Abstract
Previous studies have suggested that plasminogen activator inhibitor 1 (PAI-1) released from platelets convey resistance of platelet-rich blood clots to thrombolysis. However, the majority of PAI-1 in platelets is inactive and therefore its role in clot stabilization is unclear. Because platelets retain mRNA and capacity for synthesis of some proteins, we investigated if platelets can de novo synthesize PAI-1 with an active configuration. PAI-1 mRNA was quantified with real-time polymerase chain reaction and considerable amounts of PAI-1 mRNA were detected in all platelet samples. Over 24 hours, the amount of PAI-1 protein as determined by an enzyme-linked immunosorbent assay increased by 25% (P = .001). Metabolic radiolabeling with 35S-methionine followed by immunoprecipitation confirmed an ongoing PAI-1 synthesis, which could be further stimulated by thrombin and inhibited by puromycin. The activity of the newly formed PAI-1 was investigated by incubating platelets in the presence of tissue-type plasminogen activator (tPA). This functional assay showed that the majority of the new protein was in an active configuration and could complexbind tPA. Thus, there is a continuous production of large amounts of active PAI-1 in platelets, which could be a mechanism by which platelets contribute to stabilization of blood clots. (Blood. 2004;104:3943-3948)
Introduction
Vascular injury complicated by thrombus formation plays a key role in the pathogenesis of arterial diseases such as myocardial infarction and ischemic stroke.1 Clinical studies have shown that in a substantial proportion (15%-30%) of patients with myocardial infarction, a spontaneous reperfusion occurs during the first hour after the vascular injury as a consequence of activation of the endogenous fibrinolytic system.2-4 On the other hand, in a similar proportion of patients no clot lysis can be achieved despite pharmacologic thrombolytic therapy.5 The mechanisms behind this variability in resistance of arterial thrombi to lysis are not fully understood.
Clot lysis in vivo results primarily from the enzymatic activation of the fibrinolytic system, and stimulated secretion of tissuetype plasminogen activator (tPA) from the vascular endothelium is the key event in this activation.6 Its activity in plasma is regulated by specific inhibitors, the most important of which is plasminogen activator inhibitor 1 (PAI-1). Arterial clots contain 2- to 3-fold more PAI-1 than venous clots,7,8 and the relative PAI-1 content determines their resistance to thrombolysis.9 It is likely that the major part of the PAI-1 in thrombi has been released from activated platelets because platelets contain large amounts of the inhibitor in their α-granules.10 The hypothesis that PAI-1 released from platelets is an important determinant of thrombolysis resistance is supported by in vitro clot lysis studies on platelets from a patient with complete loss of PAI-1 expression11 as well as studies on thrombi generated in the Chandler loop.12 Also, studies in transgenic mice have suggested that PAI-1 not only influences the resistance to thrombolysis but also the rate of progression of thrombus formation following vascular injury.13
On the other hand, the pathophysiologic importance of the platelet PAI-1 pool for inhibition of the fibrinolytic system has been difficult to reconcile with the fact that the majority of PAI-1 in platelets exists in a predominantly inactive or latent form. Previous studies have shown that only 5% to 10% of the PAI-1 present in the platelets is in an active configuration that could complex-bind and thereby inhibit tPA.14-16 Although platelets lack nuclear DNA they retain mRNA from the megakaryocyte,17-19 and it has been shown that platelets retain the ability for protein synthesis and can synthesize at least some proteins.20-22 We have recently shown that mRNA extracted from platelets can be accurately quantified by real-time polymerase chain reaction (PCR).23 In the present work, we provide direct experimental evidence that platelets contain large amounts of PAI-1 mRNA that is translationally active. Because the major part of the newly synthesized PAI-1 was found to be active, this could provide a mechanism by which platelet-rich clots maintain resistance to fibrinolysis.
Materials and methods
Reagents
Two different types of medium 199 were used, M199 without phenol red from Gibco BRL, Life Technology (Paisley, United Kingdom) and M199 without phenol red and methionine purchased from JHR Biosciences (Lenexa, KS). The RiboGreen RNA Quantitation Kit was from Molecular Probes (Leiden, The Netherlands). All PCR consumables were purchased from Applied Biosystems (Foster City, CA). Human aortic endothelial cells (HAECs) were purchased from Clonetics, BioWhittaker (Walkersville, MD). 35S-labeled methionine was from Amersham Biosciences (Buckinghamshire, United Kingdom). Trizol was from Gibco BRL, Life Technology. Monoclonal antibody against human PAI-1 (ab-1) was from Calbiochem (Darmstadt, Germany) and MAI-12 as well as single chain tPA were obtained from Biopool (Umeå, Sweden). Immunoprecipitation was performed with a protein-G immunoprecipitation kit purchased from Sigma (St Louis, MO). Supplies for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were from Bio-Rad (Hercules, CA). PAI-1 antigen enzyme-linked immunosorbent assay (ELISA) was purchased from Chromogenix (Mölndal, Sweden). LeucoCOUNT was from Becton Dickinson (Erembodegem-Aalst, Belgium). All other reagents were from Sigma: prostaglandin E1 (PGE1), piperazine diethanesulfonic acid (PIPES), puromycin, thrombin, and Triton X-100.
Research subjects
Platelets were collected from 21 apparently healthy research subjects (platelet counts 150-350 × 109/L) recruited from among hospital employees. The research subjects were not allowed to take aspirin or nonsteroidal anti-inflammatory drugs 10 days prior to the investigation. The protocols were approved by the Ethics Committee of Göteborg University.
Platelet isolation
Human blood was drawn in acid-citrate-dextrose (ACD; 1.5 mL/8.5 mL blood) containing 100 nM PGE1. Platelet-rich plasma (PRP) was prepared by centrifugation at 150g for 20 minutes. The PRP was recentrifuged at 150g for 10 minutes and then pelleted at 800g for 15 minutes. The supernatant was discarded and the platelet pellet was resuspended in PIPES/saline/glucose (5 mM PIPES, 145 mM NaCl, 4 mM KCl, 50 μM Na2HPO4, 1 mM MgCl2, and 5.5 mM glucose) containing 100 nM PGE1. Finally the platelets were pelleted (800g, 15 minutes) and the supernatant was discarded.
Analysis of leukocyte contamination
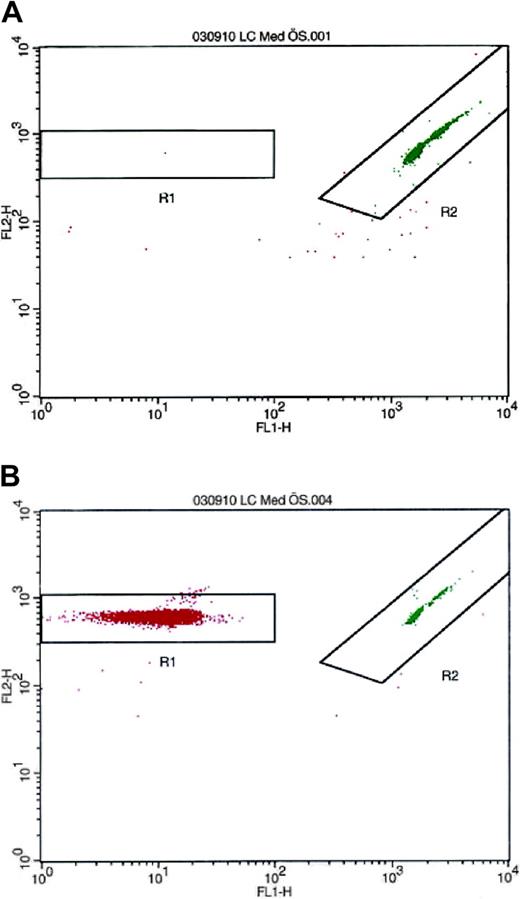
Because monocytes are known to synthesize PAI-1,24-26 separate experiments were performed to rule out the possibility that these cells could be the source of PAI-1 in the platelet suspension. To analyze the number of contaminating leukocytes, the isolated platelets were resuspended in platelet-free plasma to a concentration of 300 × 109/L. The PRP was analyzed by flow cytometry on a FACSCalibur (Becton Dickinson) with LeucoCOUNT according to the manufacturer's instructions. In these studies, the contamination rate was invariably fewer than 3 leukocytes in 10 million platelets (Figure 1).
Detection and enumeration of residual white blood cells in PRP with flow cytometry after LeucoCOUNT treatment. (A) PRP prepared by repeated centrifugation contained 2 to 3 contaminating leukocytes per 10 million platelets. (B) Buffy coat-contaminated PRP was used as positive control.
Detection and enumeration of residual white blood cells in PRP with flow cytometry after LeucoCOUNT treatment. (A) PRP prepared by repeated centrifugation contained 2 to 3 contaminating leukocytes per 10 million platelets. (B) Buffy coat-contaminated PRP was used as positive control.
Quantitative reverse transcriptase real-time PCR
RNA extraction and reverse transcription. Total RNA was extracted from platelets incubated in M199 for 0, 3, 8, and 24 hours using Trizol according to manufacturer's instructions. Total RNA was also extracted from human umbilical vein endothelial cells (HUVECs) explanted by incubation with 0.1% collagenase for 12 minutes at 37°C. The cells were rinsed out with phosphate-buffered saline (PBS) and after pelleting (10 minutes at 260g) the RNA was extracted using Trizol. RNA from purchased HAECs was also extracted using Trizol. Total RNA concentrations were determined using the RiboGreen RNA Quantitation Kit according to the manufacturer's instructions. The spectrofluorometric measurements were made with FLU-Ostar Galaxy (BMG Labtechnologies, Offenburg, Germany).
Reverse transcription of 0.2 μg total RNA was carried out in a total volume of 20 μL reaction mixture, consisting of 5 mM MgCl2, 1 mM dNTP mix, 50 mM KCl, 10 mM Tris (tris(hydroxymethyl)aminomethane)-HCl, pH 8.3, 2.5 μM random hexamer, 1 U/μL RNase inhibitor, and 2.5 U/μL Moloney murine leukemia virus (MuLV) reverse transcriptase. Samples were incubated at 20°C for 10 minutes, at 42°C for 15 minutes, at 99°C for 5 minutes, and finally at 5°C for 5 minutes.
Relative quantification was performed on an ABI PRISM 7700 Sequence Detector (Perkin-Elmer Applied Biosystems, Foster City, CA).
PCR conditions. Oligonucleotide primers and TaqMan probes were designed using the Primer Express version 1.0 software (Applied Biosystems). Each primer pair was selected so that the amplicon spanned an exon junction to preclude amplification of genomic DNA. Glyceraldehyde phosphate dehydrogenase (GAPDH) and cyclophilin were selected as endogenous controls to correct for potential variation in RNA loading or efficiencies of the reverse transcription or amplification reaction (Table 1).
Oligonucleotide primers and probes used for real-time quantitative PCR
Gene and oligonucleotide . | Sequence . | Position . |
|---|---|---|
| PAI-1 | ||
| Sense primer | 5′-ggc tga ctt cac gag tct ttc a-3′ | 11616-11637 |
| Antisense primer | 5′-ttc act ttc tgc agc gcc t-3′ | 11758-11780 |
| Probe | 5′ - (FAM) acc aag agc ctc tcc acg tcg cg (TAMRA) -3′ | 11782-11800 |
| Cyclophilin | ||
| Sense primer | 5′-gta cta tta gcc atg gtc aac ccc-3′ | 1648-1671 |
| Antisense primer | 5′-cag tca aag gag acg cgg cc-3′ | 1711-1727, 4178-4180 |
| Probe | 5′ - (FAM) cgt cga cgg cga gcc ctt g (TAMRA) -3′ | 1692-1710 |
| GAPDH | ||
| Sense primer | 5′-cca cat cgc tca gac acc at-3′ | 2171-2190 |
| Antisense primer | 5′-cca ggc gcc caa tac g-3′ | 2195-2217, 3850-3854 |
| Probe | 5′ - (FAM) aag gtg aag gtc gga gtc aac gga (TAMRA) -3′ | 3857-3872 |
Gene and oligonucleotide . | Sequence . | Position . |
|---|---|---|
| PAI-1 | ||
| Sense primer | 5′-ggc tga ctt cac gag tct ttc a-3′ | 11616-11637 |
| Antisense primer | 5′-ttc act ttc tgc agc gcc t-3′ | 11758-11780 |
| Probe | 5′ - (FAM) acc aag agc ctc tcc acg tcg cg (TAMRA) -3′ | 11782-11800 |
| Cyclophilin | ||
| Sense primer | 5′-gta cta tta gcc atg gtc aac ccc-3′ | 1648-1671 |
| Antisense primer | 5′-cag tca aag gag acg cgg cc-3′ | 1711-1727, 4178-4180 |
| Probe | 5′ - (FAM) cgt cga cgg cga gcc ctt g (TAMRA) -3′ | 1692-1710 |
| GAPDH | ||
| Sense primer | 5′-cca cat cgc tca gac acc at-3′ | 2171-2190 |
| Antisense primer | 5′-cca ggc gcc caa tac g-3′ | 2195-2217, 3850-3854 |
| Probe | 5′ - (FAM) aag gtg aag gtc gga gtc aac gga (TAMRA) -3′ | 3857-3872 |
A standard curve was obtained through amplification of the 3 target gene cDNAs in 2-fold template dilution series of RNA from 1:4 through 1:128. For all mRNAs, straight inverse correlations were observed between threshold cycle (CT) values and the amount of applied cellular RNA (R2 = 0.991, 0.991, and 0.986 for PAI-1, GAPDH, and cyclophilin, respectively).
For amplification of PAI-1, GAPDH, and cyclophilin, 2 μL cDNA diluted 1:4 was added to the PCR mixture consisting of TaqMan Universal PCR Master Mix, 15 pmol of each primer, and 10 pmol probe in a final volume of 50 μL. Thermal cycling conditions were 2 minutes at 50°C and 10 minutes at 95°C followed by 50 cycles of 2-step PCR consisting of 15 seconds at 95°C and 1 minute at 60°C. All samples were amplified in triplicate.
Metabolic radiolabeling and immunoprecipitation
Platelets were prepared as described (see “Platelet isolation”) and the platelets were resuspended in 1 mL methionine-free medium 199 and incubated for 2 hours followed by addition of 50 μCi (1.85 MBq) 35S-methionine. For each experimental point 5 × 108 platelets were used. Platelets were gently rocked alone or in the presence of thrombin (0.1 U/mL), tPA (1, 10, or 100 ng/mL), or puromycin (1 mM) for designated times. After each time period the platelets were lysed in the medium (50 mM Tris-HCl, 50 mM NaCl, 1 mM MgCl2, 1 mM EDTA [ethylenediaminetetraacetic acid], and 0.1% Triton X-100) for 30 minutes on ice. The soluble protein fractions, that is, both intracellular and released, were immunoprecipitated with anti-human PAI-1 antibodies using protein G-agarose beads according to the manufacturer's instructions. The beads were washed, placed in Laemmli sample buffer, and boiled for 5 minutes to elute the immunoprecipitated 35S-labeled proteins. The proteins were separated on a SDS-PAGE gel and then analyzed by autoradiography.
Assay of PAI-1 antigen
To study the amount of PAI-1 synthesized by platelets during 24 hours, platelets were isolated and prepared as described (see “Platelet isolation”) and incubated at 37°C in M199. After 24 hours the platelets were lysed as described and the PAI-1 antigen level at 0 and 24 hours was determined by ELISA. To study the release rate for PAI-1, platelets were prepared and incubated as described and after 0, 3, 8, and 24 hours the platelets were pelleted (800g, 15 minutes), the supernatant was removed, and lysis buffer was added. PAI-1 antigen levels were analyzed by ELISA, in platelet lysate as well as in relisate (supernatant).
Statistical methods
Standard statistical methods were used. Parametric methods (t test) were used after log-transformation for evaluation of the changes in PAI-1 concentration and significance tests were considered significant at P less than .05 (2-tailed test).
Results
Detection and quantification of PAI-1 mRNA
On the average, 0.2 to 0.6 ng total RNA was retrieved per million platelets. PAI-1 mRNA expression in platelets was then analyzed with real-time PCR and compared with the levels of the 2 endogenous control genes, GAPDH and cyclophilin. Substantial amounts of PAI-1 mRNA were consistently detected in all platelet samples. At baseline, the expression level of PAI-1 mRNA was approximately 6% of GAPDH and 7% of cyclophilin.
To get an estimation of the relative expression level of PAI-1 in platelets, we compared PAI-1 mRNA levels with those of HUVECs and HAECs. The total amount of RNA isolated from HUVECs and HAECs was approximately 5 μg per million cells. The expression of PAI-1 mRNA in platelets was approximately 60 times lower than in HUVECs and 45 times lower than in HAECs. When normalized to the endogenous control genes, PAI-1 normalized to GAPDH was 18-fold lower and PAI-1 normalized to cyclophilin was 26 times lower in platelets than in HUVECs. The corresponding relative expression levels were 17 and 12 in HAECs, respectively.
The degradation rate of PAI-1 mRNA compared to GAPDH and cyclophilin was analyzed by incubation of platelets for 0, 3, 8, and 24 hours followed by mRNA extraction and analysis of mRNA levels for the 3 genes by real-time PCR. As shown in Figure 2, the degradation rate of PAI-1 mRNA was found to be 4-fold higher than that of GAPDH and 8-fold higher than that of cyclophilin.
The relative degradation rate of platelet PAI-1 mRNA in comparison to the 2 endogenous reference genes, GAPDH and cyclophilin. mRNA extracted from platelets incubated 0, 3, 8, and 24 hours was determined by real-time PCR. The degradation rate of PAI-1 mRNA was 4-fold higher than that of GAPDH and 8-fold higher than that of cyclophilin.
The relative degradation rate of platelet PAI-1 mRNA in comparison to the 2 endogenous reference genes, GAPDH and cyclophilin. mRNA extracted from platelets incubated 0, 3, 8, and 24 hours was determined by real-time PCR. The degradation rate of PAI-1 mRNA was 4-fold higher than that of GAPDH and 8-fold higher than that of cyclophilin.
De novo synthesis
To estimate the PAI-1 protein synthesis rate, platelets were incubated and the total amount of PAI-1 antigen was analyzed by ELISA at baseline and after 24 hours. In fresh platelets, the average content of PAI-1 was 1.00 ± 0.33 ng (mean and SD) per million platelets. After 24 hours of incubation, the concentration of PAI-1 increased in 16 of 18 samples with individual responses ranging between 2% and 52%. On the average, the PAI-1 content increased by 25% to 1.25 ± 0.54 ng (mean and SD) per million platelets (P = .001).
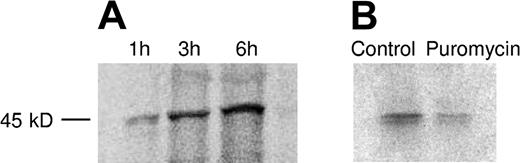
To confirm that there was an ongoing de novo synthesis of PAI-1 in platelets metabolic radiolabeling was performed. Following 35S-methionine incorporation for 1, 3, and 6 hours, immunoprecipitation was performed, which yielded a protein of the expected molecular mass of approximately 45 kD. The protein was detected both with the MAI-12 and PAI-1 (ab-1) antibodies. Figure 3A shows the results of the immunoprecipitation with PAI-1 (ab-1) of the 35S-labeled PAI-1. The increasing amount of radioactive PAI-1 over time confirms that there is an ongoing synthesis of PAI-1 in platelets. Following control immunoprecipitating without antibodies, no protein was detected (data not shown).
Metabolic radiolabeling and immunoprecipitation of platelet PAI-1. (A) Isolated platelets were incubated in the presence of 35S-methionine for 1, 3, and 6 hours. Platelet lysate and medium were immunoprecipitated with PAI-1 (ab-1) and the precipitated protein was separated with SDS-PAGE and subsequently examined by autoradiography. This yielded a protein of the expected molecular mass (approximately 45 kD), and the increasing amount of radioactive PAI-1 over time confirmed that there is an ongoing synthesis. (B) To inhibit protein synthesis, platelets were incubated for 6 hours in the presence of 1 mM puromycin, resulting in attenuated PAI-1 expression.
Metabolic radiolabeling and immunoprecipitation of platelet PAI-1. (A) Isolated platelets were incubated in the presence of 35S-methionine for 1, 3, and 6 hours. Platelet lysate and medium were immunoprecipitated with PAI-1 (ab-1) and the precipitated protein was separated with SDS-PAGE and subsequently examined by autoradiography. This yielded a protein of the expected molecular mass (approximately 45 kD), and the increasing amount of radioactive PAI-1 over time confirmed that there is an ongoing synthesis. (B) To inhibit protein synthesis, platelets were incubated for 6 hours in the presence of 1 mM puromycin, resulting in attenuated PAI-1 expression.
To inhibit protein translation, puromycin was added at a concentration of 1 mM. As shown in Figure 3B, this resulted in a partial inhibition of protein synthesis.
PAI-1 activity
To investigate to what extent the newly synthesized PAI-1 was active, a functional assay was performed in which platelets were incubated with 35S-methionine in the presence of increasing tPA concentrations. PAI-1 as well as the tPA/PAI-1 complex was detected by immunoprecipitating with the MAI-12 antibody. Figure 4A shows the result of 2 platelet incubations; the first is a control without tPA present and the second is with 100 ng tPA. Incubation with a molar excess of 100 ng tPA resulted in a different migration position corresponding to the expected position of the tPA/PAI-1 complex. This finding indicated that the newly formed PAI-1 was in an active configuration. As shown in Figure 4B, addition of increasing concentrations of 1-10-100 ng tPA resulted in a gradual diminution of free PAI-1 protein.
Functional analysis of the activity of newly synthesized PAI-1. Platelets were incubated in the presence of 1, 10, and 100 ng tPA for 6 hours as described in Figure 3. (A) Addition of 100 ng tPA resulted in a different migration position corresponding to the expected position of the tPA/PAI-1 complex, indicating that newly synthesized PAI-1 is active. (B) Addition of increasing concentrations of tPA reduced the free PAI-1 protein and increased the tPA/PAI-1 complex.
Functional analysis of the activity of newly synthesized PAI-1. Platelets were incubated in the presence of 1, 10, and 100 ng tPA for 6 hours as described in Figure 3. (A) Addition of 100 ng tPA resulted in a different migration position corresponding to the expected position of the tPA/PAI-1 complex, indicating that newly synthesized PAI-1 is active. (B) Addition of increasing concentrations of tPA reduced the free PAI-1 protein and increased the tPA/PAI-1 complex.
Platelet PAI-1 release and effects of thrombin stimulation
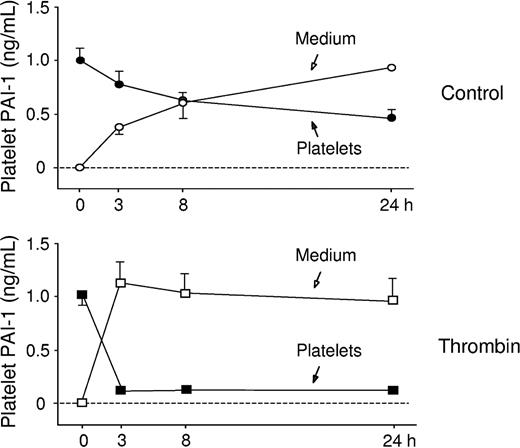
During control conditions, there was a continuous release of PAI-1 from the platelets (P < .0001), and after 24 hours of incubation 67% of platelet PAI-1 was found in the medium.
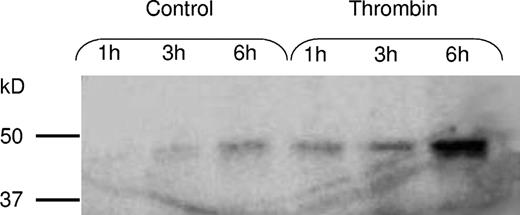
Thrombin was added to investigate if the synthesis or release rate could be stimulated by a platelet agonist. Platelets were incubated for 1, 3, and 6 hours in the presence of 0.1 U/mL thrombin. As shown in Figure 5, thrombin activation increased the rate of de novo synthesis of PAI-1. Also, the release rate of PAI-1 from the activated platelets was increased, and during thrombin stimulation almost all PAI-1 was released into the medium within the first 3 hours of incubation (Figure 6). Radiolabeling and immunoprecipitation were also performed to study the release of newly synthesized PAI-1 from platelets. Platelet lysate and relisate were studied separately and the results from this assay (data not shown) were in agreement with the findings described. This experiment showed that during thrombin stimulation almost all of the new protein was released into the medium, in contrast to the unstimulated condition where the majority of the radiolabeled PAI-1 was retained within the platelets. When performing radiolabeling studies in the presence of tPA, the PAI-1 synthesized during thrombin stimulation was found to be active.
Stimulation of PAI-1 synthesis with thrombin. To investigate whether the platelets could be stimulated to increase the synthesis rate, platelets were incubated for 1, 3, and 6 hours, as described in Figure 3, in the absence or presence of 0.1 U/mL thrombin. Thrombin activation was found to increase the rate of PAI-1 synthesis.
Stimulation of PAI-1 synthesis with thrombin. To investigate whether the platelets could be stimulated to increase the synthesis rate, platelets were incubated for 1, 3, and 6 hours, as described in Figure 3, in the absence or presence of 0.1 U/mL thrombin. Thrombin activation was found to increase the rate of PAI-1 synthesis.
Analysis of release rate of PAI-1 from activated platelets. To study the release rate of PAI-1, platelets were incubated for 0, 3, 8 and 24 hours and then pelleted and lysed. The PAI-1 concentration in medium and lysate was determined by ELISA. During basal conditions 67% of total PAI-1 was released in 24 hours. As expected, thrombin stimulated the release of PAI-1, and after 3 hours 85% of the total PAI-1 pool was found in the medium (mean ± SEM).
Analysis of release rate of PAI-1 from activated platelets. To study the release rate of PAI-1, platelets were incubated for 0, 3, 8 and 24 hours and then pelleted and lysed. The PAI-1 concentration in medium and lysate was determined by ELISA. During basal conditions 67% of total PAI-1 was released in 24 hours. As expected, thrombin stimulated the release of PAI-1, and after 3 hours 85% of the total PAI-1 pool was found in the medium (mean ± SEM).
Discussion
In the present study, we demonstrate for the first time that human platelets contain translationally active mRNA for PAI-1 and that there is a continuous de novo synthesis of PAI-1. Substantial amounts of PAI-1 mRNA were detected by real-time PCR and incorporation of radioactive methionine followed by immunoprecipitation confirmed that this mRNA was translated into protein. Over a period of 24 hours, the average total amount of PAI-1 protein in unstimulated platelets increased by 25%. Furthermore, the synthesis rate could be further stimulated when the platelets were activated by thrombin, and thrombin also induced a rapid release of PAI-1 from the activated platelets. Importantly, the newly formed protein was in an active form as shown by its ability to complex-bind and inhibit tPA.
Circulating platelets are anucleate and cannot synthesize mRNA. However, already in 1967 Booyse et al demonstrated that platelets retain megakaryocyte-derived mRNAs,17,18 and this was later confirmed by PCR amplification.19 Recently, it was shown by microarray analysis that platelets contain mRNA transcripts for a large number of genes.27 In the present study we provide evidence that mRNA for PAI-1 is present in considerable amounts in platelets. At baseline, the expression level of PAI-1 mRNA was approximately 5% to 10% of the endogenous references GAPDH and cyclophilin. Compared to its expression in HUVECs and HAECs, the relative amount of PAI-1 mRNA in platelets was between 12- and 26-fold lower, respectively.
To get an estimate of the absolute amounts of total RNA and the proportions of PAI-1 mRNA thereof, we determined the absolute amount of RNA in platelets and endothelial cells. The total RNA content was approximately 0.2 to 0.6 ng per million platelets, which is in accordance with what has been reported previously.28 The total RNA content of HUVECs and HAECs was approximately 5 μg per million cells. Thus, total RNA had a 10 000-fold lower concentration in platelets compared to endothelial cells. However, assuming a cell volume of 10 and 2000 fL for platelets and endothelial cells, respectively, the average total RNA content would be on the order of 50 times lower in platelets per unit cell volume. Because PAI-1 mRNA had a 45- to 60-fold lower concentration in platelets than in HAECs and HUVECs, this would indicate that the absolute expression of PAI-1 is on the order of 2250 to 3000 times lower in platelets per cell volume. However, we have previously found that the degradation rates of different platelet mRNA species vary substantially.23 During the 24-hour incubation period the degradation rate of PAI-1 mRNA was approximately 4-fold higher than that of GAPDH and 8-fold higher than that of cyclophilin. This finding suggests that the PAI-1 mRNA content of young, newly released platelets may be considerably higher.
Electron microscopy studies on platelets have revealed the presence of rough endoplasmic reticulum and polyribosomes,21 and platelets have the capacity to de novo synthesize proteins.21,22 The present findings clearly show that this is true also for PAI-1. At baseline, the content of PAI-1 was approximately 1.0 ng per million platelets, which is comparable to what has been reported earlier.14,15 During 24 hours of incubation, the total amount of PAI-1 increased by 25%, showing that despite its decay rate, considerable amounts of translationally active mRNA were still present. Under control conditions, there was a gradual release of PAI-1 protein into the medium, and after 24 hours 67% of the PAI-1 content was found in the medium. Metabolic radiolabeling with 35S-methionine and immunoprecipitation confirmed that there was an ongoing de novo synthesis of the protein in the platelets. Furthermore, this synthesis could be markedly attenuated by the protein synthesis blocker puromycin, which inhibits translation by prematurely terminating peptide chains. Although the present study does not permit any conclusion as to the relative contribution of PAI-1 from the megakaryocyte and from de novo synthesis in the platelet itself, it is likely that at least part of the protein has been synthesized in the megakaryocyte. Several previous studies have demonstrated the presence of PAI-1 mRNA29 and PAI-1 antigen30 in megakaryocytes and the storage of PAI-1 into α-granules.
Despite the considerable amount of PAI-1 present in the platelets, most previous studies have shown that only 5% to 10% of the platelet PAI-1 is in an active configuration.14-16 This observation has led to the inference that platelet PAI-1 is likely to be of minor importance for resistance of clots to fibrinolytic degradation. However, a few previous studies have reported the presence of unexpectedly high proportions of active PAI-1 in platelets.31,32 The existence of active PAI-1 in a platelet population with a mean age on the order of 3 to 4 days has been difficult to reconcile with the fact that under normal conditions, newly synthesized active PAI-1 spontaneously converts into an inactive form with a half-life on the order of 1 hour.33 A potential mechanism to explain the delayed inactivation has been suggested by Lang and Schleef,32 who showed that platelets have a unique mechanism for stabilization of PAI-1 in its active configuration by packaging with other, large α-granule proteins in a calcium-dependent manner.
The present observations that there is an ongoing de novo synthesis of PAI-1 provide an additional mechanism for the presence of active PAI-1 in the platelets. As shown by Wiman and coworkers, the mechanisms involved in the stability of active PAI-1 and the reaction between PAI-1 and tPA are rather complicated.34 Because it is difficult to directly determine the activity of PAI-1, we used a functional assay (determination of tPA/PAI-1 complexes) to prove that the newly formed PAI-1 was in an active configuration. The basis for this approach is that only active PAI-1 is capable of forming a stoichiometric 1:1 complex with tPA.35 This analysis confirmed that both during basal conditions and when protein synthesis was stimulated by thrombin, the new PAI-1 was active.
As expected, thrombin stimulated platelet PAI-1 synthesis in a time-dependent manner and also induced a rapid release of PAI-1 from the platelets. Radiolabeling studies in the presence of tPA confirmed that almost all of the PAI-1 synthesized during thrombin stimulation was active. Thus, even if thrombin may also cause release of inactive PAI-1 from the α-granules these observations indicate that thrombin will increase both the absolute and relative amount of active PAI-1 in the relisate. This mechanism may make more active PAI-1 available in a thrombin-rich environment, such as during formation of a blood clot.
Taken together, our observations suggest that the contribution of platelets for clot stabilization may be related to their capacity for synthesis of active PAI-1. Indeed, we observed that there was a substantial variation in the synthesis rate of PAI-1 among individuals. In view of the rather rapid decay of PAI-1 mRNA in the platelets, it is conceivable that at least partly this variation may be due to differences in the age distribution of the platelet population. Another interesting possibility is that the synthesis is related to the 4G/5G promoter polymorphism at the PAI-1 locus because individuals who are homozygous for the 4G allele have significantly higher plasma and platelet PAI-1 concentrations as well as higher levels of active PAI-1.31,36,37 However, the present study was not large enough to investigate the relative importance of the platelet age distribution and a putative influence of the polymorphism on PAI-1 synthesis.
In conclusion, the present study shows that platelets contain considerable amounts of translationally active mRNA for PAI-1 and that there is an ongoing de novo synthesis of PAI-1. The majority of the newly formed protein is in an active configuration, and its production and release could be further stimulated by thrombin. The continuous production of large amounts of active PAI-1 in platelets could be a mechanism by which platelets contribute to stabilization of blood clots.
Prepublished online as Blood First Edition Paper, August 17, 2004; DOI 10.1182/blood-2004-04-1439.
Supported by the Swedish Research Council (grant no. 9046), the Heart Lung Foundation, and the Faculty of Medicine, Göteborg University.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal