS (fluorescence activated cell sorting) Vantage (Becton Dickinson, Mountain View, CA). To compare CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPC differentiation potential, pre-B cells were depleted to less than 10% from CD34+ HPCs with CD19 mAb-coated magnetic Dynabeads (Dynal, Oslo, Norway) prior to sorting.
For phenotypic analysis, CD34+ cells were incubated for 30 minutes at 4°C with mAbs (1:100 final) in PBS, 2% FCS, washed, and analyzed with a FACscalibur (Becton Dickinson). Cells were labeled with PE-, PECy5-, or APC-conjugated CD34 mAb clone 581 and the following mAbs: CD10-PECy5, CD19-FITC (clone HIB19) or CD38-APC (clone HIT2; BD Pharmingen), CD127-PE (clone R34.34; Beckman Coulter). Isotype-matched FITC-, PE-, PECy5- and APC-conjugated irrelevant mAbs were from BD Pharmingen and Beckman Coulter.
Assessment of NK cell and BL differentiation potential in bulk cultures
NK cell differentiation was assessed by growing sorted CD34+ HPC subsets at 37°C in a humidified 5% CO2 atmosphere, in RPMI 1640, 10% FCS, 1% glutamine, 1% antibiotics (GIBCO BRL, Life Technologies, Cergy-Pontoise, France), with human recombinant cytokines: 50 ng/mL stem cell factor, 50 ng/mL Flt-3 ligand (FL; both from R&D Systems, Abingdon, United Kingdom), 100 U/mL interleukin 2 (IL-2), 20 ng/mL IL-7 and IL-15 (all from Tebu, Le Perray en Yvelines, France). Cultures were conducted for 3 weeks in 24- or 48-well plates (Dutscher) with or without murine MS5 stromal cells,27 with half changes and cytokine addition every 6 to 7 days. Before culture initiation, MS5 cells were seeded in gelatin-coated plates and grown for 24 to 48 hours in complete alpha-Minimum Essential Media (MEM; GIBCO BRL), 10% FCS, 1% antibiotics. BL differentiation was assessed by growing cells for 2 weeks in the MS5 cell-coated plates, in RPMI 1640, 3% FCS, 1% glutamine, 1% antibiotics, 50 μM β-mercaptoethanol, plus 50 ng/mL stem cell factor (SCF), 100 ng/mL thrombopoietin (TPO), and 20 ng/mL IL-7 (both from Tebu). Cells were collected and labeled with CD56-PE (clone B159) and CD19-FITC (clone HIB19) (both from BD Pharmingen) or CD19-PE (clone J4.119; Coulter Immunotech, Marseilles, France) mAbs to detect NK cells and BLs, respectively.
Clonogenic assays
Erythroid and granulomacrophagic potential of each CD34+ HPC subset was assessed by seeding 500 cells in 35-mm duplicate dishes (Dutscher), in 1.1 mL complete Methocult GF+ H4435 medium (Stem Cell Technologies, Meylan, France), supplemented with IL-3, IL-6, granulocyte colony-stimulating factor (G-CSF), SCF, granulocyte-macrophage colony-stimulating factor (GM-CSF), and erythropoietin (EPO). Dishes were incubated at 37°C in humidified 5% CO2. Erythroid burst-forming colonies (BFU-E), granulocyte (CFU-G/GM) and macrophage (CFU-M) colony-forming units were counted under an inverted microscope on culture day 14, as reported.28
Limiting-dilution assays and single-cell cultures
To assess NK cell progenitor frequency, cells were seeded at 300, 100, 30, 10, 3, and 1 cells/well into MS5 cell-coated 96-well plates (ATGC; Marne la Vallée, France) and cultured for 3 weeks in RPMI 1640, 10% human AB serum (Etablissement Français du Sang, Paris, France), 5% FCS, 50 μM β-mercaptoethanol, supplemented with SCF, FL, IL-2, IL-7, IL-15 as described in “Clonogenic assays.” BL progenitors were assessed by growing the cells, as described in “Assessment of NK cell and BL differentiation potential in bulk cultures,” with SCF, TPO, and IL-7, in RPMI 1640, 3% FCS, 50 μM β-mercaptoethanol. Cultures lasted 2 to 3 weeks with half medium changes and fresh cytokines added every 6 to 7 days. Plates were examined weekly and cell-containing wells were scored under the microscope. At the end of cultures, BLs and NK cells were identified by CD19 and CD56 expression. The maximum likelihood estimate of NK cell or BL precursors was calculated according to the single-hit Poisson model. For single-cell suspension cultures, cells were individually plated by FACS onto MS5-coated 96-well plates and cultured in RPMI 1640, 10% human AB serum, 5% FCS, 50 μM β-mercaptoethanol, supplemented with SCF, TPO, IL-2, and IL-7. Cell-containing wells were scored on days 19 to 21, and NK cells, BLs, and macrophagic cells were detected by labeling with CD56, CD19, and CD14 (clone M5E2; BD Pharmingen) mAbs.
Assessment of TL differentiation potential
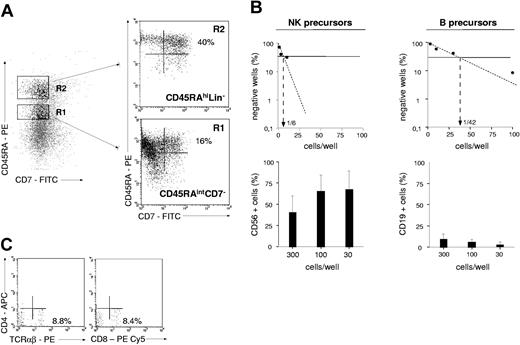
TL differentiation potential of sorted CD34+ HPC subsets was first assessed in fetal thymic organ cultures (FTOCs) essentially performed as reported.29,30 Briefly, thymic lobes were collected from days 14 to 15 NOD/SCID (nonobese diabetic/severe combined immunodeficiency) mice embryos. Hanging drops were prepared in Terasaki plates (Polylabo, Strasbourg, France) by adding 25 μL RPMI 1640, 10% human AB serum, 5% FCS, 50 ng/mL SCF, 5 ng/mL IL-2, and 20 ng/mL IL-7, with 0.2 to 2.5 × 104 cells/lobe (2-5 lobes/condition). Plates were immediately inverted and incubated for 48 hours in a humidified incubator before lobes were removed, washed, transferred onto floating nucleopore filters (Millipore SA, Molsheim, France) in 6-well plates, and cultured for 2 to 4 weeks. Lobes were subsequently removed, human cells were recovered by mechanical disruption, pooled, and labeled with CD45-FITC, CD4-PECy5, CD8-APC or CD8-PE-Cy5, and T-cell receptor αβ (TCRαβ)-PE (all from Beckman Coulter) and/or with CD4-APC (BD Pharmingen), before FACS analysis. Alternatively, cells were cocultured onto OP9 murine stromal cells expressing murine Notch ligand Delta-like-1 (OP9-DL1), which were produced by retroviral-mediated gene transfer with a bicistronic Delta-like-1-IRES-EGFP construct kindly provided by A. Cumano (Institut Pasteur, Paris, France).31 OP9-DL1 cells were seeded in gelatin-coated 48-well plates and cultured for 24 to 48 hours before initiating cocultures in Dulbecco modified Eagle medium (DMEM; GIBCO BRL), 10% FCS, 1% antibiotics. Cocultures of sorted CD34+ HPCs and OP9-DL1 cells were conducted for 2 to 4 weeks in RPMI 1640, 10% human AB serum, 5% FCS, 50 μM β-mercaptoethanol, supplemented with SCF, FL, IL-2, IL-7, IL-15. Media and cytokines were renewed every 6 to 7 days.
In vitro generation of CD45RAhiCD7hi and CD45RAhiLin-CD10+ HPCs
Purified CD34+ HPCs were labeled with CD34-APC, CD45RA-PE, CD10-PECy5, FITC-conjugated CD7 and CD19 mAbs as previously described. CD45RAintCD7- and CD45RAhiLin- populations were sorted and cultured onto MS5 cells in 48-well plates (Dutscher) in RPMI 1640, 10% FCS, 1% glutamine, 1% antibiotics, 50 μM β-mercaptoethanol. Cultures were conducted for 4 to 6 days without media change in the presence of 50 ng/mL FL, with or without 50 ng/mL recombinant human thymic stromal lymphopoietin (TSLP; R&D Systems).
Microarray sample preparation
Total RNA from CD45RAintCD7- (biologic triplicates), CD45RAhiCD7+ (biologic triplicates), and CD45RAhiLin-CD10+ HPC populations were extracted using the RNeasy Mini Kit (Qiagen, Hildren, Germany). Only biologic duplicates were available for CD45RAhiLin-CD10+ HPCs; one sample obtained from an individual sorting experiment, the other corresponding to a total RNA pool of 3 independent experiments. RNA quality control and quantification were monitored by spectrophotometry and capillary electrophoresis using the RNA 6000 Pico LabChip Kit and the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA samples (starting amounts, 15-25 ng) were subjected to double amplification using the GeneChip Eukaryotic Small Sample Target Labeling Assay Version II (Affymetrix, Santa Clara, CA). The resulting cRNAs were controlled and quantified using the Agilent 2100 Bioanalyzer. Amplification efficiency was monitored by hybridization to probe sets designed to 3′, middle, and 5′ regions of housekeeping genes (Test3 Array; Affymetrix). cRNA (10 μg) was hybridized to Affymetrix HG-U133A GeneChip arrays according to the manufacturer's protocol. Quantitative scanning was performed using an Agilent GeneArray Scanner 2500A (Agilent Technologies). Probe set annotations for Affymetrix HG-U133A GeneChip arrays were downloaded from the Affymetrix website and updated on August 18, 2003. Affymetrix GeneChip probe level data and background correction were obtained using the log scale robust multiarray analysis based on a log scale linear additive model available in Affy package 1.3.6 under Bioconductor/R 1.7.132 (http://www.bioconductor.org; http://www.r-project.org). Quantile normalization, “perfect match-only” model, and calculation of expression measures using median polish were considered, corresponding to the rma function available in the Affy package.33
Microarray data analysis
HPC populations were first compared 2 by 2: CD45RAintCD7- versus CD45RAhiCD7+, CD45RAhiLin-CD10+ versus CD45RAintCD7-, and CD45RAhiLin-CD10+ versus CD45RAhiCD7+. Because of the limited number of replicates, the local pooled error (LPE) test was used for this purpose.34 Criteria used to consider genes as differentially expressed were the following: a fold change (FC) more than 1.3, a raw P value less than .05 according to the LPE test, an adjusted P value less than .10 (false discovery rate < 10%) according to Benjamini and Hochberg's procedure for multiple comparison adjustment,35 and a minimal average signal intensity for each considered probe sets above 70 in at least 1 of the 2 compared groups. To identify genes discriminating the 3 HPC populations, the classification analysis module of Array Miner 4.1 (Optimal Design, Brussels, Belgium), ClassMarker, was used. For this purpose, the complete dataset of the 22 283 probe sets obtained with the robust multiarray average (rma) function of Affy package was introduced in ClassMarker and filtered according to the following criteria: maximal threshold, 16 000; minimal fold change, 2.0; minimal absolute change, 100. Filtered data (n = 3640 probe sets) were subsequently introduced in a cross-validation analysis, and only probe sets with a signal-to-noise ratio more than 2.00 were considered significantly associated with a cell population.
Results
Identification of UCB lymphoid progenitors according to CD45RA and CD7 expression levels
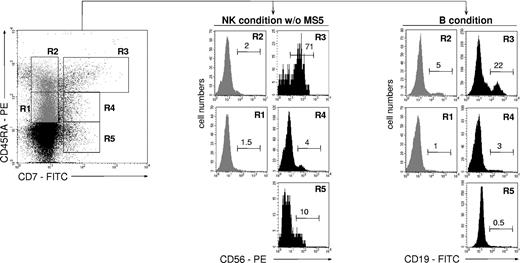
To compare the lymphoid potential of UCB CD34+CD45RAhiCD7+ HPCs26 with that of 4 other CD34+ cell populations, CD34+ cells were sorted according to CD45RA and CD7 expression levels and tested for NK cell and BL potentials (Figure 1). Only the CD45RAhiCD7+ HPCs exhibited then a strong capacity to generate NK cells, with 70% ± 18% (mean ± SD, n = 3) CD56+ cells after 2-week culture with SCF, FL, IL-2, IL-7, and IL-15 (referred to as NK condition without MS5 thereafter), albeit with limited expansion (6- to 30-fold). These data were confirmed by culturing the CD45RAhiCD7+ cells with the same cytokine combination onto MS5 cells (referred to as NK condition thereafter).36 Adding the stromal layer increased growth rates to 112- to 130-fold, with up to 95% CD56+ NK cells (data not shown). In parallel, the 5 cell populations were cultured for 2 weeks on MS5 cells with SCF, TPO, and IL-7 to assess their differentiation capacity into BLs (referred to as B condition thereafter).37 Again, the CD45RAhiCD7+ HPCs displayed the greatest potential, newly generated CD19+ BLs representing then 27% ± 17% (n = 6) of cells relative to equal 5% yields for the other populations. These data indicate that, of all the populations tested, only CD45RAhiCD7+ HPCs display mixed NK cell and BL differentiation potential.
NK cell and BL differentiation potential of UCB CD34+ HPC populations. Purified UCB CD34+ cells were sorted into CD45RAintCD7- (R1), CD45RAhiCD7- (R2), CD45RAhiCD7+ (R3), CD45RAintCD7+ (R4), and CD45RA-CD7+ (R5) populations and cultured for 21 days with SCF, FL, IL-2, IL-7, and IL-15 (NK condition without MS5). Alternatively, cells were seeded onto MS5 cells with SCF, TPO, and IL-7 and cultured for 14 days (B condition). At the end of the cultures, cells were harvested and labeled with CD56-PE and CD19-FITC mAbs before FACS analysis: histograms and the indicated percentages of specifically labeled cells are based on control mAb labeling. In vitro-generated NK cells were homogeneously CD8+CD56+, whereas a minority of CD10+CD19+ BLs coexpressed CD20. At culture initiation none of the populations tested contained CD56+ cells; CD34+CD19+ pre-B cells represented 6% ± 2% (n = 3) of sorted CD45RAhiCD7- (R2) cells. Data are from 1 of 2 experiments.
NK cell and BL differentiation potential of UCB CD34+ HPC populations. Purified UCB CD34+ cells were sorted into CD45RAintCD7- (R1), CD45RAhiCD7- (R2), CD45RAhiCD7+ (R3), CD45RAintCD7+ (R4), and CD45RA-CD7+ (R5) populations and cultured for 21 days with SCF, FL, IL-2, IL-7, and IL-15 (NK condition without MS5). Alternatively, cells were seeded onto MS5 cells with SCF, TPO, and IL-7 and cultured for 14 days (B condition). At the end of the cultures, cells were harvested and labeled with CD56-PE and CD19-FITC mAbs before FACS analysis: histograms and the indicated percentages of specifically labeled cells are based on control mAb labeling. In vitro-generated NK cells were homogeneously CD8+CD56+, whereas a minority of CD10+CD19+ BLs coexpressed CD20. At culture initiation none of the populations tested contained CD56+ cells; CD34+CD19+ pre-B cells represented 6% ± 2% (n = 3) of sorted CD45RAhiCD7- (R2) cells. Data are from 1 of 2 experiments.
CD45RA-CD7-, CD45RAintCD7-, CD45RAhiCD7-, and CD45RAhiCD7+ HPCs were then tested for the capacity to form erythroid and granulomonocytic colonies (Table 1). After 2-week culture in methylcellulose with IL-3, IL-6, G-CSF, SCF, GM-CSF, and EPO, only CD45RA-CD7- HPCs displayed significant erythroid potential, which was consistent with their capacity to generate megakaryocytes in liquid cultures with SCF and TPO (data not shown). Conversely, only the CD34+CD45RA+ HPC populations comprised macrophage precursors. Of note, the CD45RAintCD7- HPCs were highly enriched in CFU-G/GM. These data confirm a report that erythroid potential segregates with the CD45RA- fraction of UCB CD34+ HPCs,38 and they indicate that acquisition of lymphoid potential by CD45RAhiCD7+ HPCs correlates with a decreased capacity to form both erythroid and granulocytic colonies.
Assessment of erythroid and granulomacrophagic potential of UCB CD34+CD45RA+/−CD7+/− HPC populations
. | CD34+HPC populations . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | CD45RA−CD7− . | . | CD45RAintCD7− . | . | CD45RAhiCD7− . | . | CD45RAhiCD7+ . | . | |||||||
. | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | |||||||
| BFU-E | 94 | 96 | 21 | 19 | 5 | 3 | 5 | 9 | |||||||
| CFU-G/GM | 69 | 74 | 113 | 104 | 59 | 72 | 35 | 38 | |||||||
| CFU-M | 5 | 5 | 21 | 19 | 27 | 45 | 29 | 35 | |||||||
. | CD34+HPC populations . | . | . | . | . | . | . | . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | CD45RA−CD7− . | . | CD45RAintCD7− . | . | CD45RAhiCD7− . | . | CD45RAhiCD7+ . | . | |||||||
. | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | |||||||
| BFU-E | 94 | 96 | 21 | 19 | 5 | 3 | 5 | 9 | |||||||
| CFU-G/GM | 69 | 74 | 113 | 104 | 59 | 72 | 35 | 38 | |||||||
| CFU-M | 5 | 5 | 21 | 19 | 27 | 45 | 29 | 35 | |||||||
Results are from 2 independent experiments. Exp indicates experiment.
CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs differ in the capacity to generate NK cells and BLs
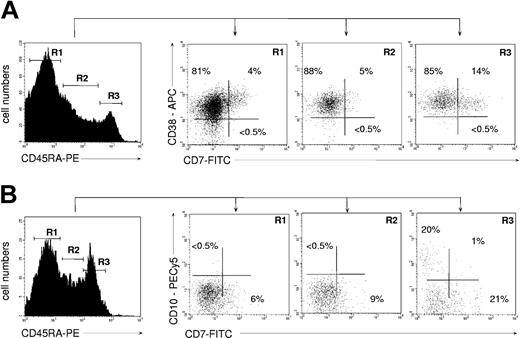
Inasmuch as the above data suggested that CD45RAhiCD7+ HPCs may correspond to a CLP population, we examined their ontogenetic relationship with previously reported CD45RAhiLin-CD10+ and CD38-CD45RA+CD7+ candidate CLPs.24,39 FACS analysis of UCB CD34+ HPCs showed that the CD45RAhiCD7+ HPCs were homogeneously CD38+ (Figure 2A). In our hands, no CD7-expressing cell was detected among UCB CD34+CD38- HPCs, precluding further analysis of the CD38-CD45RA+CD7+ population.24 Inasmuch as CD38-CD45RA+CD7+ HPCs have been shown to lack granulomonocytic potential, it can be assumed that they do not overlap with CD45RAhiCD7+ HPCs. Quadruple CD34-PECy5, CD45RA-PE, CD7-FITC, and CD10-PECy5 mAb labeling showed that only a minority of CD34+CD45RAhi cells coexpressed CD7 and CD10 (Figure 2B). That the CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPC populations were independent was confirmed by the fact that only the latter HPCs homogeneously expressed CD127/IL-7R-α, which was barely detectable on CD45RAhiCD7+ HPCs (data not shown). Interestingly, based on CD7, CD19, CD38, and CD127 expression, UCB CD45RAhiLin-CD10+ HPCs were undistinguishable from their postnatal BM homologues.25 Finally, CD45RAhiLin-CD10+ HPCs were found to display similar granulomacrophagic and erythroid potentials than CD45RAhiCD7+ HPCs in methylcellulose assays (data not shown).
Immunophenotype of UCB CD34+ HPCs according to CD45RA expression levels. (A) CD38 and CD7 expression by CD45RA- (R1), CD45RAint (R2), and CD45RAhi HPCs (R3). (B) CD10 and CD7 expression by CD45RA- (R1), CD45RAint (R2), and CD45RAhi HPCs (R3). Prior to labeling, the CD34+ HPCs were depleted to less than 10% CD19+ pre-B cells. In all cases cells were gated based on expression of CD34 marker (data not shown). Data are from 1 representative experiment of 4.
Immunophenotype of UCB CD34+ HPCs according to CD45RA expression levels. (A) CD38 and CD7 expression by CD45RA- (R1), CD45RAint (R2), and CD45RAhi HPCs (R3). (B) CD10 and CD7 expression by CD45RA- (R1), CD45RAint (R2), and CD45RAhi HPCs (R3). Prior to labeling, the CD34+ HPCs were depleted to less than 10% CD19+ pre-B cells. In all cases cells were gated based on expression of CD34 marker (data not shown). Data are from 1 representative experiment of 4.
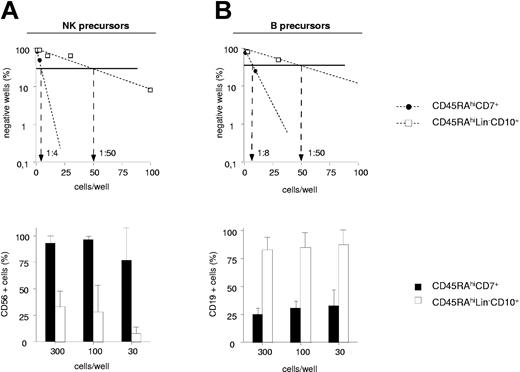
UCB CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs were then compared for the capacity to generate NK cells and BLs in limiting-dilution assays (Figure 3A-B). Because these populations display low cloning efficiency in the absence of feeder cells (data not shown), assays were performed on MS5 cells. In 2 independent experiments, NK cell precursor frequency varied between 1:4 and 1:10 among CD45RAhiCD7+ HPCs relative to 1:50 and 1:100 for CD45RAhiLin-CD10+ HPCs. The 2 populations also markedly differed regarding the intrinsic capacity to generate NK cells. As shown in Figure 3A, CD56+ NK cell percentages were always equal to 77% in wells seeded with 300 to 30 CD45RAhiCD7+ HPCs relative to 35% in CD45RAhiLin-CD10+ HPC cultures. Because of greater overall cloning efficiency under the B condition, 5- to 16-fold greater BL precursor frequency was also noted among CD45RAhiCD7+ HPCs (1:8 and 1:10 versus 1:50 and 1:60), but CD45RAhiLin-CD10+ HPCs displayed stronger BL potential, with 75% CD19+ cells in wells seeded with 300 to 30 cells versus less than 40% in those seeded with CD45RAhiCD7+ HPCs (Figure 3B).
NK cell and BL differentiation potential of CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs. Comparative analysis of CD45RAhiLin-CD10+ and CD45RAhiCD7+ HPCs in limiting-dilution assay cultures under the NK condition (A) or the B condition (A). Positive wells were scored and FACS analyzed after 2 (B condition) or 3 (NK condition) weeks of culture. Upper panels show NK cell (A) and BL precursor (B) frequencies; lower panels display mean percentages + SD of CD56+ and CD19+ cells per well. Differences of CD56+ and CD19+ cell percentages in wells from CD45RAhiLin-CD10+ versus CD45RAhiCD7+ HPC cultures seeded with 300 to 10 and 300 to 30 cells per well under the NK and B condition, respectively, were statistically significant (P = .01; Student t test). Results are representative of 1 experiment of 2 under each culture condition.
NK cell and BL differentiation potential of CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs. Comparative analysis of CD45RAhiLin-CD10+ and CD45RAhiCD7+ HPCs in limiting-dilution assay cultures under the NK condition (A) or the B condition (A). Positive wells were scored and FACS analyzed after 2 (B condition) or 3 (NK condition) weeks of culture. Upper panels show NK cell (A) and BL precursor (B) frequencies; lower panels display mean percentages + SD of CD56+ and CD19+ cells per well. Differences of CD56+ and CD19+ cell percentages in wells from CD45RAhiLin-CD10+ versus CD45RAhiCD7+ HPC cultures seeded with 300 to 10 and 300 to 30 cells per well under the NK and B condition, respectively, were statistically significant (P = .01; Student t test). Results are representative of 1 experiment of 2 under each culture condition.
To confirm these data, we established a single-cell clonal assay allowing for simultaneous differentiation of NK cells and BLs (referred to as B/NK condition thereafter). After 3-week culture, 43% NK cell clones, 24% mixed BL/NK cell clones, and only 7% BL clones were recovered from positive wells seeded with CD45RAhiCD7+ HPCs (Table 2). Again, CD45RAhiLin-CD10+ HPCs gave an inversed pattern with 63% BL clones, 9% NK clones, and no mixed BL/NK cell clones. Double-negative CD19-CD56- clones, noted irrespective of the population used, comprised a majority of CD14+ cells (data not shown).
Clonal analysis of BL and NK cell differentiation potential of CD45RAhiCD7+ and CD45RAhiLin−CD10+ HPCs
. | CD34+HPC populations . | . | |
|---|---|---|---|
. | CD45RAhiCD7+ . | CD45RAhiLin−CD10+ . | |
| No. cells seeded | 418 | 148 | |
| Total no. of clones | 58 | 11 | |
| NK clones, % | 43 | 9 | |
| B clones, % | 7 | 63 | |
| Mixed B/NK clones, % | 24 | 0 | |
| CD19−CD56− clones, % | 26 | 28 | |
. | CD34+HPC populations . | . | |
|---|---|---|---|
. | CD45RAhiCD7+ . | CD45RAhiLin−CD10+ . | |
| No. cells seeded | 418 | 148 | |
| Total no. of clones | 58 | 11 | |
| NK clones, % | 43 | 9 | |
| B clones, % | 7 | 63 | |
| Mixed B/NK clones, % | 24 | 0 | |
| CD19−CD56− clones, % | 26 | 28 | |
Cells were seeded individually in MS5 cell-precoated wells and cultured for 3 weeks with SCF, TPO, IL2, and IL7. Clones were then harvested, labeled with CD56-PE and CD19-FITC mAbs, and analyzed by FACS. Data are pooled from 2 independent experiments.
These findings indicate that CD45RAhiCD7+ HPCs are enriched in progenitors with strong NK potential, whereas CD45RAhiLin-CD10+ HPCs comprise a majority of BL progenitors.
CD45RAhiCD7+ HPCs display enhanced TL differentiation potential
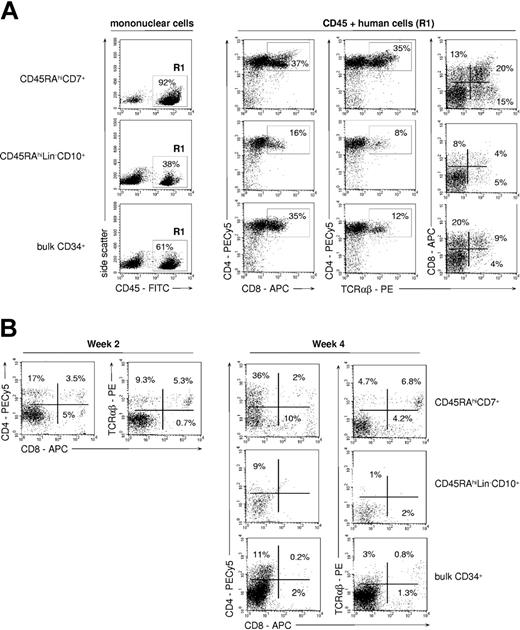
FTOCs were used to assess whether the differences noted between CD45RAhiLin-CD10+ and CD45RAhiCD7+ HPCs extended to the capacity to generate TLs (Figure 4A). Only CD45RAhiCD7+ HPCs displayed strong TL potential, with CD4+CD8+/- TCRαβ+ TLs representing about 35% of human cells recovered from thymic lobes. Only few mature CD4+CD8- TCRαβhi TLs were recovered from cultures initiated with CD45RAhiLin-CD10+ or bulk CD34+ HPCs (< 5% of human cells). Of note, CD45RAintCD7- cells lacked detectable TL potential (data not shown). Accordingly, human cell yields were 6- to 7-fold greater in thymic lobes seeded with CD45RAhiCD7+ HPCs than with CD45RAhiLin-CD10+ HPCs or bulk CD34+ cells. The difference was even more pronounced when considering CD4+CD8+ TCRαβ- thymocytes (9- to 15-fold increase) and CD4+CD8- TCRαβhi mature TLs (35- to 50-fold increase).
TL differentiation potential of CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs. (A) FTOC assay: sorted CD45RAhiCD7+, CD45RAhiLin-CD10+, and bulk CD34+ cells were cultured for 4 weeks in NOD/SCID mouse fetal thymic lobes before cells were recovered and analyzed by FACS. Only CD45+ (R1) human cells were further analyzed. Percentages of positive cells are indicated; data are from 1 of 5 experiments. (B) Coculture with OP9-DL1 cells: CD34+ HPC populations were cultured for 2 or 4 weeks onto OP9-DL1 cells with SCF, FL, IL-2, IL-7, and IL-15, before FACS analysis. Percentages of positive cells are indicated; data are from 1 of 4 experiments.
TL differentiation potential of CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs. (A) FTOC assay: sorted CD45RAhiCD7+, CD45RAhiLin-CD10+, and bulk CD34+ cells were cultured for 4 weeks in NOD/SCID mouse fetal thymic lobes before cells were recovered and analyzed by FACS. Only CD45+ (R1) human cells were further analyzed. Percentages of positive cells are indicated; data are from 1 of 5 experiments. (B) Coculture with OP9-DL1 cells: CD34+ HPC populations were cultured for 2 or 4 weeks onto OP9-DL1 cells with SCF, FL, IL-2, IL-7, and IL-15, before FACS analysis. Percentages of positive cells are indicated; data are from 1 of 4 experiments.
To examine whether the T-lineage potential of CD45RAhiCD7+ HPCs correlated with a functional Notch signaling pathway, the same populations were assayed in cultures onto OP9-DL1 cells.31 As expected, only CD45RAhiCD7+ cells displayed then significant TL potential, albeit with limited 3- to 5-fold expansion (Figure 4B). After 2-week culture, 3.5% CD4+CD8+ TCRαβ- thymocytes and 15% TCRαβhi TLs were detected, with up to 11% mature TCRαβhi TLs on culture week 4. Of note, TCRαβhi TLs were there predominantly CD4-CD8+.31 CD45RAintCD7- and CD45RAhiLin-CD10+ HPCs did not differentiate into TLs on OP9-DL1 cells, bulk CD34+ HPCs expressing only marginal TL potential. Thus, only the CD45RAhiCD7+ HPCs respond to Notch ligand Delta-like 1 ligation.
Gene expression profiling of CD45RAhiCD7+ and CD45RAhiLin-CD10+ progenitors
DNA microarrays were used to determine CD45RAhiCD7+, CD45RAhiLin-CD10+, and CD45RAintCD7- HPC gene expression profiles. CD45RAintCD7- and CD45RAhiCD7+ HPCs displayed very close transcriptional profiles. Only 166 probe sets, representing 101 genes (complete coding sequences [cds] or mRNA sequences) and 62 consensus sequences, were differentially expressed in the 2 populations. The complete lists of these genes may be found in supporting information Tables S1 and S2 at the Blood website (see the Supplemental Materials link at the top of the online article). As expected from their biologic phenotype, TL lineage-committed genes CD7, CCR9, TRG, TRGC2, TRGV9, DNTT/TDT as well as the NK cell lineage-committed gene TYROBP/DAP12 were expressed to higher levels in CD45RAhiCD7+ cells. Relative to CD45RAintCD7- HPCs, CD45RAhiCD7+ cells also overexpressed BL lineage-committed genes BLNK, CD24, IGJ, CD10/MME, VPREB1. The coregulated serprocidins (AZU1, CTSG, PRTN3) as well as MPO, CSF1R, LYZ, ELA2, HCK, S100A8, CCL3, and CEBPD, which are usually expressed in myeloid cells, were also differentially expressed in CD45RAhiCD7+ cells. Comparatively, CD45RAintCD7- HPCs displayed a more immature gene transcription profile with selective expression of stem cell-associated transcription factors TAL1, GATA3, HLF, and MLLT3.40,41
CD45RAhiLin-CD10+ HPCs were more distantly related to CD45RAhiCD7+ (1200 probe sets differentially expressed) and CD45RAintCD7- (1368 probe sets differently expressed) HPCs, with 857 common probe sets being differentially expressed in both comparisons, ie, CD45RAhiLin-CD10+ versus CD45RAintCD7- HPCs and CD45RAhiLin-CD10+ versus CD45RAhiCD7+ HPCs (Table S3). They expressed higher levels of transcripts for B-cell receptor components (IGLL1, CD79A, CD79B), BL-specific surface markers (CD10, CD19, CD22), or signaling molecules (BLK, BLNK), as well as transcription factors involved in BL lineage commitment (E2A/TCF3, PAX5, POU2AF1), which confirms that they correspond to prototypic pro-B cells. Regarding genes expressed in the TL or NK cell lineages, CD45RAhiLin-CD10+ HPCs displayed lower expression of TRB, TRG, TRGC2, and TRGV9. The myeloid “signature” of CD45RAhiLin-CD10+ HPCs was less clear with only low expression of myeloid lineage-affiliated transcripts.
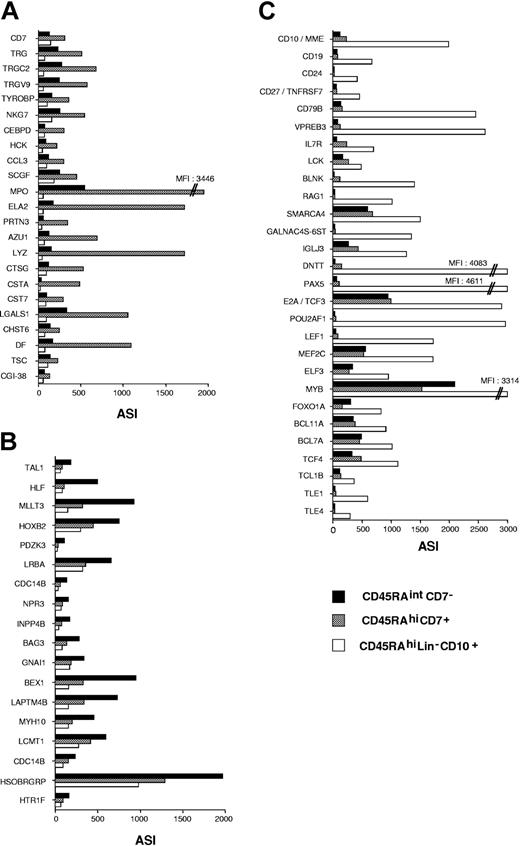
Class prediction analysis extended and validated CD45RAhiCD7+, CD45RAhiLin-CD10+, and CD45RAintCD7- HPC gene transcription profiles. Among the 381, 27, and 27 probe sets that discriminated the 3 populations, it confirmed that AZU1, CCL3, CD7, CTSG, ELA2, LYZ, MPO, PRTN3, TRG, TRGC2, TRGV9, NKG7, and TYROBP were CD45RAhiCD7+ specific, whereas HLF, TAL1, HOXB2, and MLLT3 were among CD45RAintCD7--specific genes, and CD10, CD19, CD79B, VPREB1, VRPEB3, RAG1, SMARCA4, GALNAC4S-6ST, IGLJ3, E2A/TCF3, PAX5, POUF2A1, TLE1, BCL7A, BCL11A, IL7R, SOCS2, and BLNK were among CD45RAhiLin-CD10+-specific genes (Figure 5A-C).
Supervised analysis of CD45RAhiCD7+, CD45RAintCD7-, and CD45RAhiLin-CD10+ HPC gene expression profiles. Genes discriminating between the 3 cell populations were selected by using the classification analysis module of Array Miner 4.1 (see “Materials and methods”) and were plotted according to average signal intensity (ASI). Relative expression levels of genes differentially expressed in (A) CD45RAhiCD7+, (B) CD45RAintCD7-, and (C) CD45RAhiLin-CD10+ HPCs. Among the 381 probe sets differentially expressed in CD45RAhiLin-CD10+ HPCs, only those corresponding to genes underlying polarization toward the B lineage are presented.
Supervised analysis of CD45RAhiCD7+, CD45RAintCD7-, and CD45RAhiLin-CD10+ HPC gene expression profiles. Genes discriminating between the 3 cell populations were selected by using the classification analysis module of Array Miner 4.1 (see “Materials and methods”) and were plotted according to average signal intensity (ASI). Relative expression levels of genes differentially expressed in (A) CD45RAhiCD7+, (B) CD45RAintCD7-, and (C) CD45RAhiLin-CD10+ HPCs. Among the 381 probe sets differentially expressed in CD45RAhiLin-CD10+ HPCs, only those corresponding to genes underlying polarization toward the B lineage are presented.
The 3 populations were finally examined for expression of known TL or NK cell markers, cytokine receptors, lymphoid signaling pathways, or transcription factors (data not shown). This disclosed that, whereas CD45RAhiCD7+ HPCs expressed low TRB levels, they lacked CD3 polypeptides, TRA, TRD, and PTCRA. Accordingly, RAG1 and RAG2 expression was restricted to CD45RAhiLin-CD10+ pro-B cells that also expressed IL7R and protein-tyrosine kinase LCK to higher levels. In addition, CD45RAhiLin-CD10+ cells selectively expressed transcription factor LEF1 as well as dominant-negative helix-loop-helix protein ID3.42 As to Notch signaling pathway, the 3 populations expressed Notch 1, Notch 2, and RBPSUH to similar levels, whereas DLK1 and HRY/HES1 were up-regulated in CD45RAhiCD7+ and CD45RAintCD7- HPCs.
Altogether, these results confirm the respective polarization of CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs toward the T/NK or B lineages, respectively, and they indicate that active transcription of TRG locus coincides with acquisition of TL potential.
In vitro generation of CD45RAhiCD7+ HPCs
The proximity of CD45RAhiCD7+ and CD45RAintCD7- HPCs disclosed by microarray analysis led us to investigate their ontogenetic relationship. Because CD45RAintCD7- HPCs lack NK potential when cultured in the absence of MS5 feeders (2% of CD56+ cells; Figure 1), we first examined whether coculture onto MS5 cells affected their capacity to generate NK cells. Hence, these cells were grown with SCF, FL, IL-2, IL-7, and IL-15, with or without MS5 cells. After 3 weeks, CD56+ cells represented 20% to 30% of total cells with, versus less than 5% without, MS5 cells, indicating that CD45RAintCD7- HPCs acquire substantial NK cell differentiation potential under these conditions. Whether CD45RAintCD7- cells represented precursors of CD45RAhiCD7+ HPCs was then examined. CD45RAintCD7- (R1) and CD45RAhiLin- (R2) HPC populations were thus sorted and cultured onto MS5 cells in the presence of FL (Figure 6A). After 4 to 6 days under these conditions, CD45RAintCD7- cells had become uniformly CD45RAhi and 16% ± 11% were CD7+ (n = 7), whereas CD45RAhiCD7+ cells represented up to 40% of CD45RAhiLin-CD7- HPC-derived cells (27% ± 19%; n = 4). In vitro-generated CD45RAhiCD7+ cells were then sorted and assessed for their lymphoid potential in limiting-dilution (Figure 6B). As for UCB-isolated counterparts, in vitro-generated CD45RAhiCD7+ cells expressed higher NK cell than BL potential, and they displayed substantial TL potential in coculture with OP9-DL1 cells (Figure 6C). Altogether these data indicate that CD45RAhiCD7+ HPCs differentiate from CD45RAintCD7- via a stepwise process.
Lymphoid differentiation potential of in vitro-generated CD45RAhiCD7+ HPCs. (A) Generation of CD45RAhiCD7+ HPCs from CD45RAintCD7- and CD45RAhiLin- HPCs: based on sorting gates set on CD34+CD7-CD19- cells, CD45RAintCD10- (ie, CD45RAintCD7-) and CD45RAhiCD10- (ie, CD45RAhiLin-) cells were sorted and cultured onto MS5 cells with FL for 4 to 6 days. Cells were gated based on high CD34 expression and analyzed by FACS for CD45RA and CD7. Percentages of labeled cells are indicated; data are from 1 of 3 experiments. (B) Analysis of NK cell and BL differentiation potentials of in vitro-generated CD45RAhiCD7+ HPCs: CD45RAintCD7- HPC-derived CD45RAhiCD7+ cells were sorted on culture day 6 and seeded in limiting-dilution under the NK or B conditions. Upper panels show NK cell and BL progenitor frequencies; lower panels display mean percentages and SD of CD56+ and CD19+ cells in the corresponding replicate cell-containing wells. Data are from 1 experiment of 2. (C) TL potential of CD45RAintCD7- HPC-derived CD45RAhiCD7+ cells: sorted CD45RAhiCD7+ cells were cocultured for 3 weeks with OP9-DL1 cells, as indicated in the legend of Figure 4B, before FACS analysis. Percentages of labeled cells are indicated; data are from 1 of 2 experiments.
Lymphoid differentiation potential of in vitro-generated CD45RAhiCD7+ HPCs. (A) Generation of CD45RAhiCD7+ HPCs from CD45RAintCD7- and CD45RAhiLin- HPCs: based on sorting gates set on CD34+CD7-CD19- cells, CD45RAintCD10- (ie, CD45RAintCD7-) and CD45RAhiCD10- (ie, CD45RAhiLin-) cells were sorted and cultured onto MS5 cells with FL for 4 to 6 days. Cells were gated based on high CD34 expression and analyzed by FACS for CD45RA and CD7. Percentages of labeled cells are indicated; data are from 1 of 3 experiments. (B) Analysis of NK cell and BL differentiation potentials of in vitro-generated CD45RAhiCD7+ HPCs: CD45RAintCD7- HPC-derived CD45RAhiCD7+ cells were sorted on culture day 6 and seeded in limiting-dilution under the NK or B conditions. Upper panels show NK cell and BL progenitor frequencies; lower panels display mean percentages and SD of CD56+ and CD19+ cells in the corresponding replicate cell-containing wells. Data are from 1 experiment of 2. (C) TL potential of CD45RAintCD7- HPC-derived CD45RAhiCD7+ cells: sorted CD45RAhiCD7+ cells were cocultured for 3 weeks with OP9-DL1 cells, as indicated in the legend of Figure 4B, before FACS analysis. Percentages of labeled cells are indicated; data are from 1 of 2 experiments.
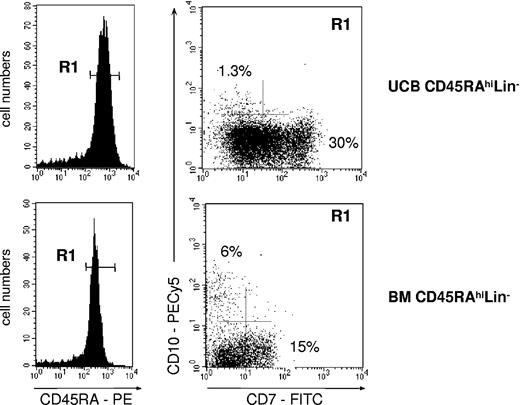
Whether CD45RAhiLin-CD10+ HPCs differentiated from CD45RAintCD7- and CD45RAhiLin- HPCs was also examined, but neither population generated CD10+ cells when cultured under the conditions described earlier—that is, onto MS5 cells with FL (data not shown). Both populations were then cultured with a combination of 1 to 4 cytokines (SCF, FL, TPO, IL-7) onto MS5 or S17 murine cells,37,43,44 αSM-56, or SV40-56 human stromal cells,45 with similar negative results. Finally, CD45RAhiLin- HPCs isolated from the UCB or adult BM were cultured onto MS5 stromal layer cells in the presence of FL and TSLP46 (Figure 7). After 4 to 6 days, CD45RAhiLin-CD10+ HPCs represented 0.5% to 1% of CD34+ cells derived from UCB CD45RAhiLin- HPCs, whereas they reached 6% in cultures of adult BM cells. Of note, secondary-sorting experiments confirmed that, like their primary counterparts, in vitro-generated CD45RAhiLin-CD10+ HPCs displayed predominant BL differentiation potential (data not shown).
Generation of CD45RAhiLin-CD10+ from UCB and BM CD34+CD45RAhiLin- cells. CD34+CD45RAhiLin- cells were sorted by FACS from UCB or BM CD34+ HPCs and cultured for 4 to 5 days onto MS5 cells in the presence of FL and TSLP. The recovered cells, gated based on CD34 expression (not shown), were then analyzed by FACS. Data are from 1 of 3 experiments.
Generation of CD45RAhiLin-CD10+ from UCB and BM CD34+CD45RAhiLin- cells. CD34+CD45RAhiLin- cells were sorted by FACS from UCB or BM CD34+ HPCs and cultured for 4 to 5 days onto MS5 cells in the presence of FL and TSLP. The recovered cells, gated based on CD34 expression (not shown), were then analyzed by FACS. Data are from 1 of 3 experiments.
Altogether, these data show that CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs emerge independently from CD45RAhiLin- precursors, and they indicate that BM and cord blood CD45RAhiLin- HPCs differ in their capacity to generate CD45RAhiLin-CD10+ HPCs.
Discussion
Here, we found that, among 6 UCB CD34+ HPC populations identified on the basis of CD45RA, CD7, and CD10 expression, only CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs displayed substantial lymphoid potential. Using semisolid assays, we show in addition that, although both populations had a reduced capacity to generate erythroid colonies, they nonetheless retained substantial granulomonocytic differentiation potential. The results presented in this study also confirm that acquisition of CD45RA marker correlates with loss of erythromegakaryocytic potential as well as with the capacity to generate macrophage colonies,38 arguing thus for an early partition of erythromegakaryocytic and granulomonocytic progenitors. Although conflicting with the current model of hematopoiesis, these findings are consistent with a previous report that murine fetal liver Lin- c-kit+Sca-1hi HPCs generate myeloid cells, TLs, and BLs but not erythroid cells or megakaryocytes.9
As to the early stages of lymphopoiesis, the current paradigm postulates a unique founder CLP population from which TLs, BLs, and NK cells differentiate.47 To date, most studies of human ELPs are based on the identification of populations endowed with the capacity to generate NK cells, TLs, and BLs and reduced granulomonocytic and erythroid potentials.3,24,25 However, this should not be considered an absolute criterion given that even early CD34+CD1a- postnatal thymocytes retain the capacity to generate macrophages and dendritic cells.48,49 Here, at variance with previous reports,24,25 we used a double comparative approach based on quantification of each population's intrinsic capacity to generate NK cells and BLs in limiting dilution and in mixed B/NK clonal assays, and on direct comparison of their lymphoid potentials. This led to the observation that CD45RAhiCD7+ HPCs are highly enriched in NK cell precursors and display strong T potential, whereas CD45RAhiLin-CD10+ HPCs comprise a majority of pro-B cells. Gene profiling of CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPC populations confirmed their biologic phenotypes by showing that CD45RAhiCD7+ HPCs selectively express T/NK cell lineage-affiliated genes, whereas CD45RAhiLin-CD10+ HPCs display a typical B signature. Surprisingly, gene expression profiles also revealed that both CD45RAhiLin-CD10+ and CD45RAintCD7- populations retained expression of globin genes despite a drastically reduced erythroid potential (data not shown), suggesting that overlap between the diverse hematopoietic differentiation programs may have been underestimated.50 Taken as a whole, these findings indicate that, although they retain some degree of multipotency, CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs are endowed with either T/NK or B lineage-biased lymphoid differentiation potential. Thus, neither population fulfills the defining criteria of prototypic CLPs, ie, the overall similar capacity to generate NK cells, TLs, and BLs together with lack of myeloid potential. However, because MS5 stromal cells have been previously shown to antagonize BL differentiation from mouse ELPs,11 the hypothesis that the clonal B/NK assay used in this study could lead to underestimation of the BL differentiation potential of CD45RAhiCD7+ HPCs should also be considered. Although such inhibitory effect has not been reported for human hematopoietic progenitors, one cannot exclude that prototypic CLPs could nonetheless be present among CD45RAhiCD7+ HPCs.
Regarding the ontogenetic relationship of the CD34+CD45RA+ cell populations examined here, we provide direct evidence that CD45RAhiCD7+ HPCs differentiate from CD45RAintCD7- precursors via a stepwise process. These results are consistent with the immature gene expression profile of CD45RAintCD7- HPCs characterized by selective expression of stem cell transcription factors,40 and with their mixed granulomonocytic and lymphoid differentiation potential. That CD45RAintCD7- cells actually correspond to lymphomyeloid precursors is further supported by the observation that BL/NK-containing clones derived from single CD45RAintCD7- HPCs seeded under the B/NK condition always comprised a significant fraction of CD19-CD56- myeloid cells (R. Haddad, unpublished data).Although both differentiate from CD45RAhiLin- HPCs, the limited efficiency of CD45RAhiLin-CD10+ HPC in vitro generation does not allow conclusions regarding their ontogenetic relationship with CD45RAhiCD7+ HPCs. That CD45RAhiCD7+ HPCs represented precursors of CD45RAhiLin-CD10+ HPCs was also tested, but culture onto murine or human stromal layers supplemented with diverse cytokine combinations never yielded CD45RAhiLin-CD10+ HPCs from CD45RAhiCD7+ HPCs (R.H. and B.C., unpublished data, 2003). For this reason, our working hypothesis is that the 2 populations differentiate independently from CD45RAhiLin- precursors in response to signals provided by soluble factors such as FL or TSLP, or by stromal cell surface counter-receptors.
In conclusion, the results presented in this study show that UCB CD45RAhiCD7+ and CD45RAhiLin-CD10+ HPCs correspond to multipotent ELPs polarized toward either the T/NK or B lineage, arguing thus for an early partition of TL/NK and BL precursors.51
Prepublished online as Blood First Edition Paper, August 26, 2004; DOI 10.1182/blood-2004-05-1845.
Supported by the Institut National de la Santé et de la Recherche Médicale, the Ministère de la Recherche Scientifique, the Association pour la Recherche sur le Cancer, the Comité de Paris de la Ligue Nationale Contre le Cancer, the Comité Leucémie de la Fondation de France, Eurocord III (QLRT-2001-01918), and the Société Française d'Hématologie.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Pr J. Milliez (service de Gynécologie-Obstétrique, Hôpital Saint-Antoine) for help, Pr. J. Oury (Maternité, Hôpital Robert Debré, Paris, France) for the gift of cord blood samples, and Pr E. Gluckman and Dr P. Richard (service de Greffe de Moelle Osseuse, Hôpital Saint-Louis, Paris, France) for providing BM samples. We thank Dr A. Cumano for the gift of OP9-DL1 stromal cells and Dr D. Chalmers (IMSERM EPI-119, Besançon, France) for the gift of αSM-56 and SV40-56 human stromal cells. We thank M. Yagello, F. Jourquin, N. Brunel, and B. Dumas for technical assistance, and Pr F. Sigaux (INSERM U462, Hôpital Saint-Louis, Paris, France) for critical discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal