Abstract
Truncated granulocyte colony-stimulating factor receptors (G-CSF-Rs) are implicated in severe congenital neutropenia (SCN) and the consecutive development of acute myeloid leukemia (AML). Mice expressing G-CSF-R truncation mutants (gcsfr-d715) show defective receptor internalization, an increased signal transducer and activator of transcription 5 (STAT5)/STAT3 activation ratio, and hyperproliferative responses to G-CSF treatment. We determined whether a lack of negative feedback by suppressor of cytokine signaling (SOCS) proteins contributes to the signaling abnormalities of G-CSF-R–d715. Expression of SOCS3 transcripts in bone marrow cells from G-CSF–treated gcsfr-d715 mice was approximately 60% lower than in wild-type (WT) littermates. SOCS3 efficiently suppressed STAT3 and STAT5 activation by WT G-CSF-R in luciferase reporter assays. In contrast, while SOCS3 still inhibited STAT3 activation by G-CSF-R–d715, STAT5 activation was no longer affected. This was due mainly to loss of the SOCS3 recruitment site Tyr729, with an additional contribution of the internalization defects of G-CSF-R–d715. Because Tyr729 is also a docking site for the Src homology 2–containing protein tyrosine phosphatase-2 (SHP-2), which binds to and inactivates STAT5, we suggest a model in which reduced SOCS3 expression, combined with the loss of recruitment of both SOCS3 and SHP-2 to the activated receptor complex, determine the increased STAT5/STAT3 activation ratio and the resulting signaling abnormalities projected by truncated G-CSF-R mutants.
Introduction
Granulocyte colony-stimulating factor (G-CSF) receptor is the major regulator of neutrophil production, both under steady-state conditions and during stages of bacterial infections.1-3 G-CSF exerts its activity via a receptor (G-CSF-R) of the hematopoietin receptor superfamily.4,5 Typical of this class of receptors, G-CSF-R has no intrinsic kinase activity but recruits cytoplasmic tyrosine kinases of both the Janus kinase (Jak) and Src kinase families and activates signal transducer and activator of transcription (STAT) proteins.6-11 G-CSF activates STATs 1, 3, and 5.12-14 Whereas the contribution of STAT1 to G-CSF responses remains unclear, STAT3 has been implicated in G-CSF–mediated growth arrest preceding differentiation, while activation of STAT5 has been linked to proliferation and survival signaling.15-17 Four tyrosine residues (Tyr704, Tyr729, Tyr744, and Tyr764) in the G-CSF-R carboxy-terminus are involved in the recruitment of signaling molecules, such as the adapter molecules growth factor receptor–bound protein 2 (Grb2) and Src homology and collagen protein (Shc) of the p21Ras–mitogen-activated protein (MAP) kinase pathway, and the Src homology 2–containing protein tyrosine phosphatase-2 (SHP-2).18-20 In addition, activation of STAT3 depends on its recruitment to the G-CSF-R via tyrosines 704 or 744.20-22 At higher G-CSF concentrations, STAT3 can also be activated in a tyrosine-independent way via the G-CSF-R C-terminus.22,23 In contrast, activation of STAT1 and STAT5 is achieved via the membrane-proximal region of G-CSF-R and does not require receptor tyrosine residues.13,17
In approximately 20% of patients suffering from severe congenital neutropenia (SCN), G-CSF-R mutations are found that result in the expression of a G-CSF-R with a truncated C-terminus.24,25 These patients have an increased risk of developing acute myeloid leukemia (AML).25,26 Activation of a G-CSF-R mutant truncated at amino acid 715 (G-CSF-R–d715) causes a hyperproliferative response in 32D cells, without induction of neutrophilic differentiation.27 Mice with a targeted G-CSF-R–d715 mutation show various degrees of neutropenia, and their myeloid precursors react to G-CSF administration with hyperproliferation, resulting in a sustained neutrophilia.28,29 Interestingly, transgenic (Tg) mice overexpressing G-CSF-R mutants truncated at amino acids (aa's) 718 and 731 demonstrated increased susceptibility to infection with Staphylococcus aureus, suggesting that production of functional neutrophils is compromised in these animals.30 Indeed, these Tg mice had only one third of the peripheral neutrophil levels of wild-type (WT) controls, and their bone marrow showed increased percentages of immature myeloid cells.
Functional analysis of truncated receptors revealed that a number of properties are altered compared with WT G-CSF-R. Ligand-induced internalization of G-CSF-R–d715 is severely affected owing to the loss of 2 distinct motifs in the receptor C-terminus that are important for internalization.27,31,32 G-CSF-R–d715 also has a somewhat reduced ability to activate STAT3, possibly owing to the loss of the STAT3 recruitment site Tyr744 and the receptor C-terminus.22,31 In contrast, activation of STAT5 is strongly increased and is sustained after removal of G-CSF, suggesting a prominent role for the C-terminus in mediating negative feedback on STAT5 activation.17,27,31 Although the defective internalization properties of truncated G-CSF-R forms contributed significantly to their sustained signaling function, it was also clear that this did not fully explain these findings.27,32 In particular, the differential effects of receptor truncations on the kinetics of STAT3 versus STAT5 activation remained unclear.
Suppressor of cytokine signaling (SOCS) proteins are involved in the down-regulation of signaling from a number of hematopoietic growth factor receptors, including G-CSF-R.33-36 A conserved SH2 domain and a C-terminal SOCS box are characteristic for the SOCS family (reviewed in Krebs and Hilton,37 Kile and Alexander,38 and Alexander39 ). The expression of most SOCS genes is controlled by STAT transcription factors.40-45 SOCS proteins therefore act in a classical negative feedback loop to suppress cytokine signaling. Three distinct inhibitory mechanisms have been linked to SOCS proteins. Cytokine-inducible SH2-containing protein (CIS), founding member of the family, inhibits activation of STAT5 by competing for STAT5 recruitment to phosphotyrosine motifs in, for example, the growth hormone receptor (GHR) and the erythropoietin receptor (EpoR).46,47 SOCS1 and SOCS3, on the other hand, directly suppress Jak kinase activity by means of a kinase inhibitory region (KIR).48-50 Upon recruitment to the signaling complex via the SH2 domain of SOCS, the KIR mediates inhibition by blocking access of both adenosine triphosphate (ATP) and substrate to their binding sites in the catalytic groove of Jak2.49-53 Finally, SOCS proteins are also thought to down-regulate signaling via SOCS box–mediated targeting of signaling proteins for proteasomal degradation.54,55 An important difference between SOCS1 and SOCS3 relates to how they are recruited into activated receptor complexes. Whereas the SH2 domain of SOCS1 has a high affinity for, for example, phosphorylated Tyr1007 in the Jak homology 1 (JH1) domain of Jak2, the affinity of the SH2 domain of SOCS3 for this residue is much lower.49,52,56 Instead, SOCS3 is recruited with high affinity to phosphotyrosine-based motifs in certain receptors and then subsequently inhibits Jak activity via its KIR.34,35,48,57-59 Importantly, for a number of cytokine receptors, it has been established that the protein tyrosine phosphatase SHP-2 and SOCS3 dock to identical tyrosine-based motifs with comparable affinities.57-60
Studies in SOCS-deficient mice have demonstrated major physiologic roles for SOCS proteins in controlling the levels of cytokine signaling in both nonhematopoietic and hematopoietic cells.61-72 G-CSF induces the expression of SOCS1, SOCS2, SOCS3, and CIS in hematopoietic cells,35,73 but only SOCS1 and SOCS3 appeared to inhibit G-CSF–induced STAT activation.74 In 2 independent studies, Tyr729 of G-CSF-R was identified as the major recruitment site for SOCS3.35,36 Recently, it was reported that SOCS3 is a key negative regulator of G-CSF–induced neutrophil production in vivo.75,76
In the present study, we investigated to what extent and by which mechanism(s) truncation of the G-CSF-R C-terminus, as found in SCN, affects the negative feedback regulation of G-CSF signaling by SOCS3. We show that truncation of the G-CSF-R completely relieved the inhibitory effects of SOCS3 on activation of STAT5. In striking contrast, the suppressive effects of SOCS3 on G-CSF-R–d715–induced activation of STAT3 were hardly affected by the truncation. These findings provide a new mechanistic explanation for the increased ratio of STAT5/STAT3 activation in gcsfr-d715 mice, which has previously been linked to the shift in the proliferation/differentiation balance in the myeloid progenitor cell compartment found in these animals.31
Materials and methods
Expression constructs
The constructs of human G-CSF-R WT, d715, Tyr729Phe, d735, d749-769, d715-735, and the green fluorescent protein (GFP)–G-CSF-R fusions in the pBabe vector77 have been described before18,20,32,36 (Figure 1A). The d735Phe and d749-769Phe mutants were generated from mutant Tyr729Phe by, respectively, introduction of a stop codon or deletion of aa's 749-769 with a site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene, La Jolla, CA). Expression constructs of myc-tagged SOCS1 and SOCS3 in pcDNA3 were a gift from A. Yoshimura.56 For expression of SHP-2, hemagglutinin (HA)–tagged human SHP-2 was cloned into the EcoRI-XhoI sites of expression vector pSG5.78 For expression of STAT5, pME18S-STAT5B was used.79
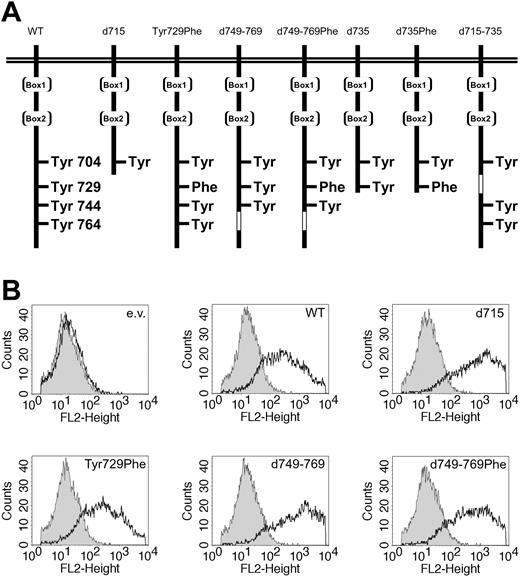
Expression of WT and mutant G-CSF receptors. (A) Schematic representation of the intracellular domain of the G-CSF receptor and mutants. Boxes 1 and 2 represent subdomains conserved in the hematopoietin receptor superfamily. The open box indicates the deleted region in G-CSF-R–d749-769 and G-CSF-R–d715-735. (B) Flow cytometric analysis of the expression levels of the different G-CSF-R forms used in luciferase reporter assays. Open histograms indicate cells stained with biotinylated G-CSF-R antibodies and phycoerythrin-conjugated streptavidin (SA-PE); shaded histograms, cells stained with SA-PE only; and e.v., cells transfected with empty vector.
Expression of WT and mutant G-CSF receptors. (A) Schematic representation of the intracellular domain of the G-CSF receptor and mutants. Boxes 1 and 2 represent subdomains conserved in the hematopoietin receptor superfamily. The open box indicates the deleted region in G-CSF-R–d749-769 and G-CSF-R–d715-735. (B) Flow cytometric analysis of the expression levels of the different G-CSF-R forms used in luciferase reporter assays. Open histograms indicate cells stained with biotinylated G-CSF-R antibodies and phycoerythrin-conjugated streptavidin (SA-PE); shaded histograms, cells stained with SA-PE only; and e.v., cells transfected with empty vector.
Bone marrow cells and isolation of RNA
WT and gcsfr-d715 mice28 were stimulated daily for 4 days with G-CSF or received solvent only. Each experimental group contains 2 mice of each genotype. Bone marrow cells were isolated, resuspended in TRIzol RNA extraction reagent (Invitrogen, Breda, The Netherlands), snap frozen, and stored at –80°C.28 For the in vitro stimulation, bone marrow cells were harvested from 2 WT and 2 gcsfr-d715 mice that had not been treated with G-CSF. Cells were cultured for 1 hour in Hanks balanced salt solution (HBSS; Invitrogen) with 5% fetal calf serum (FCS); nonadherent cells were taken and starved for 4 hours in RPMI (Invitrogen) plus 0.5% bovine serum albumin (BSA). Cells were stimulated for the indicated periods with G-CSF (100 ng/mL) and resuspended in Trizol. RNA was isolated according to the manufacturer's instructions and subsequently treated with DNAse to remove genomic DNA. DNAse treatment of 5 μg RNA was performed in DNAse buffer (40 mM Tris-HCl [tris(hydroxymethyl)aminomethane–HCl], pH 7.5; 6 mM MgCl2; 2 mM CaCl2) with 10 U DNAse I (Stratagene) for 1 hour at 37° C.
Quantitative reverse-transcriptase PCR (RT-PCR)
To generate cDNA, 1 μg RNA was denatured at 65° C for 5 minutes followed by 10 minutes on ice. After addition of first-strand buffer (250 mM Tris-HCl, pH 8.3; 375 mM KCl; 15 mM MgCl2) with deoxynucleoside triphosphates (dNTPs; 1 mM final concentration), dithiothreitol (DTT; 1 mM final concentration), 4 μg random hexamers (Amersham Pharmacia, Uppsala, Sweden), 40 U RNasin, and 200 U Superscript II reverse transcriptase (Invitrogen), the reaction was incubated at 42° C for 2 hours. The cDNA was diluted 1:10, 1:30, and 1:60 for SOCS1, SOCS3, and ribonuclease inhibitor, respectively, before polymerase chain reaction (PCR) amplification. Primers used for amplification for SOCS1 were as follows: FTMSOCS1, 5′-tggtagcacgcaaccaggtg; and RTMSOCS1, 5′-tggcgaggacgaagacgag. Primers used for SOCS3 were FTMSOCS3, 5′-tcaagaccttcagctccaa; and RTMSOCS3, 5′-tcttgacgctcaacgtgaag. Primers for murine ribonuclease inhibitor were forward 5′-tccagtgtgagcagctgag, and reverse 5′-tgcaggcactgaagcacca. For the quantitative real-time PCR, Taqman technology was used (Model 7900 sequence detector; PE Applied Biosystems, Foster City, CA). The reactions were performed in a 25 μL vol of a mixture containing 2 μL of the respective cDNA dilution, primers at 5 μM, and 12.5 μL of 2 × SYBR green PCR Master mix (PE Applied Biosystems) containing Amplitaq Gold DNA polymerase, reaction buffer, dNTP mix with uridine 5′-triphosphate (UTP), and the double-stranded DNA–specific fluorescence dye SYBR green I. The PCR program used was 1 cycle of 2 minutes at 50° C, 1 cycle of 10 minutes at 95° C, 45 cycles of denaturation for 15 seconds at 95° C, annealing for 30 seconds at 62° C, and extension for 30 seconds at 62° C. To determine the expression levels, samples were tested in duplicate, and the average values of the threshold cycle (Ct) were used for quantification. To quantify the relative expression of SOCS1 and SOCS3, the Ct values were normalized for endogenous reference (ΔCt = CtSOCS – Ctribonuclease inhibitor) and compared with a calibrator, by means of the ΔΔCt method (Ct = CtSample – CtCalibrator). As calibrator for G-CSF stimulation in vivo, we used the expression in WT bone marrow of unstimulated mice. As calibrator for the in vitro stimulation, we used expression after 4 hours of starvation.
Luciferase assays
Luciferase assays were performed as described previously.36 In short, HEK293 cells, grown in 24-well plates, were transfected by the calcium phosphate precipitation method with a mixture of the following plasmids. For STAT5 luciferase experiments, we used expression vector pME18S-STAT5, a β-casein–derived STAT5 luciferase reporter plasmid, a β-galactosidase expression plasmid pRSVLacZ, a pBabe construct with WT or mutant G-CSF-R (Figure 1A), and different amounts of pCDNA3 with myc-tagged SOCS or empty pcDNA3 (Invitrogen). For the STAT3 luciferase experiments pME18S STAT5 was replaced by pcDNA3 vector, and an m67-derived STAT3 luciferase reporter was added instead of the STAT5 reporter. A volume of 100 μL calcium phosphate precipitate in a total of 2 μg DNA was added to each well. With the exception of SOCS3, 400 ng DNA for each construct was added per well. Different amounts of pcDNA3-SOCS3 were added, supplemented with empty pcDNA3 vector up to 400 ng. After 24 hours, the cells were starved overnight in Dulbecco minimum essential medium (DMEM) plus penicillin/streptomycin (pen/strep) plus 0.1% BSA. The next day, the cells were stimulated with 250 ng/mL G-CSF for 6 hours, lysed, and assayed for luciferase activity with the use of Steady-Glo reagents (Promega, Madison, WI). In parallel, the transfection efficiency was determined by means of lacZ staining. Luciferase activity levels were corrected for transfection efficiency with the use of β-galactosidase expression levels. All experiments were performed in triplicate. Fold induction by G-CSF was calculated and set at 100% in the absence of SOCS. For inhibitor studies, the Src inhibitor PP-2, the Jak inhibitor WHI-P154 (Calbiochem, San Diego, CA), or dimethyl sulfoxide (DMSO) as a solvent control was added to the cells 1 hour prior to G-CSF stimulation. Unless stated otherwise, data were analyzed by means of analysis of variance (ANOVA). To compare G-CSF-R expression levels, transfected cells were stained with biotinylated mouse antihuman G-CSF-R antibody (Pharmingen, San Diego, CA) followed by SA-PE (Caltag Laboratories, Burlingame, CA), and analyzed by flow cytometry (FACSCalibur; Becton Dickinson, Sunnyvale, CA). As shown in Figure 1B, G-CSF-R expression is detectable with increased (less than 1 log unit) expression of the internalization-defective mutants (G-CSF-R–d715, d749-769, and d749-769Phe). As previously reported, this difference is due mainly to decreased spontaneous internalization of these mutants in the absence of G-CSF in nonmyeloid cells.32
Immunoprecipitations and Western blotting
Phoenix E cells (a gift from G. Nolan, Stanford, CA) were transfected with G-CSF-R, SHP-2, and, in the case of the SHP-2-STAT5 coimmunoprecipitations, with the STAT5 expression construct as well. After 24 hours, the medium was replaced by DMEM plus 0.1% BSA. The next day, cells were stimulated for 10 minutes with G-CSF, washed twice with cold phosphatebuffered saline (PBS), and lysed in lysis buffer containing 20 mM Tris-HCl, pH 8.0; 137 mM NaCl; 10 mM EDTA (ethylenediaminetetraacetic acid), 100 mM NaF, 1% Nonidet P40 (NP40), 10% glycerol, 2 mM Na3VO4, and 1 mM Pefablock SC, 50 μg/mL aprotinin, 50 μg/mL leupeptin, 50 μg/mL bacitracin, and 50 μg/mL iodoacetamide as protease inhibitors. Immunoprecipitations with anti-HA antibody and protein G-sepharose beads (Sigma, Zwijndrecht, The Netherlands) and subsequent Western blotting were performed as described previously.7 Antibodies used were mouse anti-HA (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti–SHP-2 (Santa Cruz Biotechnology), mouse anti–green fluorescent protein (Roche, Almere, The Netherlands), and rabbit anti-STAT5B (Santa Cruz Biotechnology).
Results
Reduced G-CSF–induced SOCS3 expressionin gcsfr-d715 mice
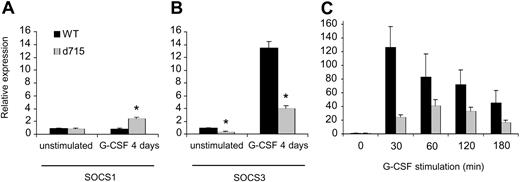
SOCS proteins are under the transcriptional control of STATs.41,43,44,78 Because G-CSF-R–d715 shows altered activation of STAT3 and STAT5 compared with WT G-CSF-R,27,31 we first investigated expression of SOCS1 and 3 in WT versus gcsfr-d715 mice under steady-state conditions and after daily treatment of the animals with G-CSF for 4 days. As shown in Figure 2A, steady-state levels of SOCS1 in gcsfr-d715 mice and their wild-type littermates were similar. Also after G-CSF treatment, SOCS1 transcript levels in wild-type and gcsfr-d715 mice had not changed dramatically. A slight (2.5-fold) increase in SOCS1 mRNA levels in gcsfr-d715 mice compared with wild-type animals was noted (Figure 2A). In contrast, SOCS3 expression was strongly (greater than 12-fold) induced by G-CSF in WT mice, while expression levels in gcsfr-d715 mice reached only about 30% of these levels both after G-CSF stimulation and in steady state (Figure 2B). In addition, we isolated bone marrow cells from untreated WT and d715 animals and stimulated them with G-CSF in vitro. As shown in Figure 2C, stimulation with G-CSF gives a strong induction of SOCS3 mRNA. Again, SOCS3 transcript levels are reduced in G-CSF-R–d715 cells upon stimulation with G-CSF. This demonstrates that the reduced up-regulation of SOCS3 mRNA also occurs outside the bone marrow compartment and is due to altered signaling in the G-CSF-R–d715 cells. These results establish that SOCS3 is the principal SOCS protein induced by G-CSF and that C-terminal truncation of the G-CSF-R results in a significantly reduced ability of the receptor to induce SOCS3.
Reduced SOCS3 but not SOCS1 expression in gcsfr-d715 mice. WT mice and gcsfr-d715 littermates were injected with vehicle or G-CSF for 4 consecutive days. (A-B) RNA was isolated, and SOCS1 (A) and SOCS3 (B) transcript levels were measured by quantitative RT-PCR. Expression of ribonuclease inhibitor was used for normalization of the data. SOCS levels were expressed relative to untreated WT mice. Data shown are mean + standard error of the mean (SEM) of 3 experiments. *Difference between WT and G-CSF-R–d715 is significant; P < .05 by Student t test. (C) SOCS3 expression in bone marrow cells deprived of growth factor for 4 hours and then stimulated with G-CSF in vitro for the indicated times. SOCS3 levels were expressed relative to growth factor–deprived cells. Data shown are mean + SEM of 4 experiments; differences between WT and G-CSF-R–d715 are significant (Student t test, P < .05).
Reduced SOCS3 but not SOCS1 expression in gcsfr-d715 mice. WT mice and gcsfr-d715 littermates were injected with vehicle or G-CSF for 4 consecutive days. (A-B) RNA was isolated, and SOCS1 (A) and SOCS3 (B) transcript levels were measured by quantitative RT-PCR. Expression of ribonuclease inhibitor was used for normalization of the data. SOCS levels were expressed relative to untreated WT mice. Data shown are mean + standard error of the mean (SEM) of 3 experiments. *Difference between WT and G-CSF-R–d715 is significant; P < .05 by Student t test. (C) SOCS3 expression in bone marrow cells deprived of growth factor for 4 hours and then stimulated with G-CSF in vitro for the indicated times. SOCS3 levels were expressed relative to growth factor–deprived cells. Data shown are mean + SEM of 4 experiments; differences between WT and G-CSF-R–d715 are significant (Student t test, P < .05).
G-CSF-R truncation relieves the suppressive effects of SOCS3 on G-CSF–induced STAT5 but not STAT3 activation
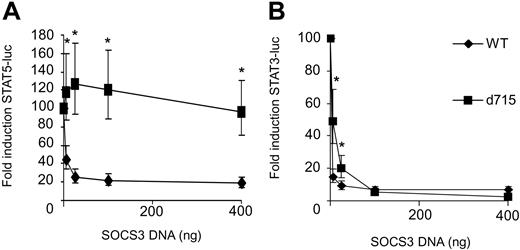
We next studied the consequences of SOCS3 expression on G-CSF–induced activation of STAT5 and STAT3 luciferase reporter constructs in HEK293 cell transfectants expressing either WT G-CSF-R or G-CSF-R–d715. G-CSF–induced STAT5 activity by WT G-CSF-R was dose dependently inhibited by SOCS3 (Figure 3A). In contrast, STAT5 activity induced by G-CSF-R–d715 was not affected, even at the highest SOCS3 expression levels. This finding can be reconciled with recent reports showing that Tyr729 in G-CSF-R is the major docking site for SOCS3, a residue that is lacking in G-CSF-R–d715.35,36 Surprisingly, STAT3 activation by G-CSF-R–d715 remained highly sensitive to inhibition by SOCS3 (Figure 3B). These data establish that Tyr729, while essential for down-regulation of STAT5, is dispensable for the inhibitory effects of SOCS3 on G-CSF–induced STAT3 activation.
STAT5 activation by G-CSF-R–d715 is insensitive to inhibition by SOCS3, whereas STAT3 activation is not. HEK293 cells transfected with STAT5 or STAT3 luciferase reporter constructs were stimulated with G-CSF for 6 hours and assayed for luciferase activity. G-CSF–induced STAT5 (A) or STAT3 (B) luciferase reporter activity in the absence of SOCS3 was set at 100%. Data are expressed as mean + 95% confidence interval of 4 independent experiments. *Differences between WT and G-CSF-R–d715 are significant; P < .01.
STAT5 activation by G-CSF-R–d715 is insensitive to inhibition by SOCS3, whereas STAT3 activation is not. HEK293 cells transfected with STAT5 or STAT3 luciferase reporter constructs were stimulated with G-CSF for 6 hours and assayed for luciferase activity. G-CSF–induced STAT5 (A) or STAT3 (B) luciferase reporter activity in the absence of SOCS3 was set at 100%. Data are expressed as mean + 95% confidence interval of 4 independent experiments. *Differences between WT and G-CSF-R–d715 are significant; P < .01.
Differential effects of SOCS3 on G-CSF-R–d715–mediated STAT3 and STAT5 activation are not due to distinct involvement of upstream tyrosine kinases
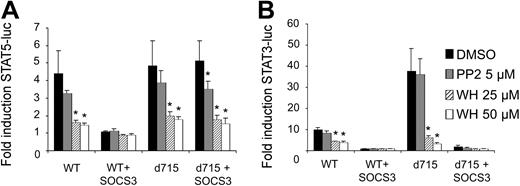
G-CSF-R activates the Janus kinases Jak1, Jak2, and Tyk2, but also the Src kinases Lyn and Hck.6-8,10,11 Both the Jak and Src kinases can phosphorylate STAT proteins in hematopoietic cells.81,82 Importantly, SOCS proteins differentially affect these kinases: in contrast to Jak kinases, the Src kinase Lyn is insensitive to SOCS-mediated inhibition.83 We considered a possible scenario in which Jak and Src kinases are differentially involved in the activation of STAT3 and STAT5. In that hypothetical context, G-CSF–induced STAT5 activation by G-CSF-R–d715 would become insensitive to SOCS because, as a result of the truncation of the receptor C-terminus, involvement of Src activity in the activation of STAT5 might become prevalent. To investigate this possibility, we performed the STAT reporter experiments in the presence of the Jak inhibitor WHI-P154 or the Src inhibitor PP-2.84,85 Both STAT3- and STAT5-induced luciferase activity was inhibited by WHI-P154, but not by PP-2, indicating that Jak, but not Src-kinase activity is essential for G-CSF–induced activation of both STAT3 and STAT5 by WT G-CSF-R (Figure 4). Notably, this remained essentially unchanged when activation was induced via G-CSF-R–d715. Although PP-2 slightly reduced STAT5 activation, G-CSF-R–d715 signaling was still completely dependent on Jak activity. These results thus exclude the possibility that, in the case of G-CSF-R–d715, the loss of SOCS-mediated inhibition of STAT5 signaling is caused by altered involvement of tyrosine kinases (eg, Lyn instead of Jak) as a consequence of the receptor truncation. We have previously published data from electrophoretic mobility shift assays (EMSAs) showing that activation of STAT3 by G-CSF-R–d715 is decreased compared with WT G-CSF-R when measured between 0 and 60 minutes after stimulation. This is due to the partial lack of STAT3 recruitment mechanisms.31 We observed that activation of STAT3 by G-CSF-R–d715 in the luciferase reporter assay was increased relative to WT G-CSF-R (Figure 4B). This could be directly linked to the defective receptor internalization of G-CSF-R–d715 (data not shown). Apparently, the internalization defect of G-CSF-R–d715 causes reduced off-switch of signaling, which results in increased accumulation of luciferase activity during the 6-hour time period of the experiment. However, despite this increased STAT3 activation, this signal remains fully sensitive for inhibition by SOCS3 (Figure 3B,4B).
Effects of Jak and Src inhibitors on STAT5 and STAT3 activation by WT G-CSF-R and G-CSF-R–d715. Luciferase assays were performed as in Figure 3. One hour before initiation of STAT5 (A) and STAT3 (B) luciferase reporter assays, the Src inhibitor PP-2 or the Jak inhibitor WHI-P154, dissolved in DMSO, was added to the cells at the concentrations indicated. Solvent control cells were treated with DMSO only. Data are expressed as mean + SEM of 3 independent experiments. *Difference with DMSO-treated control of same group is significant; P < .05.
Effects of Jak and Src inhibitors on STAT5 and STAT3 activation by WT G-CSF-R and G-CSF-R–d715. Luciferase assays were performed as in Figure 3. One hour before initiation of STAT5 (A) and STAT3 (B) luciferase reporter assays, the Src inhibitor PP-2 or the Jak inhibitor WHI-P154, dissolved in DMSO, was added to the cells at the concentrations indicated. Solvent control cells were treated with DMSO only. Data are expressed as mean + SEM of 3 independent experiments. *Difference with DMSO-treated control of same group is significant; P < .05.
Defective G-CSF-R internalization reduces sensitivity to SOCS3 only when combined with the loss of Tyr729
G-CSF-R–d715–induced STAT5 activation was considerably more resistant to the suppressive effects of SOCS3 than was a full-length G-CSF-R mutant lacking the SOCS3 docking site Tyr729 (eg, G-CSF-R–Tyr729Phe) (Figure 5). This could be suggestive of a second, tyrosine-independent mechanism of SOCS3 recruitment via the G-CSF-R C-terminus or relate to defective ligand-induced internalization of G-CSF-R–d715.27,31,86 To directly address this issue, we compared the SOCS3 sensitivity of WT G-CSF-R and G-CSF-R–d715 with that of G-CSF-R–d749-769 (Figure 1A). This mutant is as defective in internalization as G-CSF-R–d715, owing to the lack of 2 internalization domains.32 As shown in Figure 5, SOCS3 inhibits G-CSF-R–d749-769 as effectively as WT G-CSF-R, indicating that the loss of receptor internalization per se does not alleviate STAT5 inhibition by SOCS3. However, mutation of SOCS3 recruitment site Tyr729 in this internalization-defective mutant (G-CSF-R–d749-769Phe) resulted in a complete loss of SOCS3-mediated STAT5 inhibition. These results provide evidence for 2 distinct mechanisms of SOCS3 recruitment to the G-CSF-R, one that is independent of internalization (via Tyr729) and one that requires internalization. Possibly this latter mechanism involves direct interaction of SOCS3 to Jaks. Both of these mechanisms are disrupted in G-CSF-R–d715 (Figure 5).
Reduced internalization of truncated G-CSF-R mutants alleviates inhibition by SOCS3 only in the absence of Tyr729. Comparison of G-CSF-R mutants with normal (WT and Tyr729Phe) and defective (d715, d749-769, and d749-769Phe) internalization kinetics for sensitivity to SOCS3-mediated inhibition of STAT5 luciferase reporter activity. STAT5 luciferase reporter assay was performed as described in Figure 3. Data are expressed as mean of at least 2 independent experiments with triplicate measurements. *Difference between WT and d749-769 versus all other G-CSF-R mutants is significant, P < .01. #Difference between Tyr729Phe and d749-769, P = 0.07. ϕDifference between TyrY729Phe versus d715 and d749-769Phe is significant; P < .02.
Reduced internalization of truncated G-CSF-R mutants alleviates inhibition by SOCS3 only in the absence of Tyr729. Comparison of G-CSF-R mutants with normal (WT and Tyr729Phe) and defective (d715, d749-769, and d749-769Phe) internalization kinetics for sensitivity to SOCS3-mediated inhibition of STAT5 luciferase reporter activity. STAT5 luciferase reporter assay was performed as described in Figure 3. Data are expressed as mean of at least 2 independent experiments with triplicate measurements. *Difference between WT and d749-769 versus all other G-CSF-R mutants is significant, P < .01. #Difference between Tyr729Phe and d749-769, P = 0.07. ϕDifference between TyrY729Phe versus d715 and d749-769Phe is significant; P < .02.
Tyr729 is a combined recruitment site for SOCS3 and SHP-2
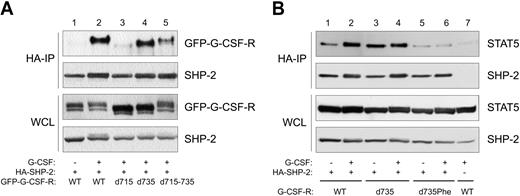
For a number of receptors, it has been demonstrated that SOCS3 recruitment sites are also SHP-2 docking sites.57-60 By combined immunoprecipitation (IP) and Western-blotting (WB), we tested whether SHP-2 binds to Tyr729 of G-CSF-R. We performed these experiments with GFP-tagged receptor constructs and used anti-GFP antibodies for immune detection. The GFP-tagged receptors were shown to behave identically to the untagged receptors with respect to proliferation, differentiation, and activation of STAT3 and STAT5.32 Stimulation with G-CSF induced coimmunoprecipitation of SHP-2 with the WT G-CSF receptor (Figure 6A, lanes 1-2). Truncation of the receptor at aa 715 resulted in a complete loss of the interaction with SHP-2 whereas truncation at aa 735 did not affect the SHP-2–G-CSF-R interaction (Figure 6, lanes 3-4). This result strongly supports the notion that Tyr729, the only tyrosine present in this region, is a binding site for SHP-2. To determine the relative contribution of Tyr729 to SHP-2 recruitment to full-length G-CSF-R, we also tested G-CSF-R mutant d715-735 (Figure 1) in coimmunoprecipitations. Although this mutant demonstrated a significantly reduced SHP-2 binding compared with WT G-CSF-R, it clearly bound more SHP-2 than did G-CSF-R–d715 (Figure 6, lane 5). Taken together, these results support the notion that Tyr729 of G-CSF-R forms a combined SOCS3 and SHP-2 recruitment site and show that an alternative mechanism of SHP-2 binding to the C-terminal region (aa's 737-813) of G-CSF-R exists. The latter mechanism possibly involves recruitment of SHP-2 via Tyr764.20
SHP-2 associates with distinct regions of G-CSF-R, and Tyr729 is required for the formation of a SHP-2–STAT5 complex. (A) HA–SHP-2 IPs of Phoenix E cells expressing GFP-tagged G-CSF-R mutants. Cells were starved overnight (–) and stimulated for 10 minutes with G-CSF. As a control, expression of GFP–G-CSF-R and SHP-2 in whole cell lysate (WCL) is shown in the lower 2 panels. (B) HA–SHP-2 IPs of Phoenix E cells expressing different G-CSF-R mutants and STAT5 were performed as described for Figure 6A. Lane 7 is an HA-IP in the absence of HA–SHP-2, demonstrating the specificity of the IP.
SHP-2 associates with distinct regions of G-CSF-R, and Tyr729 is required for the formation of a SHP-2–STAT5 complex. (A) HA–SHP-2 IPs of Phoenix E cells expressing GFP-tagged G-CSF-R mutants. Cells were starved overnight (–) and stimulated for 10 minutes with G-CSF. As a control, expression of GFP–G-CSF-R and SHP-2 in whole cell lysate (WCL) is shown in the lower 2 panels. (B) HA–SHP-2 IPs of Phoenix E cells expressing different G-CSF-R mutants and STAT5 were performed as described for Figure 6A. Lane 7 is an HA-IP in the absence of HA–SHP-2, demonstrating the specificity of the IP.
Tyr729 of the G-CSF-R is required for the formation of a SHP-2–STAT5 complex
It was recently shown that SHP-2 can interact with STAT5, resulting in dephosphorylation and inactivation of STAT5.87,88 Given the requirement of Tyr729 for recruitment of SHP-2 to the G-CSF-R, we investigated if the formation of a SHP-2–STAT5 complex would be dependent on the presence of Tyr729 of the G-CSF-R as well. As shown in Figure 6B (lanes 1-4), stimulation of the WT and G-CSF-R–d735 indeed results in coimmunoprecipitation of STAT5 with SHP-2. However, in the absence of Tyr729, formation of this complex is disrupted (lanes 5-6), demonstrating the importance of Tyr729 of the G-CSF-R for the formation of a SHP-2–STAT5 complex.
Discussion
We investigated whether, and by which mechanism(s), altered susceptibility to the inhibitory effects of SOCS proteins contributes to the hyperproliferative signaling of G-CSF-R C-terminal truncation mutants, which are frequently found in SCN patients with disease progression toward AML.24-26 We first concentrated on the regulation of expression of SOCS1 and SOCS3, which represent the SOCS family members with the most prominent negative effects on G-CSF signaling.74 Both SOCS1 and SOCS3 promoters contain STAT-binding sites,41,43,44,80 and dominant negative STAT3 blocks SOCS1 as well as SOCS3 mRNA expression, indicating that both SOCS genes are transcriptional targets of STAT3.41 STAT1 also binds to the SOCS3 promoter,44 but STAT3 is the major STAT protein involved in SOCS3 induction by G-CSF. This was demonstrated most clearly in conditional STAT3 knock-out mice that also lacked G-CSF–induced up-regulation of SOCS3.45 Because the loss of the G-CSF-R C-terminus affects both intensity and duration of the activation of STATs, we investigated SOCS1 and SOCS3 transcript levels in wild-type and gcsfr-d715 mice, both before and after 4 days of G-CSF treatment of the mice. G-CSF treatment of wild-type animals did not affect SOCS1 transcript levels. Receptor truncation resulted in a modest increase in SOCS1 expression after G-CSF stimulation, possibly owing to the increased STAT5 activation by G-CSF-R–d715.31 This is in agreement with data showing that interleukin 3 (IL-3)–induced SOCS1 expression is almost completely abrogated in the presence of dominant negative STAT5.89 SOCS3 mRNA levels, on the other hand, increased more than 12-fold in response to G-CSF. SOCS3 levels in gcsfr-d715 mice were clearly lower than in their wild-type littermates during steady state and upon stimulation with G-CSF in vivo or in vitro. These results establish that distinct regulatory mechanisms for upregulation of SOCS1 and SOCS3 expression exist in the context of the G-CSF-R. Recent studies with conditional STAT3 and SOCS3 knock-out strains demonstrated that the levels of neutrophilia in these mice in response to G-CSF treatment are remarkably similar to those observed in gcsfr-d715 mice.28,29,45,75,76 This would support the hypothesis that the loss of the negative feedback loop involving STAT3-controlled induction of SOCS3 might suffice to explain the gcsfr-d715 phenotype. However, in view of previous data showing that STAT3 activation in gcsfr-d715 mice is only moderately and temporarily reduced,31 it is conceivable that factors other than STAT3 also contribute to G-CSF–induced SOCS3 expression.
In addition to the reduced expression of SOCS3 in the gcsfr-d715 mice, the truncated G-CSF-R lacks Tyr729, which is the major recruitment site for SOCS3.35,36 While this provided an obvious second mechanism by which the truncated G-CSF-R forms bypass the suppressive effects of SOCS3, this applied to the activation of STAT5 but not STAT3, which still remained SOCS3 sensitive (Figure 3). This result provides a likely explanation for the increased ratio of STAT5/STAT3 activation by G-CSF-R–d715, which has been linked to shifting the proliferation/differentiation balance toward proliferation and extended cell survival.31 Two potential explanations, differential involvement of upstream tyrosine kinases and differential sensitivity due to altered internalization kinetics, were shown, respectively, to play no or only a limited role in this study. For efficient activation of STAT3, first G-CSF-R tyrosine phosphorylation is required, which creates STAT3 recruitment sites. This indicates that activation of STAT3 is a multistep process. It is conceivable that inhibition of STAT3 is still achieved when recruitment of SOCS3 is suboptimal owing to the lack of Tyr729. In contrast, inhibition of STAT5, which is activated through direct interaction with Jak kinases,90 may require optimal SOCS3 recruitment for complete inhibition. STAT5 activation by G-CSF-R mutant Tyr729Phe was inhibited to a considerable extent at the highest levels of SOCS3, which is probably mediated through low-affinity interaction of SOCS3 and Jaks.
Moreover, we think that the loss of recruitment of SHP-2 activity to Tyr729 also plays a major role in the sustained STAT5 activation by G-CSF-R–d715. It was shown for a number of cytokine receptors, for example, the leptin receptor and the shared cytokine receptor subunit gp130, that SOCS3 and SHP-2 dock to the same phosphotyrosine-based motif.57-60,91,92 Our data on the G-CSF-R (Figure 6) corroborate this and implicate Tyr729 as a common recruitment site for SOCS3 and SHP-2. Because SHP-2 was recently identified as a STAT5 phosphatase, a model can be envisaged in which impaired recruitment of SHP-2 is key to the loss of negative control of STAT5 activation.87,88 In this scenario, reduced recruitment of SHP-2 activity to the G-CSF-R would thus result in increased levels of active STAT5 complexes. Indeed, as shown in Figure 6B, G-CSF-R-Tyr729 is essential for formation of a SHP-2–STAT5 complex, which strongly argues for a scenario in which loss of Tyr729 not only affects SOCS3-mediated inhibition of STAT5 activity but also disrupts formation of a SHP-2–STAT5 complex that has been reported to contribute to STAT5 dephosphorylation.
In conclusion, our study has unveiled a new mechanism by which C-terminal truncation mutants of G-CSF-R, associated with leukemic progression of SCN, attain altered signaling abilities. While the altered signaling abilities were originally attributed mainly to defective internalization properties of G-CSF-R–d71527,31,86 the loss of negative feedback projected by SOCS3 and, possibly, SHP-2 via binding to Tyr729 of G-CSF-R have now been identified as additional signaling defects of truncated G-CSF-R. Our results fit into a model in which the combined loss of regulation by SOCS3 and SHP-2 contributes to the perturbed signaling by G-CSF-R–d715, resulting in an increased STAT5/STAT3 activation ratio.
Prepublished online as Blood First Edition Paper, April 6, 2004; DOI 10.1182/blood-2003-08-2913.
Supported by the Dutch Cancer Society “Koningin Wilhelmina Fonds.”
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Montserrat Blazquez-Domingo for advice and help with quantitative RT-PCR, Karola van Rooyen for help with preparation of the figures, Alexandra Klomp and Dr Joanna Prasher for assistance with animal experiments, Wim van Putten for help with statistical analysis, and Dr Akihiko Yoshimura for providing SOCS3 expression plasmids.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal