Abstract
Mutations in TERC, encoding the RNA component of telomerase, have been found in autosomal dominant dyskeratosis congenita (DC) and aplastic anemia (AA). Several polymorphisms also exist in the TERC gene, making functional testing of potential pathogenic mutations essential. Here, we have tested normal and mutant TERC molecules in 2 telomerase reconstitution assays, 1 in vitro and 1 in transfected telomerase-negative cells. We find that 2 polymorphic mutations G58A and G228A have no effect on telomerase activity in these assays, whereas 6 mutations found in DC and AA cause reduction or abolition of telomerase activity. Mutations in the pseudoknot region of the TERC molecule, C72G, 96-7ΔCT, GC107-8AG and 110-3ΔGACT reduce the catalytic activity of reconstituted telomerase, whereas mutations in the 3′ portion of the molecule C408G and a deletion of the 3′ 74 bases have normal activity in vitro but reduced intracellular activity. By analyzing second site mutations that recreate regions of secondary structure but retain the pathogenic mutations we show that mutations C72G, GC107-8AG, and C408G act by disrupting the secondary structure or folding of TERC. Finally, experiments reconstituting telomerase with both normal and mutant TERC molecules suggest the mutations act via haploinsufficiency rather than by a dominant-negative mechanism. (Blood. 2004;104:3936-3942)
Introduction
Dyskeratosis congenita (DC) is a rare, usually fatal, inherited skin and bone marrow (BM) failure syndrome that displays considerable genetic and clinical heterogeneity.1 At the genetic level X-linked recessive, autosomal dominant (AD) and autosomal recessive (AR) forms of the disease exist where the genetic basis of the X-linked and AD forms have been determined. The X-linked form of DC is due to mutations in DKC1.2 This gene encodes dyskerin, a protein that is part of the H box/ACA-motif (H/ACA) small nucleolar ribonucleoproteins (snoRNPs) responsible for pseudouridylation of specific residues of nascent ribosomal RNA molecules.3-6 Dyskerin is also part of the telomerase complex7 where its role has not been clearly defined. Autosomal dominant DC (AD-DC) is caused by mutations in TERC,8 which codes for the RNA component of telomerase. Together with telomerase reverse transcriptase (TERT), TERC forms the core of the active telomerase complex. This complex is responsible for synthesizing 6-base pair (bp) telomere repeats which form part of the nucleoprotein telomeres that cap the ends of chromosomes.9,10
Telomerase is expressed at very low levels in normal somatic cells,11 and telomeres shorten with each cell division.12 Very short telomeres can lead to the formation of chromosomal aberrations, such as end-to-end joining, and this genomic instability leads to cell death and occasionally to the development of malignancy.13,14 In keeping with this, all forms of DC are associated with very short telomeres,15 chromosomal aberrations, and an increased incidence of cancer.16 The convergence of both known genetic causes of the disease on the telomerase complex suggests that defective telomere maintenance is important in disease pathology. Yet the effect of defective rRNA production in the more severe X-linked form of the disease cannot be ruled out.17,18 DC affects tissues, such as blood and skin, that undergo constant renewal from a stem cell population, leading to the hypothesis that critically short telomeres cause cell death in the stem cell or precursor population.19,20 The most common cause of death in DC is BM failure and its consequences.16
TERC is a 451-bp RNA that acts as a template for the synthesis of the TTAGGG telomere repeat.21 Examination of TERC molecules from different vertebrates led to the deduction of a model secondary structure which consists of 4 structural domains: the pseudoknot domain, the CR4-CR5 domain, the H/ACA domain, and the CR7 domain.22 Structure function analysis of vertebrate TERC reveals that all 4 domains contribute to telomerase function, but only the pseudoknot and CR4-CR5 domains are required in in vitro reconstitution experiments,23-25 whereas the H/ACA and CR7 domains are required for TERC RNA accumulation.23 Here, we examine the activity of telomerase reconstituted in vitro and in vivo with TERC molecules that contain mutations found in patients with AD-DC and aplastic anemia (AA). Our results show that TERC mutations reduce telomerase activity in vitro and in vivo to different extents and that they do so either by directly reducing the catalytic activity of telomerase or indirectly by lowering the stability or assembly of the complex. We find no evidence for a dominant negative effect with any of the mutations, and in 3 cases we show that the mutations act by disrupting the secondary structure rather than via the primary sequence of the molecule.
Materials and methods
Plasmid constructs and mutagenesis
The wild-type (WT) TERC gene was cloned 3′ of a T7 promoter within a pUC19 backbone as described by Autexier et al26 to produce the pUC T7 WT TERC vector. The TERT cDNA, a kind gift from Dr R. Weinberg was ligated into pBluescript via SalI and EcoRI digest to produce the pBS T7 WT TERT vector. A 1.3-Kb human genomic fragment containing the WT TERC gene was placed into a pCMV 3.1-backbone (Invitrogen, Carlsbad, CA) using EcoRI and HindIII digests to produce the p3.1- WT TERC vector. The TERT cDNA was transferred into a pCMV 3.1+ backbone using EcoRI and SalI digests to produce the p3.1+ WT TERT vector. All the TERC mutations were either produced directly through polymerase chain reaction (PCR) of patient genomic DNA or used a 2-step PCR approach: forward and reverse primers were made which were complementary to each other and contained the mutation to be induced. Two PCR products were then produced by using these 2 mutation primers and 2 flanking primers containing unique restriction sites. The flanking primers were then used to join the 2 PCR fragments together. The resulting PCR products from either genomic DNA amplification or 2-step PCR was then digested and ligated into the appropriate vectors. The sequence of the final constructs was verified.
In vitro telomerase reconstitution
The pUC T7 TERC vectors were cleaved with BamHI and transcribed by T7 RNA polymerase (T7 Ribomax; Promega, Madison, WI). The resulting RNA was purified by electrophoresis through a 5% polyacrylamide gel containing 1 × TBE (tris [tris(hydroxymethyl)aminomethane]-borate-EDTA [ethylenediaminetetraacetic acid]) and 8 M urea followed by electroelution. To check the quantification of each RNA sample produced, 100 ng of each purified TERC RNA was analyzed by Northern blot on a 1.6% gel probed with a 32P-labeled TERC probe. Once normalized, 100 ng of each purified TERC RNA was placed into a rabbit reticulocyte lysate (RRL) master mix (Promega) containing 250 ng pBS T7 TERT plasmid/reaction and incubated at 30°C for 90 minutes. The resulting RRL lysate was then diluted 1:50, 1:100, 1:200, and 1:400 in 1 × CHAPS (Intergen, Atlanta, GA) buffer and used in the telomere repeat amplification protocol (TRAP) assay.
Telomerase reconstitution in transfected WI-38 VA13 cells
Confluent WI-38 VA13 cells were plated out at 4 × 106/2 10-cm2 plates in antibiotic-free Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS; Sigma, St Louis, MO). A total of 8μg of each p3.1-TERC plasmid was mixed with 8 μg p3.1+TERT plasmid and 8 μg phRL-TK (luciferase internal control; Promega) in 1.5 mL serum-free DMEM. This was mixed in a 1:1.25 ratio with lipofectamine 2000 (Invitrogen), split between the 2 plates, and incubated at 37°C and 5% CO2 overnight. After 24 hours, the medium was changed to DMEM containing 10% FCS, and the cells were collected 48 hours after transfection. The pooled cells were split into 1 × CHAPS buffer for TRAP analysis and 1 × lysis buffer for luciferase analysis. The resulting TRAP lysates were then assayed for protein concentration by using a DC protein assay (Bio-Rad, Hercules, CA); normalized to 650 ng/μL; and were diluted 1:10, 1:40, and 1:160 using 1 × CHAPS buffer prior to TRAP analysis.
TRAP analysis
Lysates from the transcription/translation system or from the transfected cells were analyzed by using the TRAPeze telomerase detection kit (Intergen). The TS primer was labeled at 37°C for 20 minutes and heat denatured at 85°C for 5 minutes. The PCR was then run for 28 cycles with an annealing temperature of 59°C. The resulting TRAP products were then diluted into TRAP loading dye and run on a 12.5% acrylamide-0.5 × TBE gel, dried, and exposed to x-ray film. The percentage figures shown for the TRAP activities are approximate values derived from comparing TRAP ladders from serial dilutions from mutant samples with those from WT samples. The relative values were consistent in at least 4 separate experiments.
Luciferase assay
The luciferase lysates were analyzed by using the Renilla luciferase assay protocol (Promega). Each lysate was diluted 1:10 and then 1:1000 in 1 times lysis buffer and 20 μL of the final dilution was measured (in duplicate) in a luminometer following manufacturer's recommendations.
Results
TERC mutations used in this study
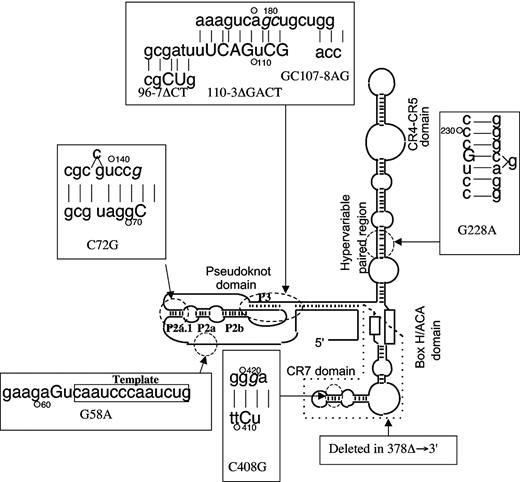
In this study we have analyzed the effect of 8 separate mutations found in the human TERC gene (Figure 1; Table 1). The mutation 378Δ → 3′ is a deletion of the last 74 bp from the 3′ end of TERC that was found to cause AD-DC in the large DCR101 family and through which TERC was first identified as the gene whose mutation underlies AD-DC.8 The pseudoknot mutation GC107-8AG and the CR7 domain mutation C408G were also found to segregate with AD-DC disease.8 The AD-DC-causing mutation 96-7ΔCT34 was found in a 40-year-old woman who had developed BM failure in her 20s and had gone on to develop the mucocutaneous features typical of DC. In addition she had severe osteoporosis and developed squamous cell carcinoma of the skin, both conditions frequently found in DC. Her mother had developed AA in her 60s, an example of the disease anticipation seen in families in which TERC mutations are segregating.34 The template mutation G58A and the pseudoknot mutations C72G and 110-3ΔGACT have been found in patients presenting with AA,30 whereas the hypervariable paired region mutation G228A was found in a cell line derived from a human tumor that maintains telomere length by a recombination mechanism ALT (alternative lengthening of telomeres).35 Recently, the mutations G58A and G228A have been found at polymorphic frequency in African Americans.31,32
The secondary structure of TERC, showing the mutations used in this study. The secondary structure domains are indicated. In the boxes the mutated bases are indicated by capital letters. The bases complementary to mutations C72G, GC107-8AG, and C408G are shown in italics. The secondary structure elements in the pseudoknot domain referred to in the text are indicated. In this in vivo system, the telomerase components must be translocated to the nucleolus and assembled into the functional complex for any activity to be detected.
The secondary structure of TERC, showing the mutations used in this study. The secondary structure domains are indicated. In the boxes the mutated bases are indicated by capital letters. The bases complementary to mutations C72G, GC107-8AG, and C408G are shown in italics. The secondary structure elements in the pseudoknot domain referred to in the text are indicated. In this in vivo system, the telomerase components must be translocated to the nucleolus and assembled into the functional complex for any activity to be detected.
TERC mutations investigated in this study
. | . | . | . | . | Telomerase activity . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | In vitro . | . | In vivo . | . | . | . | |||||
| Mutation . | Disease . | History of index case at presentation . | Domain . | References* . | This paper, % . | Fu and Collins27 . | This paper, % . | Fu and Collins27 . | Comolli et al37 . | Ly et al29, % . | |||||
| G58A | DC, NSAA, AA, MDS, WT | Various individuals with usually no family history. | Near template | Vulliamy et al30 Wilson et al31 Yamaguchi et al32 Fogarty et al33 | 85 | +++ WT | 100 | +++ 5-fold | — | 100 | |||||
| Normally asymptomatic | |||||||||||||||
| C72G | NSAA | 9-y-old boy with no family history. | Pseudoknot stem P2a. 1 | Vulliamy et al30 | 0 | +++ WT | 5 | ++ 25-fold | — | Low | |||||
| Thrombocytopenia and severe osteoporosis | |||||||||||||||
| 96-7δCT | DC | 40-y-old woman with AD-DC family history. BM failure, mucocutaneous features, osteoporosis, and skin carcinoma | Pseudoknot stem P2b | Vulliamy et al34 | 0 | — | 0 | — | — | — | |||||
| GC107-8AG | DC | 27-y-old woman with AD-DC family history. | Pseudoknot stem P3 | Vulliamy et al8 | 50 | + 25-fold | 5 | + 125-fold | > 100-fold | — | |||||
| Mucocutaneous features | |||||||||||||||
| 110-3δGACT | NSAA | 20-y-old woman with AD-DC family history. | Pseudoknot stem P3 | Vulliamy et al30 | 0 | ++ 5-fold | 0 | — > 125-fold | — | Low | |||||
| BM failure and elphic appearance | |||||||||||||||
| C408G | DC | 12-y-old girl with family history. | CR7 domain stem P8b | Vulliamy et al8 | 100 | +++ WT | 25 | + 125-fold | — | — | |||||
| BM failure, mucocutaneous features, and liver disease | |||||||||||||||
| 378δ → 3′ | DC | 13-y-old boy with AD-DC family history. | Box H/ACA domain | Vulliamy et al8 | 85 | — | 5 | — | — | — | |||||
| BM failure, mucocutaneous features | |||||||||||||||
| G228A | ALT, DC, AA, WT | Patients with AA and African-American control subjects | CR4-5 domain stem P4.1 | Wilson et al31 Yamaguchi et al32 Bryan et al28 | 100 | — | 100 | — | — | — | |||||
. | . | . | . | . | Telomerase activity . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | In vitro . | . | In vivo . | . | . | . | |||||
| Mutation . | Disease . | History of index case at presentation . | Domain . | References* . | This paper, % . | Fu and Collins27 . | This paper, % . | Fu and Collins27 . | Comolli et al37 . | Ly et al29, % . | |||||
| G58A | DC, NSAA, AA, MDS, WT | Various individuals with usually no family history. | Near template | Vulliamy et al30 Wilson et al31 Yamaguchi et al32 Fogarty et al33 | 85 | +++ WT | 100 | +++ 5-fold | — | 100 | |||||
| Normally asymptomatic | |||||||||||||||
| C72G | NSAA | 9-y-old boy with no family history. | Pseudoknot stem P2a. 1 | Vulliamy et al30 | 0 | +++ WT | 5 | ++ 25-fold | — | Low | |||||
| Thrombocytopenia and severe osteoporosis | |||||||||||||||
| 96-7δCT | DC | 40-y-old woman with AD-DC family history. BM failure, mucocutaneous features, osteoporosis, and skin carcinoma | Pseudoknot stem P2b | Vulliamy et al34 | 0 | — | 0 | — | — | — | |||||
| GC107-8AG | DC | 27-y-old woman with AD-DC family history. | Pseudoknot stem P3 | Vulliamy et al8 | 50 | + 25-fold | 5 | + 125-fold | > 100-fold | — | |||||
| Mucocutaneous features | |||||||||||||||
| 110-3δGACT | NSAA | 20-y-old woman with AD-DC family history. | Pseudoknot stem P3 | Vulliamy et al30 | 0 | ++ 5-fold | 0 | — > 125-fold | — | Low | |||||
| BM failure and elphic appearance | |||||||||||||||
| C408G | DC | 12-y-old girl with family history. | CR7 domain stem P8b | Vulliamy et al8 | 100 | +++ WT | 25 | + 125-fold | — | — | |||||
| BM failure, mucocutaneous features, and liver disease | |||||||||||||||
| 378δ → 3′ | DC | 13-y-old boy with AD-DC family history. | Box H/ACA domain | Vulliamy et al8 | 85 | — | 5 | — | — | — | |||||
| BM failure, mucocutaneous features | |||||||||||||||
| G228A | ALT, DC, AA, WT | Patients with AA and African-American control subjects | CR4-5 domain stem P4.1 | Wilson et al31 Yamaguchi et al32 Bryan et al28 | 100 | — | 100 | — | — | — | |||||
Results from recent studies investigating some of the mutants studied here are presented. DC indicates dyskeratosis congenita; NSAA, nonsevere aplastic anemia; AA, aplastic anemia; MDS, myelodysplastic syndrome; WT, wild type/normal. Percentage figures refer to percentage of wild-type activity, figures given as “fold” represent fold decrease over wild type and figures given as n+ represent comparative values in which WT is “++++” in vivo and “+++” in vitro.—indicates not applicable.
References in addition to the current paper.
Pseudoknot mutations affect telomerase activity directly
Two separate assay systems were used to reconstitute telomerase with mutant and WT TERC molecules. In the in vitro system a standard amount of purified TERC RNA was added to a rabbit reticulocyte lysate (RRL) coupled transcription/translation system in the presence of a plasmid containing the TERT coding sequence under the control of a T7 promoter. In this system it has been previously found that the TERT protein produced and the TERC RNA added to the system were sufficient to reconstitute active telomerase.26 The telomerase activity produced was measured by the TRAP assay in which a labeled primer is extended by the reverse transcriptase activity of the resulting telomerase complex, and the extended product is amplified by PCR. This system has been found to be dependent on added TERC but requires only the 5′ 159 bp for activity and the 5′ 354 bp for full activity.24,26,36 In the second in vivo assay we took advantage of the cell line WI-38 VA13, a cell line which does not express any TERT or TERC and which consequently produces no telomerase activity.35 After cotransfection of plasmids encoding TERT and TERC under the control of the cytomegalovirus (CMV) promoter, telomerase activity is produced.
In the RRL system, the pseudoknot mutations C72G, 96-7ΔCT, and 110-3ΔGACT all showed very low or undetectable levels of telomerase activity, indicating that the mutant RNAs are unable to form a catalytically active telomerase (Figure 2A). Similarly after cotransfection into WI-38 VA13 cells, no telomerase activity could be detected by the extremely sensitive TRAP assay (Figure 2B). The pseudoknot mutation GC107-8AG behaved differently from the others, having substantial activity (∼50% of wild type) in the RRL assay (Figure 2A) but lower activity (∼5% of wild type) in the transfected cells (Figure 2B).
Detection of telomerase activity of the 8 TERC mutations through TRAP analysis. Detection of telomerase activity is shown in vitro (A) and in vivo (B). The arrows denote the start of the TRAP ladder (TRAP), the corresponding internal control (IC), the amount of activity in relationship to WT TRAP levels (%), and the control of the assay where an equal amount of RNA was loaded (RNA) or the levels of luciferase was measured (Luc) to control for the corresponding TRAP experiment. The mutations are shown above the panels. Serial dilutions of each lysate were assayed as described in “Materials and methods.” The percentage figures shown for the TRAP activities are approximate values derived from comparing TRAP ladders from serial dilutions from mutant samples with those from WT samples.
Detection of telomerase activity of the 8 TERC mutations through TRAP analysis. Detection of telomerase activity is shown in vitro (A) and in vivo (B). The arrows denote the start of the TRAP ladder (TRAP), the corresponding internal control (IC), the amount of activity in relationship to WT TRAP levels (%), and the control of the assay where an equal amount of RNA was loaded (RNA) or the levels of luciferase was measured (Luc) to control for the corresponding TRAP experiment. The mutations are shown above the panels. Serial dilutions of each lysate were assayed as described in “Materials and methods.” The percentage figures shown for the TRAP activities are approximate values derived from comparing TRAP ladders from serial dilutions from mutant samples with those from WT samples.
CR7 domain mutation reduces telomerase activity indirectly
The CR7 mutation C408G disrupts a conserved stem in the 3′ portion of the TERC molecule. When this mutant is used to reconstitute telomerase in the RRL system (Figure 2A), no decrease in telomerase activity is seen when compared with WT levels. In the transfection assay however, this mutant showed decreased telomerase activity (∼25% of wild type) measured by the TRAP assay (Figure 2B). Because the catalytic function of this mutation in the RRL assay is normal, the decrease seen in transfected cells must reflect inefficiency in assembly of the telomerase complex or simply unstable RNA. Although reduction in C408G RNA accumulation has been reported,27 mutant RNA from primary cells from a patient was detected by PCR.8
Functional telomerase assembled with a 3′ deletion mutant of TERC
The 378Δ → 3′ mutation is a large 3′ deletion removing the last 74 bp of the TERC gene. In Epstein-Barr virus (EBV)-transformed cell lines derived from patients with this mutation only normal-sized TERC RNA was detected, suggesting RNA transcribed from this deletion mutant is unstable in vivo.8 Nevertheless, we were curious to know whether the truncated RNA had any residual activity in our 2 reconstitution systems. In the RRL system, the deletion mutant had full activity, demonstrating again that the normal 3′ end of TERC is not required for telomerase activity in this in vitro system (Figure 2A). In the transfected cells, we detected some residual telomerase activity in the cell lysate from cells cotransfected with this mutant and TERT (Figure 2B). Although the activity was very low, it was consistently present and higher than 2 of the pseudoknot mutants in the same system (Figure 2B).
Polymorphic mutations have normal activity
The mutation G58A was initially discovered in 2 patients with idiopathic AA and a patient with constitutional AA.30 Subsequently, the mutation has been found to be present at polymorphic frequencies in African Americans.31,32 The mutation G228A was initially found in the TERC gene in GM847,28 a human cell line which maintains telomere length by using the alternative lengthening of telomeres (ALT) pathway, suggesting that the endogenous TERC may be non-functional. This mutation too has recently been found in healthy African Americans.31,32 Despite appearing to be polymorphic, we were interested to see whether these mutations caused any decrease in telomerase activity in our 2 assay systems, as they could then be considered as candidate genes that might confer susceptibility to AA or other BM failure conditions. However, in both our assay systems these 2 mutations showed full activity (Figure 2A-B).
No evidence for a dominant-negative effect
The dominant nature of the BM failure caused by TERC mutations leads to the startling conclusion that 50% (or more) of the normal levels of TERC, and presumably of telomerase activity, is insufficient to maintain correctly functioning stem cells that can populate the developing blood system and continue to provide mature blood cells during adult life. This certainly appears to be the case with the 378Δ → 3′ mutation where only RNA from the WT allele is detected in patient cells.8 The implications of this are that the levels of TERC must be finely regulated, perhaps because too much would be more permissive for the development of a malignant phenotype and too little may lead to early senescence. Alternatively, the TERC mutations may exert a dominant-negative effect, reducing the cellular telomerase activity by more than 50%, and, thus, not implying such an exquisite control of cellular TERC levels. It was important, therefore, to test for a dominant-negative effect of the TERC mutations by assaying the activity of WT TERC in the presence of an equal amount of the mutant molecule. We did this in both the in vitro reconstitution system and in the transfected WI-38 VA13 cells. In both systems we found no evidence for a dominant negative effect (Figure 3A-B).
Detection of telomerase activity of the 8 TERC mutations mixed with WT TERC. Detection of telomerase activity is shown in vitro (A) and in vivo (B). The arrows denote the start of the TRAP ladder (TRAP) and the corresponding internal control (IC). For each experiment, 100 ng WT TERC RNA (A) or 4 μg WT TERC plasmid (B) was mixed with the same amount of mutant TERC, and the resulting telomerase activity was measured by TRAP assay.
Detection of telomerase activity of the 8 TERC mutations mixed with WT TERC. Detection of telomerase activity is shown in vitro (A) and in vivo (B). The arrows denote the start of the TRAP ladder (TRAP) and the corresponding internal control (IC). For each experiment, 100 ng WT TERC RNA (A) or 4 μg WT TERC plasmid (B) was mixed with the same amount of mutant TERC, and the resulting telomerase activity was measured by TRAP assay.
Disrupted secondary structure causes decreased telomerase activity
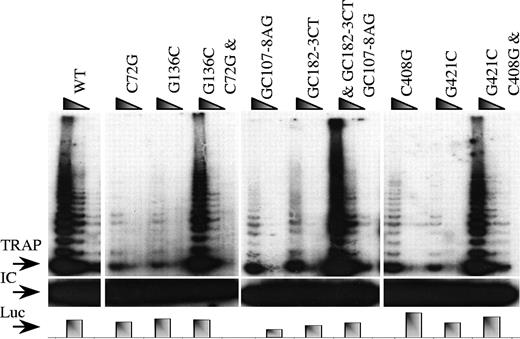
Most of the TERC mutations we have found in AD-DC or in AA occur in short double-stranded helical regions in the model of the secondary structure of TERC. The secondary structure of vertebrate telomerase is highly conserved; the model having been deduced largely from observed covariation of complementary base pairs forming the short stems that form the basis of the structure.22 The mutations we observe could affect the function of telomerase RNA in 2 ways: (1) they could disrupt the secondary structure of the molecule by preventing it from folding in the correct way or adopting a stable conformation or (2) by changing the primary sequence the mutation could prevent an important interaction between TERC and other components of the telomerase complex. To decide between these 2 alternatives, we made mutations in the bases that form a base pair with the original mutated bases such that the second site mutations disrupt the same base pairings as the pathogenic mutation but when they are in cis within the same TERC molecule as the pathogenic mutations (double mutation) the helical stem is reconstructed. The telomerase activity conferred by the second mutations and by TERC molecules containing the double mutations specific to mutations C72G (G136C), GC107-8AG (GC182-3CT), and C408G (G421C) were tested in the in vivo reconstitution system. In each case the double-reconstituted mutations, with an intact secondary structure, showed WT levels of telomerase activity while both single mutations had greatly reduced levels (Figure 4). Essentially the same results were found in the in vitro system (data not shown). These results strongly suggest that these mutations exert their effect on telomerase activity by altering the secondary structure of TERC.
In vivo detection of telomerase activity of 3 TERC mutations with their corresponding complementary mutations and the double TERC mutations, in which the secondary structure is repaired. The arrows denote the start of the TRAP ladder (TRAP), the corresponding internal control (IC), and the control of the assay where an equal amount of RNA was loaded (RNA) or the levels of luciferase was measured (Luc) to control for the corresponding TRAP experiment.
In vivo detection of telomerase activity of 3 TERC mutations with their corresponding complementary mutations and the double TERC mutations, in which the secondary structure is repaired. The arrows denote the start of the TRAP ladder (TRAP), the corresponding internal control (IC), and the control of the assay where an equal amount of RNA was loaded (RNA) or the levels of luciferase was measured (Luc) to control for the corresponding TRAP experiment.
Discussion
Our results and those of others27,29,37 show that most of the mutations found in TERC in patients with AD-DC or AA demonstrably decrease telomerase activity in functional assays. In Table 1 we present our analysis of these mutations along with a comparison of our results with those from other laboratories where these are available. The TRAP assay used to measure telomerase activity is PCR based and thus can be considered semiquantitative, and the results are difficult to compare between laboratories. Moreover, the protocols for quantitation differ in the different laboratories. Nevertheless, there is remarkably good agreement about the effect of those mutations analyzed multiple times, the minor disagreements between our and other results are discussed in the next paragraphs.
The discovery of 2 polymorphisms in the TERC gene, which do not appear to cause a reduction in telomerase activity, underlines the importance of performing functional assays on newly discovered mutations before concluding they are responsible for DC or AA. The base change G58A is found in 4% to 10% Africans31,32 and the G228A mutation is found in about 2% Africans. Base 58 lies 3 bp downstream from the highly conserved template region and could be important for telomerase activity, but evidently this is not the case. Similar results to ours for this mutant were obtained in another laboratory,29 whereas a third group observed a 5-fold reduction in in vivo activity with this mutant.27 The base change G228A was originally described in a cell line that uses the ALT pathway of telomere maintenance,28,35 which led us to suspect it may affect telomerase activity. However, our finding that TERC molecules with G228A have WT levels of activity is in keeping with the low conservation of the hypervariable paired region of TERC, which includes base 228. We cannot exclude the possibility that these polymorphic mutations cause a slight decrease in telomerase activity that our assays cannot detect. This might constitute an interesting source of genetic variation but further studies will be needed to clarify this issue.
The pseudoknot region binds to TERT and is essential for telomerase activity.23,25 Mutations in the pseudoknot region have a drastic effect on telomerase activity in vitro and in the in vivo transfection assay. The mutations C72G, 96-7ΔCT, and 110-3ΔGACT all showed very low or undetectable levels of telomerase activity in both assays. These mutations are all in double-stranded helical regions of the pseudoknot region of TERC, being located in 3 of the 4 stems that form its structure. C72G disrupts a base pair that appears to stabilize the stem P2a.1. This part of the pseudoknot domain is not conserved in all vertebrates, but the base pairing involving this base is found in all mammalian TERC molecules.22,29 Changing the other partner in the base pair, G144C, also leads to very low telomerase activity, whereas the double mutant, in which the secondary structure is restored, has normal activity. Similar results have been recently reported by another laboratory.29 These data demonstrate the importance of the P2a.1 stem for mammalian TERC function. Interestingly, the C72G mutation was also tested in another laboratory27 with similar results to ours, except those workers found almost wild-type telomerase levels in vitro. Their in vitro reconstitution assay involves incubating purified TERC and TERT together, whereas in our assay TERT is synthesized in the presence of purified TERC. Similar minor differences in the behavior of pseudoknot mutants 110-3ΔGACT and GC107-8AG are also evident between these 2 studies, implying that incubating the purified molecules together may lead to differences in conformation of the pseudoknot compared with synthesizing TERT in the presence of TERC.
The mutation 96-7ΔCT deletes 2 bp that are highly conserved and that form part of the stem P2b. Stem P2b is predicted by universal covariation of 4 of the 9 bp in the stem and more than 90% conservation of another 4 of the base pairs. The base pairs involving 96C and 97U are not universally conserved, but base 97U is found in 100% of vertebrate TERC molecules, implying that primary sequence as well as secondary structure may be important in this part of the pseudoknot. This conclusion is supported by detailed mutational analysis of the pseudoknot region of TERC.29 The 4 base deletion 110-3ΔGACT of bases that form the stem P3 removes highly conserved bases in this stem, 2 of which are always base paired in vertebrate TERC and 2 of which are 100% conserved and base paired more than 90% of the time.22 The lack of telomerase activity in this mutant, along with the conservation, points to the importance of stem P3 in telomerase activity. The stem P3 has been postulated to be involved in a molecular switch whereby residues 107 to 115 may alternatively pair with residues 174 to 183 as shown in Figure 1 or with residues 99 to 106.37 Switching between the alternative states is proposed to be essential for telomerase activity, and the hypothesis has been supported by structural studies.38,39 A second mutation in stem P3 is the double base change GC107-8AG which has been suggested to stabilize the alternative conformation and thus inhibit telomerase activity. In our experiments this mutation was the only one in the pseudoknot region which gave a TERC molecule with substantial activity in vitro but lower activity in vivo, possibly because the switch between alternative conformations requires the fully assembled telomerase complex and not merely the core components TERC and TERT. Together our results show the cluster of TERC mutations in the pseudoknot domain disrupts the highly conserved secondary structure of this region which is essential for the catalytic activity of telomerase. TERC mutants 110-3ΔGACT and GC107-8AG have been analyzed in other laboratories with similar results to ours.27,29,37
The 2 mutations, C408G and 378Δ → 3′, not in the pseudoknot domain both showed normal activity in the in vitro system. This result was expected because only the 5′ 354 bp of TERC is required in this system for full activity.36 After transfection of WI-38 VA13 cells, the C408G mutation gave telomerase activity that was about 25% of WT TERC, whereas the 378Δ → 3′ mutant gave telomerase activity that was barely detectable. It has been proposed that the steady state of these mutant TERC molecules is reduced when compared with WT TERC levels, suggesting that these 2 mutant RNAs have decreased stability. It is surprising that any telomerase activity is detected after transfection of the 378Δ → 3′ mutant because it implies that some telomerase complex is correctly assembled in the absence of the normal 3′ domain of the TERC molecule. It is possible that some telomerase activity is generated in transfected cells in a nonphysiologic manner because of the overexpression of TERC and TERT. The C408G gives some 25% of WT activity in the transfection experiment. Again the telomerase activity we find may be falsely high because of overexpression of the mutant TERC. A decrease in the stability of TERC in vivo is likely to lead to substantially reduced levels of telomerase activity given the extremely long half-life of the molecule.40 We note, however, that in cells from patients with the C408G mutation, mutant RNA was detected by reverse transcriptase (RT)-PCR.8 The 408C base is part of a stem structure next to the terminal loop in the CR7 domain. This part of the molecule has previously been found to be necessary for RNA accumulation.23,27 Recently, the CR7 domain has been found to be important in the formation of TERC dimers.41 The effect of the C408G mutation was reversed by recreating the stem by making the compensatory G421C mutation, showing that the integrity of the CR7 stem loop structure, rather than the primary sequence is crucial for telomerase RNA accumulation and possibly dimer formation.
The dominant nature of DC disease caused by TERC mutations is interesting. Simply it implies that either mutant TERC molecules exert a dominant-negative effect on the telomerase complex, reducing telomerase activity to less than 50% of WT levels or 50% or greater of WT telomerase levels in some somatic cells can lead to the devastating effects of DC. The fact that telomerase is a complex of multiple protein subunits, where its core components operate as a dimer,42,43 encourage the idea that dominant-negative mutations are likely. Previously, we noted that no aberrant-sized RNA was detected in the DC patients with the 378Δ → 3′ mutation, suggesting these patients must produce 50% normal levels of TERC; therefore, this mutation acts through haplo-insufficiency of TERC, rather than a dominant-negative effect.8 This is in contrast with the situation in laboratory mice in which early generations of mice completely lacking Terc show no aberrant phenotype.44 Later generations show progressive telomere shortening and a variety of defects, such as reduced proliferative capacity of blood cells, and increased tumor incidence that are reminiscent of patients with DC.19,44-46 The lag of several generations in the appearance of these abnormalities is most likely because laboratory mice have long telomeres and several generations in the absence of telomerase are required before they reach a critical length.47 TERC mutations have also been found in patients presenting with AA.30,32 In some of these cases there were indications, such as an affected relative or a second disease site, that the disease was constitutional. Yet in other families there were no such indications, and the parents of the affected patient, although carrying the mutation, was not affected. Similarly in the AD-DC cases the disease appears to become more severe in later generations.15 These observations might suggest that this form of DC shows a novel form of anticipation in which later generations are more severely affected because they inherit shortened telomeres as well as defective telomerase.20,34 Here we tested 8 different mutations by cotransfecting with WT TERC and looking for a reduction in the telomerase levels compared with WT alone. The fact that no reduction in telomerase activity was found in either detection system supports the notion that DC is commonly caused by haploinsufficiency of TERC. Our results confirm and extend those of Fu and Collins.27 It is possible that telomerase levels are crucial in maintaining stem cell populations and that with lower than optimum telomerase levels stem cell pools become depleted, leading to the failure of tissues that need to be renewed by stem cell activity. The fact that telomerase activity levels are so finely controlled prompts questions about the roles of other genes that influence telomerase levels and their role in stem cell maintenance, cancer susceptibility, and aging. Because the TERC gene appears to also have a high ratio of polymorphisms to coding sequence (2 in 451 bp at present), it is important to check that mutations suspected of being pathogenic do affect telomerase activity.31,32,35
Prepublished online as Blood First Edition Paper, August 19, 2004; DOI 10.1182/blood-2004-05-1829.
Supported by the Wellcome Trust.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Courtney Smith and Anne Rigby for their assistance with this project, as well as Carol Heath and Alex Mentzer for their help and advice. We thank patients and clinicians for supporting the Dyskeratosis Congenita Registry.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal