Abstract
Patients with Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ ALL) have poor prognosis despite intensive therapeutic intervention. Recently, imatinib, a BCR-ABL tyrosine kinase inhibitor, has been proven to be an effective treatment for Ph+ ALL, but nearly all patients rapidly acquire resistance. High-dose imatinib administration might overcome this resistance; however, systemic toxicities would likely limit this approach. Therefore, a new delivery system allowing for the specific targeting of imatinib is urgently needed. Because almost all Ph+ ALL cells express CD19 on their surface, we have developed an immunoliposome carrying anti-CD19 antibody (CD19-liposomes). The internalization efficiency of the CD19-liposomes approached 100% in all Ph+ ALL cells but was very low in CD19– cells. The cytocidal effect of imatinib-encapsulated CD19-liposomes (imatinib-CD19-liposomes) on Ph+ ALL cell lines and primary leukemia cells from patients with Ph+ ALL was much greater than that of imatinib with or without control liposomes. Importantly, the imatinib-CD19-liposomes did not affect the colony formation of CD34+ hematopoietic cells, even at inhibitory concentration of free imatinib. Taken together, these data clearly demonstrate that the imatinib-CD19-liposomes induced specific and efficient death of Ph+ ALL cells. This new therapeutic approach might be a useful treatment for Ph+ ALL with fewer side effects than free imatinib.
Introduction
Despite the successes of high-dose chemotherapy followed by hematopoietic stem cell transplantation for the treatment of leukemia, Philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia (ALL) remains refractory to most therapeutic modalities currently available. The Philadelphia chromosome includes one of several forms of fusion genes between bcr and c-abl, and these fusions play a substantial role in the pathogenesis of chronic myelogenous leukemia (CML) and ALL.1 The p190 bcr-abl fusion gene is detected in 20% to 35% of patients with ALL, and the prognosis of these patients is particularly poor.2-4 Allogenic stem cell transplantation is the only curative therapeutic option today. Long-term survival rates are between 35% and 65% in cases of first complete remissions, but survival rates quickly decline in cases of second and third remissions.4-9 Additionally, the relapse rate for Ph+ ALL remains quite high even with transplantation.4-9 Recently, targeted molecular therapy has been introduced to treat such refractory cases, and a BCR-ABL tyrosine kinase inhibitor, imatinib (Glivec, STI571), proved to be a useful agent for Ph+ ALL as well as CML.10 However, in contrast to patients with CML, most of the patients with Ph+ ALL quickly become resistant to imatinib.10,11 In a phase 2 clinical trial examining cases of relapsed or refractory Ph+ ALL, only 6% of patients maintained a complete response for more than 4 weeks, in contrast to an overall response rate of 29%.12 Moreover, the median estimated time to progression and overall survival of these patients were only 2.2 and 4.9 months, respectively.12 These results suggest that monotherapy with imatinib at a standard dosage (600 mg/d or less) is not sufficient to control Ph+ ALL in the long term.
Although there are a number of reasons for the development of imatinib resistance,11,13-16 such as mutation11,16 or overexpression of the bcr-abl gene,14,15,17 administration of high-dose imatinib has been suggested as a means to overcome imatinib resistance.17,18 In practice, the dose escalation of imatinib, however, is limited by various toxicities such as hematopoietic suppresion.19-21 Imatinib has been reported to inhibit not only BCR-ABL but also other tyrosine kinases, including ABL-related gene (ARG),22 c-KIT, and the platelet-derived growth factor receptors.23,24 The inhibition of these enzymes likely gives rise to the toxicity of imatinib.25 Therefore, a selective delivery system for imatinib to Ph+ ALL cells is needed.
Liposomes, small synthetic lipid bilayers, have been recently developed as a tumor-specific delivery system for antitumor agents, and immunoliposomes, liposomes with attached antibody or receptor ligands that allow for the specific targeting of a cell, hold particular promise. In a previous study that used a monoclonal antibody specific for pulmonary endothelial cells, it was demonstrated that “pendant-type liposomes” that combine antibody at the terminus of a straight-chain polyethylene glycol (PEG) polymer are most efficiently taken into target cells.26 In addition, we have reported previously that the antitumor effects of pendant-type transferrin-binding PEG-liposomal cisplatin against a gastric cancer cell line was greater than that of unencapsulated cisplatin in vitro and in vivo.27 As for hematologic malignancies, doxorubicin-encapsulated liposomes coated with anti-CD19 and folate receptor type-β exert cytotoxic effects on lymphoma and acute myelogenous leukemia (AML) cell lines, respectively.28,29 CD19 is expressed in most Ph+ ALL cells2,30 but not in normal systemic cells except those in the B-cell lineage,31 and it is known to undergo rapid internalization on antibody binding. Consequently, CD19 was thought to be a promising target molecule to obtain tumor-specific killing.32 Therefore, we developed anti-CD19 antibody-binding liposomes (CD19-liposomes), including imatinib (imatinib-CD19-liposomes) for the targeted killing of Ph+ ALL cells. In this study, we successfully demonstrate that CD19-liposomes are a useful tool for drug delivery, and imatinib-CD19-liposomes efficiently induced death of Ph+ ALL cells, including primary leukemia cells from patients with imatinib-resistant Ph+ ALL.
Materials and methods
Cell lines
Seven leukemia or lymphoma cell lines were used in this study. The Ph+ ALL cell line of OM9;22 was kindly provided by Dr K. Ohyashiki (Tokyo Medical College, Tokyo, Japan).33 The Ph+ ALL cell lines KOPN-30, KOPN-57, and KOPN-72 were established as reported previously.34 Leukemia cell lines consisting of SUP-B15 Ph+ ALL cell lines and K562 CML blastic crisis (CML-BC) cells, and Daudi Burkitt lymphoma cells were obtained from the American Type Culture Collection (Rockville, MD). BV173 CML-BC cells were provided by Hayashibara Laboratories (Okayama, Japan). NB4 acute promyelocytic leukemia (APL) cell lines were kindly provided by Dr M. Lanotte (Hôpital Saint-Louis, Paris, France). All cells except for SUP-B15 cells, which were cultured in Iscoves modified Dulbecco medium (IMDM) supplemented with 20% fetal bovine serum (FBS), were cultured in RPMI 1640 medium supplemented with 10% FBS.
Preparation of liposomes
Stabilized liposomes containing calcein (Dojindo Laboratories, Kumamoto, Japan) or imatinib (kindly provided by Novartis Pharmaceutical, Basel, Switzerland) were prepared by using reverse-phase evaporation followed by extrusion (Lipex Biomembranes, Vancouver, BC, Canada) through 2-stacked polycarbonate membrane filters (0.1-μm pore size; Whatman, Maidstone, United Kingdom) as described previously.26 Bare-liposomes were composed of dipalmitoyl phosphatidylcholine (DPPC) and cholesterol (CH), and PEG-liposomes were composed of DPPC, CH, and DSPE-PEG-Mal (2:1:0.06 m/m). CD19-liposomes were prepared by coupling the mouse monoclonal antihuman CD19 antibody, CLB-CD19 (kindly provided by Nichirei, Tokyo, Japan) to the DSPE-PEG-mal through the reaction of sulfhydryl residues on the antibody with the C-terminal maleimide groups of the aforementioned PEG-liposomes as described in our previous report.26 The average size of the liposomes used in this study was about 100 nm, and there were no significant differences in average size among all types of liposomes. The calcein-encapsulated liposomes used in this study were calcein-encapsulated bare-liposomes (Cal-bare-liposomes), calcein-encapsulated PEG-liposomes (Cal-PEG-liposomes), and calcein-encapsulated CD19-liposomes (Cal-CD19-liposomes), and the imatinib-encapsulated liposomes were imatinib-encapsulated bare-liposomes (imatinib-bare-liposomes), imatinib-encapsulated PEG-liposomes (imatinib-PEGliposomes), and imatinib-CD19-liposomes.
Laser-scanning confocal microscopy
Binding and internalization of the liposomes in Ph+ ALL cells were examined by laser scanning confocal microscopy. SUP-B15 cells were incubated with Cal-CD19-liposomes for 10 or 60 minutes at 37° C and washed twice with phosphate-buffered saline (PBS) followed by fixation with 1% paraformaldehyde (PFA) for 15 minutes. These cells were attached on a slide glass using Cytospin 3 (Shandon, Roncorn, United Kingdom). Thereafter, cells were incubated with 0.1% Triton X-100 (Sigma-Aldrich Fine Chemicals, St Louis, MO) in PBS for 3 minutes at room temperature (RT). Before cytoplasmic staining, cells were preincubated with 1% bovine serum albumin (BSA) in PBS for 20 minutes at RT to reduce nonspecific background staining. Finally, cells were incubated with 1 unit of rhodamine phalloidin (Molecular Probes, Eugene, OR) for 30 minutes at RT to stain cytoplasmic actin. Green fluorescence of calcein and red fluorescence of rhodamine phalloidin were analyzed, and merged images were produced by using Radiance2100 Confocal and Multi-photon Imaging Systems and LaserSharp 2000 software (Bio-Rad Laboratories, Hercules, CA).
Flow cytometry (FCM)
To examine the binding and internalization efficiency of the liposomes, 1 × 106 cells were incubated with 100 μg calcein-encapsulated liposomes in 1 mL culture medium for 60 minutes at 37° C, and cells were pelleted by centrifugation. Cell surface-bound liposomes were removed by incubation with 2 mg/mL pronase (Roche Diagnostics, Basel, Switzerland) for 30 minutes at 4° C, then cells were fixed with 1% PFA in PBS. To examine the specificity of liposome binding, cells were treated with various concentrations (0-1000 μg/mL) of CLB-CD19 monoclonal antibody for 30 minutes at 4° C before incubation with the liposomes. Thereafter, cells were analyzed by FCM with use of a FACS (fluorescence activated cell sorting) Calibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ).
Determination of viability and apoptosis
Leukemia cell lines (2 × 105) were cultured in the presence or absence of imatinib-encapsulated liposomes or free imatinib for 8 days at 37° C in a CO2 incubator. The concentrations of imatinib used in this assay were varied from 0.01 to 10 μM. To determine the viability of cells treated with imatinib-encapsulated liposomes or free imatinib, we assessed live and dead cell numbers every 2 days by trypan blue exclusion. Apoptosis of cells was determined by using the Annexin V–apoptosis detection kit I (Becton Dickinson Pharmingen, San Diego, CA) according to manufacturer's instruction. Cells were stained with fluorescein isothiocyanate (FITC)–labeled Annexin V and propidium iodide and analyzed by 2-color FCM using FACS Calibur flow cytometer.
Primary Ph+ ALL cells and cobblestone area formation assay
Primary leukemia cells were obtained from peripheral blood (PB) or bone marrow (BM) and used for the following experiments after obtaining informed consent from each patient. This study was approved by the institutional review board of The Institute of Medical Science, The University of Tokyo. Cells were purified by Ficoll-Hypaque centrifugation, and more than 98% of the cells isolated were leukemia cells. The effect of imatinib-encapsulated liposomes was examined by cobblestone area (CA)–forming assay as reported previously.35 Briefly, 1 × 104 to 5 × 104 leukemia cells were cocultured with the mouse feeder cell line, HESS-5,36 which were seeded in 6-well plates 2 weeks before coculture and formed a confluent monolayer at the start of coculture, in RPMI with 10% FBS in 5% CO2 at 37° C. At the beginning of coculture, imatinib-encapsulated liposomes or free imatinib (0.01-10 μM) were added to the medium. CAs, which consist of more than 5 leukemia cells, were directly counted under an inverted microscope with phase contrast (Nikon, Tokyo, Japan) 14 to 17 days after the start of coculture.
Preparation and culture of cord blood CD34+ cells (CD34+ CBCs)
Umbilical cord blood was drawn from the umbilical cord of healthy volunteers after obtaining informed consent and approval by the institutional review board of The Institute of Medical Science, The University of Tokyo. Mononuclear cells were purified from these samples by Ficoll-Hypaque methods as previously reported.37 These cells were processed further to isolate CD34+ CBCs by using a magnetic cell sorter (MACS) direct CD34 progenitor cell isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. After 2 rounds of isolation, more than 95% of isolated cells were CD34+, as assessed by FCM. To determine the colony-forming abilities of CD34+ CBCs, 200 cells were plated in IMDM containing 30% FBS, 1% bovine serum albumin, 1.2% methylcellulose, and cytokines, including 25 ng/mL stem cell factor (SCF; kindly provided by Amgen, Thousand Oaks, CA), 10 ng/mL interleukin-3 (kindly provided by Chugai Pharmaceuticals, Tokyo, Japan), 100 ng/mL interleukin-6 (kindly provided by Ajinomoto, Tokyo, Japan), 10 ng/mL granulocyte colony-stimulating factor (G-CSF), and 2 U/mL erythropoietin (kindly provided by Chugai Pharmaceuticals). At the start of culture, the imatinib-encapsulated liposomes or free imatinib were added at concentrations of 1.0 μM to 50 μM, and colony numbers were counted under an inverted microscope (Nikon) after 14 days.
Identification of patient mutations
To determine the sequence of the adenosine 5′-triphosphate (ATP)–binding sites in bcr-abl genes, total RNA was isolated from primary leukemia cells obtained from 4 patients as described in “Primary Ph+ ALL cells and cobblestone area formation assay” by using an RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. After treatment with DNase I (Invitrogen, Carlsbad, CA), cDNA was synthesized by using a SuperScript First-Strand Synthesis System for reverse transcribed polymerase chain reaction (RT-PCR; Invitrogen) with oligo(dT) primer. Thereafter, cDNA from 500 ng RNA was amplified by PCR with use of Platinum Pfx DNA polymerase (Invitrogen) and the following PCR primers with 35 cycles of thermal cycling at 94° C for 30 seconds, 60° C for 30 seconds, and 68° C for 30 seconds. Primer sequences were as follows: sense primer, 5′-CGACAAGTGGGAGATGGAACGC-3′ (c-abl no. 843-864); antisense primer, 5′-TTCCCAAAGCAATACTCCAAAT-3′ (c-abl no. 1418-1440). PCR products were cloned into pCR-Blunt II TOPO vectors by using Zero Blunt TOPO PCR Cloning Kit (Invitrogen), and 10 individual clones were sequenced bidirectionally by using the following sequence primers and an ABI PRISM 3700 DNA Analyzer (Applied Biosystems, Branchburg, NJ). Sequence primers were as follows: sense primer, 5′-CGCCAAGCTATTTAGGTGACACTATAG-3′; antisense primer, 5′-GAATTGTAATACGACTCACTATAGGGCG-3′.
Quantification of bcr-abl mRNA
To quantify p190-type bcr-abl mRNA, cDNA synthesized from total RNA of Ph+ ALL cells using an oligo(dT) primer prepared as described in “Identification of patient mutations” was analyzed by real-time PCR with use of an ABI PRISM 7700 Sequence Detection System (Applied Biosystems) as reported previously.35
Results
Binding and internalization of CD19-liposomes
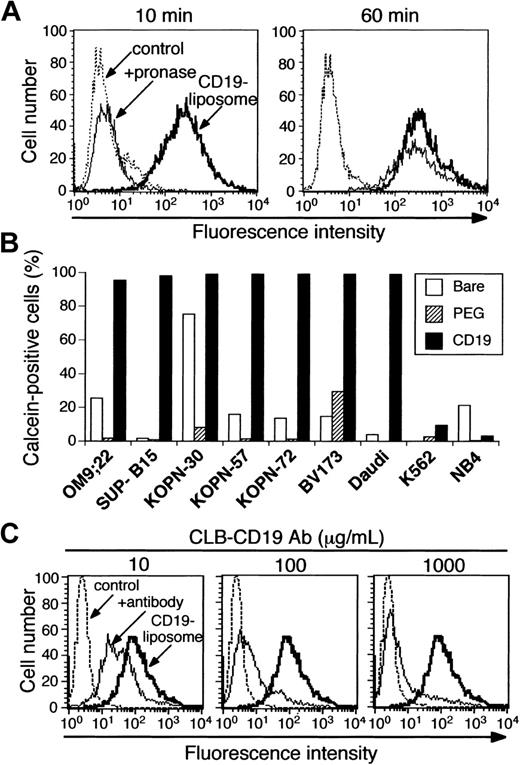
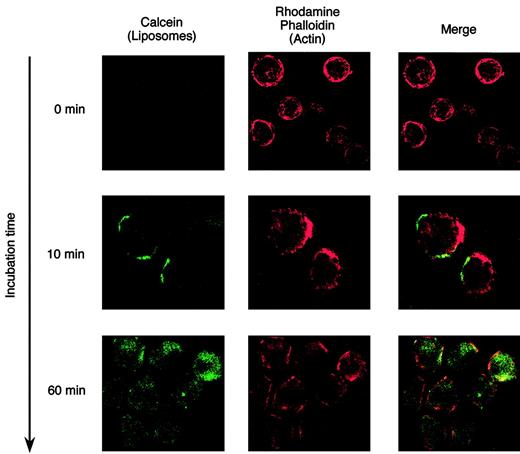
To determine the binding and internalization efficiency of CD19-liposomes, a number of cell lines were tested. All 5 of the Ph+ ALL cell lines, including OM9;22, SUP-B15, KOPN-30, KOPN-57, and KOPN-72, and control cell lines of BV173 CML-BC cells and Daudi Burkitt lymphoma cells expressed CD19, but K562 CML-BC and NB4 APL cells did not. To evaluate the binding or internalization of Cal-CD19-liposomes, cells were analyzed by FCM to detect calcein fluorescence encapsulated in those liposomes. In addition, pronase, which contains endopeptidases and exopeptidases, was used to remove the Cal-CD19-liposomes attached on the cell membrane. Namely, the calcein fluorescence was expected to disappear following treatment with pronase when those liposomes were bound but not internalized into the cells; cells that had internalized liposomes would remain fluorescent. After a 10-minute incubation, SUP-B15 cells treated with 10 μg/mL Cal-CD19-liposomes were nearly 100% positive for calcein fluorescence, but fluorescence disappeared when the cells were treated with pronase (Figure 1A). After a 60-minute incubation, almost all of the cells were calcein-positive, and the fluorescence level was maintained even after treatment with pronase (Figure 1A). These results strongly suggest that the Cal-CD19-liposomes only bound to the SUP-B15 cell surface after 10 minutes of incubation and were efficiently internalized into cells after 60 minutes. In other Ph+ ALL cell lines, BV173 cells and Daudi cells, the internalization efficiencies of Cal-CD19-liposomes were almost 100%, similar to the results obtained with SUP-B15 cells (Figure 1B). In contrast, the internalization efficiencies of Cal-bare- and Cal-PEG-liposomes were much lower than those of Cal-CD19-liposomes. Moreover, internalization of Cal-CD19-liposomes into the CD19– cell lines K562 and NB4 cells was very low (Figure 1B). To confirm the internalization efficiencies of CD19-liposomes, SUP-B15 cells incubated with Cal-CD19-liposomes were analyzed by laser scanning confocal microscopy. Internalization of Cal-CD19-liposomes would lead to cytoplasmically localized green calcein staining that would appear yellow when merged with the intracellular actin signal generated by rhodamine phalloidin. Calcein fluorescence was detected only on the cell surface after a 10-minute incubation. After a 60-minute incubation, calcein fluorescence was detected in the cytoplasm of all cells (Figure 2). In other CD19+ cells, including Ph+ ALL cell lines, similar results were obtained (data not shown).
Binding and internalization of Cal-CD19-liposomes. (A) Binding and internalization by SUP-B15 cells. Cells were incubated with Cal-CD19-liposomes at 37° C for 10 (left) or 60 (right) minutes and were analyzed by FCM to detect calcein fluorescence. In addition, cells were treated with (thin solid line) or without (bold line) pronase to remove surface-bound liposomes. When those liposomes were internalized into the cells, calcein fluorescence was expected to remain. Untreated cells were used as a negative control (broken line). (B) Internalization efficiency of liposomes by Ph+ ALL cell lines. These cell lines were incubated with Cal-bare-, Cal-PEG-, or Cal-CD19-liposomes at 37°C for 60 minutes and treated with pronase. The rate of calcein-positive cells was determined by FCM. (C) Blocking of Cal-CD19-liposome internalization into SUP-B15 cells by CLB-CD19 antibody. SUP-B15 cells were incubated with 10 (left), 100 (middle), or 1000 (right) μg/mL unlabeled CLB-CD19 antibodies at 37°C before incubation of the Cal-CD19-liposomes. Thereafter, cells were treated with pronase and assessed by FCM (thin solid line). Untreated cells (broken line) and cells not treated with CLB-CD19 antibody (bold line) were used as negative and positive controls, respectively.
Binding and internalization of Cal-CD19-liposomes. (A) Binding and internalization by SUP-B15 cells. Cells were incubated with Cal-CD19-liposomes at 37° C for 10 (left) or 60 (right) minutes and were analyzed by FCM to detect calcein fluorescence. In addition, cells were treated with (thin solid line) or without (bold line) pronase to remove surface-bound liposomes. When those liposomes were internalized into the cells, calcein fluorescence was expected to remain. Untreated cells were used as a negative control (broken line). (B) Internalization efficiency of liposomes by Ph+ ALL cell lines. These cell lines were incubated with Cal-bare-, Cal-PEG-, or Cal-CD19-liposomes at 37°C for 60 minutes and treated with pronase. The rate of calcein-positive cells was determined by FCM. (C) Blocking of Cal-CD19-liposome internalization into SUP-B15 cells by CLB-CD19 antibody. SUP-B15 cells were incubated with 10 (left), 100 (middle), or 1000 (right) μg/mL unlabeled CLB-CD19 antibodies at 37°C before incubation of the Cal-CD19-liposomes. Thereafter, cells were treated with pronase and assessed by FCM (thin solid line). Untreated cells (broken line) and cells not treated with CLB-CD19 antibody (bold line) were used as negative and positive controls, respectively.
Internalization analysis of Cal-CD19-liposomes by confocal laser-scanning microscopy. SUP-B15 cells were incubated with Cal-CD19-liposomes at 37° C for the indicated times. After washing and fixation, cells were allowed to adhere to glass slides using Cytospin 3. Thereafter, cells were stained with rhodamine phalloidin to visualize cytoplasmic actin. The prepared slides were analyzed by confocal laser-scanning microscopy. The images were acquired using a Radiance 2100 Confocal and Multi-photon Imaging System (Bio-Rad) with an Axioshop 2 plus microscope (Carl Zeiss, Jena, Germany). The type of the objective lens was Plan-Apochromat (Carl Zeiss) with a numerical aperture of 1.40. The acquisition and subsequent processing software were Laser Sharp 2000 (Bio-Rad) and Adobe Photoshop 5.0 (Adobe Systems, San Jose, CA), respectively. Original magnification × 630. Green fluorescence of calcein (left column), red fluorescence of rhodamine phalloidin (middle column), and the merged image (right column) are shown. In this assay, internalization of the Cal-CD19-liposomes will result in a yellow signal in the merged image.
Internalization analysis of Cal-CD19-liposomes by confocal laser-scanning microscopy. SUP-B15 cells were incubated with Cal-CD19-liposomes at 37° C for the indicated times. After washing and fixation, cells were allowed to adhere to glass slides using Cytospin 3. Thereafter, cells were stained with rhodamine phalloidin to visualize cytoplasmic actin. The prepared slides were analyzed by confocal laser-scanning microscopy. The images were acquired using a Radiance 2100 Confocal and Multi-photon Imaging System (Bio-Rad) with an Axioshop 2 plus microscope (Carl Zeiss, Jena, Germany). The type of the objective lens was Plan-Apochromat (Carl Zeiss) with a numerical aperture of 1.40. The acquisition and subsequent processing software were Laser Sharp 2000 (Bio-Rad) and Adobe Photoshop 5.0 (Adobe Systems, San Jose, CA), respectively. Original magnification × 630. Green fluorescence of calcein (left column), red fluorescence of rhodamine phalloidin (middle column), and the merged image (right column) are shown. In this assay, internalization of the Cal-CD19-liposomes will result in a yellow signal in the merged image.
Specific binding of CD19-liposomes to CD19+ cells
Because the internalization of Cal-CD19-liposomes was observed only in CD19+ cells, it was thought that the antibody/antigen interaction was responsible for the specificity of these results. To confirm the specificity of the interaction of Cal-CD19-liposomes with the target cells, a competition assay was performed by using free CLB-CD19 antibody. The internalization of Cal-CD19-liposomes into SUP-B15 cells was inhibited by CLB-CD19 antibody in a dose-dependent manner (Figure 1C). Similar results were obtained in other CD19+ cells (data not shown). In contrast, the slight internalization of Cal-CD19-liposomes observed in K562 cells was not inhibited by the CLB-CD19 antibody and was considered background (data not shown).
Effect of imatinib-CD19-liposomes on Ph+ ALL cells
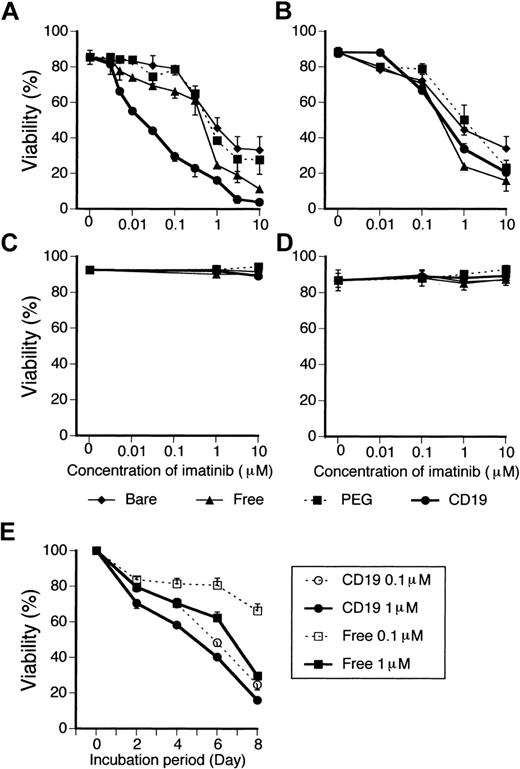
Because we confirmed the effective and specific internalization of Cal-CD19-liposomes into CD19+ cells, we encapsulated imatinib into CD19-liposomes. The 7 leukemia or lymphoma cell lines used in the above-mentioned assay were cultured with or without imatinib-bare-, imatinib-PEG-, or imatinib-CD19-liposomes or unencapsulated imatinib (free imatinib) for 8 days, and the viability of those cells was determined every 2 days. In CD19+ and BCR-ABL–positive SUP-B15 cells, all imatinib-encapsulated liposomes induced cell death in a dose-dependent manner as well as free imatinib. The induction of cell death with imatinib-CD19-liposomes was much greater than that with free imatinib (Figure 3A). Imatinib-bare- and imatinib-PEG-liposomes, however, did not induce cell death as much as free imatinib (Figure 3A). In addition, imatinib-CD19-liposomes induced cell death more rapidly than the identical concentration of free imatinib (Figure 3E). In other Ph+ ALL cell lines and BV173 CML-BC cells, which expressed both CD19 and BCR-ABL, similar results were obtained (data not shown). In contrast, in CD19– and BCR-ABL–positive K562 CML-BC cells, the induction of cell death with imatinib-CD19-liposomes was weaker than that with free imatinib or imatinib-bare-liposomes and similar to that with imatinib-PEG-liposomes (Figure 3B). Moreover, all types of imatinib-encapsulated liposomes as well as free imatinib did not affect the viability of Daudi (CD19+) and NB4 cells (CD19–), both of which do not express BCR-ABL (Figure 3C-D). To determine the differences in the cytocidal effect between imatinib-encapsulated liposomes and free imatinib more clearly, we calculated the IC50 (concentration that inhibits 50%) values as summarized in Table 1. The IC50 of imatinib-CD19-liposomes for the 5 types of Ph+ ALL cells ranged from 0.02 to 0.90 μM, concentrations approximately 4 to 27 times (average 11.3 times) lower than those of free imatinib (Table 1) and also less than those of the imatinib-bare- and imatinib-PEG-liposomes (Table 1). In contrast, in K562 cells the IC50 of imatinib-CD19-liposomes was about 4-fold higher than that of free imatinib (Table 1).
Viability of leukemia cell lines treated with imatinib-encapsulated liposomes. SUP-B15 Ph+ ALL cells (A; bcr-abl+, CD19+), CML-BC K562 cells (B; bcr-abl+, CD19–), Burkitt lymphoma Daudi cells (C; bcr-abl–, CD19+), and APL NB4 cells (D; bcr-abl–, CD19–) were cultured with or without the indicated imatinib-encapsulated liposomes. Viability of cells was evaluated 8 days after the start of culture. (E) Time course of the viability of SUP-B15 cells treated with imatinib-CD19-liposomes or free imatinib at a concentration of 0.1 or 1 μM is shown. Data represent mean ± SD of triplicate experiments.
Viability of leukemia cell lines treated with imatinib-encapsulated liposomes. SUP-B15 Ph+ ALL cells (A; bcr-abl+, CD19+), CML-BC K562 cells (B; bcr-abl+, CD19–), Burkitt lymphoma Daudi cells (C; bcr-abl–, CD19+), and APL NB4 cells (D; bcr-abl–, CD19–) were cultured with or without the indicated imatinib-encapsulated liposomes. Viability of cells was evaluated 8 days after the start of culture. (E) Time course of the viability of SUP-B15 cells treated with imatinib-CD19-liposomes or free imatinib at a concentration of 0.1 or 1 μM is shown. Data represent mean ± SD of triplicate experiments.
IC50 (μM) of imatinib-encapsulated liposomes in leukemia cell lines
. | Bare-liposome . | PEG-liposome . | CD19-liposome . | Free imatinib . |
|---|---|---|---|---|
| OM9;22 | 3.91 ± 0.78* | 5.71 ± 0.88 | 0.90 ± 0.07 | 3.19 ± 0.04 |
| SUP-B15 | 0.60 ± 0.06 | 0.52 ± 0.01 | 0.02 ± 0.001 | 0.25 ± 0.03 |
| KOPN-30 | 0.28 ± 0.03 | 3.68 ± 0.50 | 0.02 ± 0.002 | 0.53 ± 0.01 |
| KOPN-57 | 4.60 ± 0.09 | 5.37 ± 0.004 | 0.13 ± 0.03 | 0.74 ± 0.05 |
| KOPN-72 | 1.46 ± 0.03 | 5.00 ± 0.07 | 0.08 ± 0.002 | 0.65 ± 0.002 |
| BV173 | 3.28 ± 1.17 | 2.66 ± 1.26 | 0.32 ± 0.02 | 0.93 ± 0.09 |
| K562 | 0.49 ± 0.03 | 1.18 ± 0.41 | 1.46 ± 0.62 | 0.37 ± 0.02 |
| Daudi | > 10.0 | > 10.0 | > 10.0 | > 10.0 |
| NB4 | > 10.0 | > 10.0 | > 10.0 | > 10.0 |
. | Bare-liposome . | PEG-liposome . | CD19-liposome . | Free imatinib . |
|---|---|---|---|---|
| OM9;22 | 3.91 ± 0.78* | 5.71 ± 0.88 | 0.90 ± 0.07 | 3.19 ± 0.04 |
| SUP-B15 | 0.60 ± 0.06 | 0.52 ± 0.01 | 0.02 ± 0.001 | 0.25 ± 0.03 |
| KOPN-30 | 0.28 ± 0.03 | 3.68 ± 0.50 | 0.02 ± 0.002 | 0.53 ± 0.01 |
| KOPN-57 | 4.60 ± 0.09 | 5.37 ± 0.004 | 0.13 ± 0.03 | 0.74 ± 0.05 |
| KOPN-72 | 1.46 ± 0.03 | 5.00 ± 0.07 | 0.08 ± 0.002 | 0.65 ± 0.002 |
| BV173 | 3.28 ± 1.17 | 2.66 ± 1.26 | 0.32 ± 0.02 | 0.93 ± 0.09 |
| K562 | 0.49 ± 0.03 | 1.18 ± 0.41 | 1.46 ± 0.62 | 0.37 ± 0.02 |
| Daudi | > 10.0 | > 10.0 | > 10.0 | > 10.0 |
| NB4 | > 10.0 | > 10.0 | > 10.0 | > 10.0 |
Data represent mean ± SD of 3 separate experiments.
Apoptosis induction by imatinib-CD19-liposomes
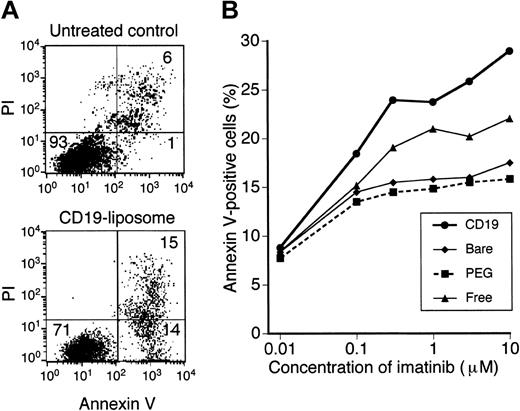
Because imatinib is known to induce apoptosis of BCR-ABL–positive cells, we assessed the ability of imatinib liposomes to induce apoptosis by Annexin V assay. In KOPN-72 cells, apoptotic cells appeared 48 hours after addition of imatinib-CD19-liposomes in a dose-dependent manner (Figure 4A-B). The Annexin V–positive rate of these cells treated with 10 μM imatinib-CD19-liposomes and free imatinib was about 29% and 22%, respectively (Figure 4A-B). However, the amount of apoptotic cells in samples treated with imatinib-bare-liposomes or imatinib-PEG-liposomes were fewer than those with free imatinib (Figure 4B). In other Ph+ ALL cell lines, apoptosis induced by imatinib-CD19-liposomes was greater than that seen in cells treated with free imatinib (data not shown). In contrast, apoptosis of K562 cells treated with imatinib-CD19-liposomes was less than that seen in cells treated with free imatinib. Apoptosis was not induced in BCR-ABL–negative cell lines even when cells were treated with free imatinib or any imatinib-encapsulated liposomes (data not shown).
Apoptosis of KOPN-72 Ph+ ALL cells treated with imatinib-encapsulated liposomes. (A) Annexin V staining of KOPN-72 cells. Cells were cultured with (bottom) or without (top) 10 μM imatinib-CD19-liposomes for 48 hours. Thereafter, cells were stained with FITC-labeled Annexin V and propidium iodide (PI) and appraised by FCM. Viable cells are located in the lower left quadrant (Annexin V– and PI-negative), early apoptotic cells are located in the lower right quadrant (Annexin V–positive and PI-negative), and dead cells are found in the upper right quadrant (Annexin V– and PI-positive). The numbers in the graphs indicate the percentage of cells in each quadrant. (B) Dose response curve of Annexin V–positive cells. KOPN-72 cells were treated with the indicated imatinib-encapsulated liposomes at various concentrations of imatinib and cultured for 48 hours. The rate of Annexin V–positive cells was determined by FCM. Data represent mean of triplicate experiments.
Apoptosis of KOPN-72 Ph+ ALL cells treated with imatinib-encapsulated liposomes. (A) Annexin V staining of KOPN-72 cells. Cells were cultured with (bottom) or without (top) 10 μM imatinib-CD19-liposomes for 48 hours. Thereafter, cells were stained with FITC-labeled Annexin V and propidium iodide (PI) and appraised by FCM. Viable cells are located in the lower left quadrant (Annexin V– and PI-negative), early apoptotic cells are located in the lower right quadrant (Annexin V–positive and PI-negative), and dead cells are found in the upper right quadrant (Annexin V– and PI-positive). The numbers in the graphs indicate the percentage of cells in each quadrant. (B) Dose response curve of Annexin V–positive cells. KOPN-72 cells were treated with the indicated imatinib-encapsulated liposomes at various concentrations of imatinib and cultured for 48 hours. The rate of Annexin V–positive cells was determined by FCM. Data represent mean of triplicate experiments.
Effect of imatinib-CD19-liposomes on primary Ph+ ALL cells
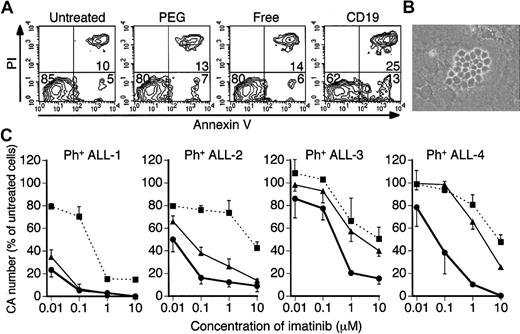
We examined the effect of imatinib-encapsulated liposomes and free imatinib on primary leukemia cells derived from Ph+ ALL patients. We used 2 samples from Ph+ ALL patients who acquired clinical resistance to imatinib, and 2 samples from Ph+ ALL patients who had not been treated with imatinib. The leukemia cells of all 4 samples were CD19+ (data not shown). We determined the sequence of the ATP-binding domain of the bcr-abl gene and found a single point mutation in both leukemia cells from patients with imatinib resistance. As shown in Table 2, a T to C change at nucleotide 904 caused a tyrosine to histidine change at amino acid 253 (all numbering based on the type Ia c-abl sequence). This point mutation is frequently reported as a cause of imatinib resistance in Ph+ ALL and CML.16,17,38 We also examined the level of bcr-abl transcript expression because increased fusion protein expression has been suggested as a possible cause of imatinib resistance as well.14,15 The amount of bcr-abl mRNA in Ph+ ALL cells from patients with imatinib resistance was 1.17% and 1.29% of glyceral-dehydes-3-phosphate dehydrogenase (GAPDH) mRNA, respectively, and those values were comparable to the values from leukemia cells from 2 patients not treated with imatinib of 1.23% and 1.15%. Initially, we examined apoptosis of these cells treated with imatinib-encapsulated-liposomes and free imatinib. The leukemia cells were cultured with several concentrations of imatinib-encapsulated liposomes or free imatinib for 48 hours and analyzed by Annexin V assay. Similar to the result of Ph+ ALL cell lines, imatinib-CD19-liposomes induced apoptosis in primary leukemia cells more efficiently than free imatinib and other imatinib-encapsulated liposomes (Figure 5A). Next, to determine the effect of imatinib-CD19-liposomes on leukemia cells possessing clonogenicity, we used a coculture system reported previously, in which a murine bone marrow stromal cell line was used as a source of feeder cells.35,39 In this coculture system, primary leukemia cells formed cell clusters on the feeder cells (CA, Figure 5B), and we counted the number of CAs on days 14 to 17 after the start of coculture. As for leukemia cells from patients not treated with imatinib (Ph+ ALL-1 and -2), a dose-dependent decrease in the number of CAs was observed when imatinib or imatinib-encapsulated liposomes were added. The inhibitory effect of the imatinib-CD19-liposomes on cell growth was much greater than that of free imatinib and other imatinib-encapsulated liposomes (Figure 5C). Moreover, CD19-liposomes inhibited CA formation of leukemia cells from patients with imatinib resistance (Ph+ ALL-3 and -4) more significantly than free imatinib and imatinib-PEG-liposomes (Figure 5C). Consequently, the IC50 of imatinib-CD19-liposomes was less than 10% that of free imatinib even when the target cells were derived from patients with imatinib resistance, whereas the IC50 of imatinib-PEG-liposomes was higher than that of free imatinib (Table 2).
IC50 of imatinib in primary Ph+ ALL cells
. | . | IC50, μM . | . | . | ||
|---|---|---|---|---|---|---|
| Sample . | Mutation in BCR-ABL . | PEG-liposome . | CD19-liposome . | Free imatinib . | ||
| Ph+ ALL-1 | WT | 5.93 ± 0.73* | < 0.01 | < 0.01 | ||
| Ph+ ALL-2 | WT | > 10.0 | 0.01 ± 0.01* | 0.04 ± 0.02* | ||
| Ph+ ALL-3 | Y253H | > 10.0 | 0.30 ± 0.06* | 3.18 ± 0.78* | ||
| Ph+ ALL-4 | Y253H | > 10.0 | 0.07 ± 0.06* | 3.34 ± 0.98* | ||
. | . | IC50, μM . | . | . | ||
|---|---|---|---|---|---|---|
| Sample . | Mutation in BCR-ABL . | PEG-liposome . | CD19-liposome . | Free imatinib . | ||
| Ph+ ALL-1 | WT | 5.93 ± 0.73* | < 0.01 | < 0.01 | ||
| Ph+ ALL-2 | WT | > 10.0 | 0.01 ± 0.01* | 0.04 ± 0.02* | ||
| Ph+ ALL-3 | Y253H | > 10.0 | 0.30 ± 0.06* | 3.18 ± 0.78* | ||
| Ph+ ALL-4 | Y253H | > 10.0 | 0.07 ± 0.06* | 3.34 ± 0.98* | ||
WT indicates wild type. Y253H indicates that the 253rd tyrosine was substituted by histidine.
Data represent mean ± SD of triplicate experiments.
Effect of imatinib-encapsulated liposomes on primary Ph+ ALL cells. (A) Apoptosis of Ph+ ALL-3 cells determined by Annexin V assay. Cells were cultured with the indicated imatinib-encapsulated liposomes or free imatinib for 48 hours. The numbers in the graphs indicate the percentage of cells in each quadrant. (B) CA formation of primary leukemia cells from Ph+ ALL-1 patients on day 17. Primary leukemia cells were cocultured with mouse stromal cells, and cobblestone areas were formed about 2 weeks after the start of coculture (original magnification × 100). The image was acquired using an inverted microscope (Diaphot TMD300; Nikon, Tokyo, Japan) with a digital camera (DXM1200, Nikon). The type of objective lens was Plan 10 DL (Nikon), and the numerical aperture was 0.30. The acquisition and subsequent processing software were ACT-1 (Nikon) and Adobe Photoshop 5.0 (Adobe Systems), respectively. (C) Inhibition of the formation of CAs derived from primary leukemia cells from 4 patients. Cells were treated with imatinib-CD19-liposomes (bold line), imatinib-PEG-liposomes (broken line), or free imatinib (thin solid line), and CAs were counted 14 to 17 days after the start of coculture. The numbers of CAs are indicated as the percentage of CA formed in each group compared with those formed by untreated primary leukemia cells. Data represent means ± SDs of triplicate experiments.
Effect of imatinib-encapsulated liposomes on primary Ph+ ALL cells. (A) Apoptosis of Ph+ ALL-3 cells determined by Annexin V assay. Cells were cultured with the indicated imatinib-encapsulated liposomes or free imatinib for 48 hours. The numbers in the graphs indicate the percentage of cells in each quadrant. (B) CA formation of primary leukemia cells from Ph+ ALL-1 patients on day 17. Primary leukemia cells were cocultured with mouse stromal cells, and cobblestone areas were formed about 2 weeks after the start of coculture (original magnification × 100). The image was acquired using an inverted microscope (Diaphot TMD300; Nikon, Tokyo, Japan) with a digital camera (DXM1200, Nikon). The type of objective lens was Plan 10 DL (Nikon), and the numerical aperture was 0.30. The acquisition and subsequent processing software were ACT-1 (Nikon) and Adobe Photoshop 5.0 (Adobe Systems), respectively. (C) Inhibition of the formation of CAs derived from primary leukemia cells from 4 patients. Cells were treated with imatinib-CD19-liposomes (bold line), imatinib-PEG-liposomes (broken line), or free imatinib (thin solid line), and CAs were counted 14 to 17 days after the start of coculture. The numbers of CAs are indicated as the percentage of CA formed in each group compared with those formed by untreated primary leukemia cells. Data represent means ± SDs of triplicate experiments.
Effect of imatinib-CD19-liposomes on CD34+ CBCs
Although imatinib was reported to induce cell death in normal human hematopoietic cells at high concentrations,25,40 our imatinib-CD19-liposomes did not affect the survival of Daudi and NB4 cells, which do not express the BCR-ABL tyrosine kinase. These results suggested that there is little nonspecific cell death in the imatinib-CD19-liposomes. To further evaluate the safety of the imatinib-CD19-liposomes, we examined colony formation of normal CD34+ CBCs after treatment with imatinib-CD19-liposomes or free imatinib. The number and composition of colonies were not affected significantly by the treatment with imatinib-CD19-liposomes or free imatinib at 10 μM. Mean ± SD of colony numbers derived from 200 cells treated with imatinib-CD19-liposomes/free imatinib/or not treated are as follows: granulocyte-macrophage colony-forming unit (CFU-GM), 36.0 ± 2.0/40.2 ± 0.2/44.0 ± 2.0; erythroid burst-forming unit (BFU-E), 34.0 ± 4.0/35.4 ± 1.4/50.2 ± 2.1; colony-forming units of mixed lineages (CFU-Mix), 16.0 ± 1.0/19.4 ± 1.3/20.2 ± 1.2; total, 86.0 ± 9.7/95.0 ± 8.6/119.6 ± 1.4. In addition, 50 μM imatinib-CD19-liposomes induced only a slight decrease in the number of colonies (CFU-GM, 30.0 ± 2.0; BFU-E, 29.9 ± 2.9; CFU-Mix, 10.5 ± 2.5; total, 70.4 ± 2.6), whereas the same concentration of free imatinib completely inhibited colony formation. These results strongly suggest that the imatinib-CD19-liposomes exert less nonspecific toxicity than free imatinib.
Discussion
Although imatinib is a breakthrough therapeutic modality for the treatment of Ph+ ALL as well as CML, a substantial portion of patients experience relapse within a short period after a response to imatinib.10-12 There have been several reports that speculate about the appearance of imatinib resistance, including genetic mutation in ATP-binding site of bcr-abl11,17,38 and enhanced expression of bcr-abl transcripts.14,15 To overcome imatinib resistance, administration of high-dose imatinib is thought to be the most promising approach, because previous in vitro reports demonstrated that treatment with high concentrations of imatinib inhibited the kinase activity of mutated BCR-ABL, which was not inhibited by lower concentration of imatinib.17,18 In clinical studies, administration of escalated doses of imatinib improved the response rate of refractory or relapsed CML cases,20,21 which suggested the usefulness of high-dose administration for Ph+ ALL cases. However, treatment of many patients with high-dose imatinib had to be discontinued or reduced because of toxicity.20,21 Therefore, we tried to develop Ph+ ALL-specific imatinib delivery systems for the purpose of minimizing nonspecific toxicity. Because CD19 is highly expressed only in cells of the B-cell lineage, including Ph+ ALL cells,2,31 and is rapidly internalized after antibody binding,32 we used CD19 as a targeting molecule. We produced PEG-liposomes coated with the anti-CD19 monoclonal antibody CLB-CD19 to encapsulate imatinib.
In this study, our Cal-CD19-liposomes were nearly completely internalized into all CD19+ cell lines tested, whereas Cal-bare- and Cal-PEG-liposomes could not efficiently be internalized (Figure 1A-B). Very high internalization efficiency of the Cal-CD19-liposomes was only observed in CD19+ cells, and underivatized anti-CD19 antibody inhibited the internalization of the Cal-CD19-liposomes in a dose-dependent manner (Figure 1B-C), suggesting that internalization of the Cal-CD19-liposomes was specifically mediated by the cell surface CD19 molecule. The internalization efficiency of our Cal-CD19-liposomes was much higher (more than 20 times of PEG-liposomes in mean fluorescence intensity by FCM) than that of other reported immunoliposomes coupled with anti-CD19 monoclonal antibody (approximately 2 times of PEG-liposomes).41 The reason for this higher internalization efficiency is thought to be due to the pendant-type immunoliposomes we used. Our previous report demonstrated that pendant-type immunoliposomes were internalized into target cells more efficiently than immunoliposomes with antibody covalently linked to the lipid of the liposome membrane.26
As a result of the higher and more selective delivery of our imatinib-CD19-liposomes, we successfully demonstrated that our imatinib-CD19-liposomes were able to induce cell death efficiently in Ph+ ALL cells and minimize toxicity to normal hematopoietic progenitor cells. Moreover, our imatinib-CD19-liposomes inhibited cobblestone area formation by primary Ph+ ALL cells more remarkably than free imatinib and imatinib-PEG-liposomes, suggesting that the imatinib-CD19-liposomes would also be effective for the treatment of clonogenic leukemic cells, namely leukemic stem cells. However, the effect of the imatinib-CD19-liposomes on the leukemic stem cells should be confirmed in vivo by using nonobese diabetic severely combined immunodeficiency (NOD-SCID) mice that received transplants with patients' CD34+ CD38– ALL stem cells.42 The inhibitory effects of the imatinib-CD19-liposomes were observed in bcr-abl–positive ALL cells with or without an amino acid mutation that causes imatinib resistance,16,17,38 suggesting that our imatinib-CD19-liposomes would be useful in the treatment of patients with Ph+ ALL who had acquired imatinib resistance. Of course, the anti-CD19 monoclonal antibody used in the CD19-liposomes should be humanized in a clinical setting in the future to decrease immunogenicity.43
To overcome the problem of resistance to imatinib, a number of approaches have been reported. Imatinib combined with other chemotherapeutic agents is one of the most investigated approaches, and cytarabine is a candidate drug that was reported to show a synergistic effect with imatinib.44 The phosphatidylinositol-3 kinase inhibitor wortmannin or LY294002,45 farnesyl transferase inhibitor SCH66336,46 third-generation bisphosphonate zoledronate,47 and proteasome inhibitor PS-34148 were reported to exert anti–bcr-abl–positive cell effects, and those inhibitory effects were synergistic with imatinib. In contrast, our imatinib-CD19-liposome is a unique preparation that can enhance the effect of imatinib itself; therefore, combination with those reagents may be a further promising strategy against Ph+ ALL.
In conclusion, we have successfully demonstrated that imatinib-CD19-liposomes induced specific and efficient death of Ph+ ALL cells. This new therapeutic approach may be useful for treatment of Ph+ ALL with greatly reduced side effects than free imatinib.
Prepublished online as Blood First Edition Paper, May 20, 2004; DOI 10.1182/blood-2004-02-0588.
Supported in part by a Grant-in-aid from the Ministry of Education, Science, Sports, and Culture, and the Ministry of Health, Labor, and Welfare of Japan. Y.B. is an awardee of a Research Fellowship of the Japan Society of the Promotion of Science for Young Scientists.
M.H., Y.S., K.T., J.O., and T. Takizawa contributed equally to this work.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Novartis Pharmaceuticals Corporation and Nichirei Corporation for providing imatinib and anti-CD19 antibody CLB-CD19. We also thank Dr K. Ohyashiki for kindly providing OM9; 22 cells. We also appreciate the hospital staff of the Research Hospital of The Institute of Medical Science, The University of Tokyo, for their support in sample collection from patients. We thank Drs N. Satoh (The University of Tokyo), Y. Nakazaki (Kyushu University), and T. Tanabe (National Institute of Advanced Industrial Science and Technology), Mr K. Takahashi, Ms N. Yusa, Ms M. Oiwa, and Ms S. Suzuki (The University of Tokyo) for technical support.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal