Abstract
Platelet factor 4 (PF4) is expressed during megakaryocytic differentiation. We previously demonstrated that the homeodomain proteins (myeloid ecotropic integra tion site 1 [MEIS1], Pbx-regulating protein 1 [PREP1], and pre-B-cell leukemia transcription factors [PBXs]) bind to the novel regulatory element tandem repeat of MEIS1 binding element [TME] and transactivate the rat PF4 promoter. In the present study, we investigated and identified other TME binding proteins in megakaryocytic HEL cells using mass spectrometry. Among identified proteins, we focused on upstream stimulatory factor (USF1) and USF2 and investigated their effects on the PF4 promoter. USF1 and 2 bound to the E-box motif in the TME and strongly transactivated the PF4 promoter. Furthermore, physiologic bindings of USF1 and 2 to the TME in rat megakaryocytes were demonstrated by the chromatin immunoprecipitation (ChIP) assay. Interestingly, the E-box motif in the TME was conserved in TME-like sequences of both the human and mouse PF4 promoters. USF1 and 2 also bound to the human TME-like sequence and transactivated the human PF4 promoter. Expressions of USF1 and 2 were detected by reverse-transcriptase–polymerase chain reaction (RT-PCR) in the human megakaryocytes derived from CD34+ cells. Thus, these studies demonstrate that the novel TME binding transcription factors, USF1 and 2, transactivate rat and human PF4 promoters and may play an important role in megakaryocytic gene expression.
Introduction
Megakaryocytes are the hematopoietic precursors of platelets. Platelets play an essential role in thrombosis and hemostasis. Platelet factor 4 (PF4) is expressed exclusively in megakaryocytes and platelets, and serves as a lineage-specific marker of megakaryocytic differentiation.1 To investigate megakaryocytic gene expression, the rat and human PF4 promoter have been studied.2-10 We previously demonstrated that GATA-1 and ETS-1 are important transcription factors for the transactivation of the PF4 gene.5 More recently, we identified the novel regulatory element termed “TME” in the PF4 promoter and demonstrated that homeodomain proteins, myeloid ecotropic integration site 1 (MEIS1) and pre-B-cell leukemia transcription factors (PBXs) (PBX1B and PBX2), bind to the TGACAG motifs in the tandem repeat of MEIS1 binding element (TME) by forming MEIS1/PBX complexes.11 Furthermore, we also demonstrated that Pbx-regulating protein 1 (PREP1), a MEIS1 homolog protein, binds to the TME, forming PREP1/PBX complexes and transactivating the PF4 promoter.12 Both MEIS1/PBX and PREP1/PBX complexes activate the PF4 promoter synergistically with GATA-1 and ETS-1, which are essential for megakaryocytic gene expression. These reports suggest that transcriptional regulation through the TME is important for megakaryocytic gene expression.
The TME contains the characteristic binding motifs of transcription factors.11 In addition to the 2 binding motifs of homeodomain proteins (TGACAG), 3 tandem E-box motifs (CAGCTG) and an ETS binding motif are included. Because it has been demonstrated that many basic helix-loop-helix (bHLH) transcription factors bind to the E-box motif and regulate gene expression, we hypothesized that E-box motifs in the TME would be important for the regulation of PF4 gene expression. Transcription factor TAL-1 is an E-box binding protein that is essential for early hematopoiesis.13,14 TAL-1 is expressed in multipotent progenitor cell lines prior to lineage commitment, and is up-regulated during erythroid/megakaryocytic differentiation and downmodulated during granulocytic/macrophage differentiation.15 Enforced TAL-1 expression stimulates primitive, erythroid, and megakaryocytic progenitors.16 From this information, it was speculated that TAL-1 is the factor binding to E-box motifs in the TME. However, our previous study indicated that TAL-1 did not bind to the TME in megakaryocytic HEL cells, although expression of TAL-1 was detected.11 We, therefore, speculated that other E-box binding proteins regulate PF4 gene expression in megakaryocytes.
In the present study, we identify many proteins included in the TME binding complex by mass spectrometry. Upstream stimulatory factor 2 (USF2), which is an E-box binding protein, is included among the identified proteins. USF1 and 2 were initially identified as binding proteins to the regulatory element of the adenovirus major late promoter.17-20 USF1 and 2 are members of the basic helix-loop-helix leucine zipper proteins and are ubiquitously expressed in a variety of tissues.17,21 These proteins preferentially bind to the E-box motif and regulate many genes.22-33 Our electrophoretic mobility shift assay (EMSA) experiments indicate that both USF1 and 2 bind to the E-box motifs in the TME. Reporter gene assays show that USF1 and 2, especially USF2, strongly activate the rat PF4 promoter. The bindings of USF1 and 2 to the TME in vivo are demonstrated by chromatin immunoprecipitation (ChIP) assay using rat megakaryocytes. In addition to the rat PF4 promoter, USF1 and 2 also regulate the human PF4 promoter through the conserved E-box motif in the human TME-like sequence. We propose that USF1 and 2 are novel E-box binding transcription factors that regulate PF4 gene expression in megakaryocytes.
Materials and methods
Identification of the TME binding proteins by mass spectrometry
Purification of the TME binding proteins by DNA affinity chromatography has been described previously.11 The obtained fractions containing the TME binding proteins were mixed with an equal volume of 20% trichloroacetic acid solution. After centrifugation, precipitated proteins were rinsed with 1 mL ethanol. All of these proteins were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and stained by SYPRO Orange (Bio-Rad, Hercules, CA). All stained bands were cut off from the gel, and proteins were reduced by dithiothreitol, alkylated by iodoacetamide (SIGMA, St Louis, MO) and digested by Trypsin that had been modified for peptide sequencing (Roche, Mannheim, Germany). We carried out this in-gel digestion according to a previous report.34 Peptide sequences were analyzed by MS/MS using LCQ-DECA (Thermo Electron, Kanagawa, Japan). Proteins were identified using SEQUEST (Thermo Electron).
Plasmid construction
The cDNAs of USF1 (GenBank accession no. XM_001638) and USF2 (GenBank accession no. NM_003367) were obtained from cDNA pooled from HEL cells by polymerase chain reaction (PCR) amplification. These PCR products were cloned into the EcoRV site in the pcDNA3 vector (Invitrogen, San Diego, CA). The DNA sequences of the inserted USF1 and 2 fragments were confirmed by DNA sequencing with an ABI PRISM 310 (Applied Biosystems, Tokyo, Japan). To make the reporter plasmid with the human PF4 promoter (hPF4luc), 1.7 kb of the 5′-flanking region of human PF4 gene was amplified by PCR using the specific primers (forward 5′-GTGGGAGAGGCAGATAAGTAGGCAA-3′ and reverse 5′-TCTGTGGCCAATGACTCCTGAGCCT-3′) and genome DNA template prepared from HEL cells. This fragment was inserted into the SmaI site of the PGV-B vector (Toyo Ink, Tokyo, Japan). Preparation of the other 3 expression vectors (pcDNA3-MEIS1, pcDNA3-PBX1B, and pcDNA3-PBX2) and the reporter plasmid of the rat PF4 promoter (PF4luc) has been described previously.5 For the preparation of PF4mut, PF4luc was digested with PvuII and the shorter fragment derived from the TME was excluded. Instead of this short fragment, the mutated double-stranded oligonucleotide prepared from 5′-CGACTGACAGCGAGCCTTCG-3′ and 5′-CGAAGGCTCGCTGTCAGTCG-3′ was cloned into the PvuII site of PF4luc. For the preparation of hPF4mut1, 2, and 3, 4 primers (wild-type [Wt] forward [Fw]: 5′-CTGGGAGGTTGGAAAGGAAACAGGA-3′, mutant [Mut] Fw: 5′-AAAGGAGGTTGGAAAGGAAACAGGA-3′, Wt reverse [Rv]: 5′-CTGCCTTGGCTGGGACCTCCAACCT-3′, and Mut Rv: 5′-AAACCTTGGCTGGGACCTCCAACCT-3′) were used. For amplifying fragments of hPF4mut constructs by PCR, each set of primers (Wt Fw and Mut Rv for hPF4mut1, Mut Fw and Wt Rv for hPF4mut2, and Mut Fw and Mut Rv for hPF4mut3) was used. hPF4luc was digested with PvuII and the shorter fragment was removed. Instead of this shorter fragment, each amplified fragment was cloned into the PvuII site. For preparing the hPF4(+485)luc construct, a fragment (–427 to +75) was amplified by PCR using primers, 5′-CTGGGAGGTTGGAAAGGAAACAGGA-3′ and 5′-GCTCGGTACCGCTGCGGCAGAGCTTCCAGCAGGAT-3′, and genome DNA template prepared from HEL cells. After digesting with PvuII and KpnI, this fragment was cloned into the PvuII-KpnI site of a 6.9-kb fragment, which was obtained by digesting hPF4luc with PvuII and KpnI. The obtained construct was digested with KpnI and used for a further cloning of a fragment (+77 to +485). This fragment was amplified by PCR using primers, 5′-ATTGGTACCTGAGCTCCGCAGCCGGGTTCTGCGC-3′ and 5′-AACGGTACCTGAGGGGGAAATGGAGAGGGTAAG-3′, and genome DNA template prepared from HEL cells. For preparing the hPF4(+485)Mut construct, the same procedure that was used for preparing hPF4mut3 was used with primers, 5′-AAAGGAGGTTGGAAAGGAAACAGGA-3′ and 5′-AAACCTTGGCTGGGACCTCCAACCT-3′.
Culture of cell lines and isolation of nuclear extracts
HEL cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 IU/mL penicillin, and 100 μg/mL streptomycin. HepG2 cells were maintained under the same conditions except that Dulbecco modified Eagle medium (DMEM) was used. The isolation method of the nuclear extract has been described previously.5 For the transient transfection assay using hPF4luc and hPF4mut constructs, HEL cells were cultured with Iscove modified Dulbecco medium (IMDM; GIBCO BRL, Grand Island, NY) supplemented with 10% fetal calf serum (FCS), 100 IU/mL penicillin, and 100 μg/mL streptomycin.
Western blotting
For Western blots, 20 μLaffinity purified fractions was electrophoresed on 10% SDS-PAGE, and the proteins were blotted onto nitrocellulose membranes. The membranes were blocked in tris(hydroxymethyl)aminomethane-buffered saline (TBS) containing 3% nonfat milk and 0.1% Tween 20 and then incubated in the same solution containing the primary antibody (USF1 [H-86] and USF2 [N-18]; Santa Cruz Biotechnology, Santa Cruz, CA). The membranes were washed and incubated in blocking solution containing the secondary antibody. The immunoblots were visualized with the enhanced chemiluminescence (ECL) Western blotting detection system (Amersham Pharmacia Biotech, Uppsala, Sweden) according to the manufacturer's instructions.
EMSA and supershift assay
The double-stranded TME fragment prepared from 2 oligonucleotides, 5′-TCCTGCTGACAGCTGCTGACAGCTGGCCTCAGCTGC-3′ and 5′-CGCAGCTGAGGCCAGCTGTCAGCAGCTGTCAGCAGGA-3′, was labeled with Klenow polymerase and used as the TME probe. Underlined sequences indicate wild-type or mutated USF binding motifs. The other probes were prepared from the following combination of 2 oligonucleotides: TMEmut, 5′-TCCTGCTGACAGCgaCTGACAGCgaGCCTtcGCTGCTTCTT-3′ and 5′-AAGAAGCAGCgaAGGCtcGCTGTCAGtcGCTGTCAGCAGGA-3′; hTME-like, 5′-CTCCCAGCCAAGGCAGCTGCCCAGAGCCTT-3′ and 5′-CAAGGCTCTGGGCAGCTGCCTTGGCTGGGA-3′; hTMEmut, 5′-TCCCAGCCAAGGtttCTGCCCAGAGCCTT-3′ and 5′-AAGGCTCTGGGCAGaaaCCTTGGCTGGGA-3′; hE-box, 5′-TGGTAGTTGCAGCTGGGAGGTTGGA-3′ and 5′-TCCAACCTCCCAGCTGCAACTACCA-3′; and hE-boxmut, 5′-TGGTAGTTGCAGaaaGGAGGTTGGA-3′ and 5′-TCCAACCTCCtttCTGCAACTACCA-3′. These fragments were labeled by T4 polynucleotide kinase and used as probes. The EMSAprocedure was similar to that described previously.11 The binding reaction was carried out at 4° C for 45 minutes. In the case of the supershift assay, 6 μg nuclear extract of HEL cells was incubated with the target antibody at room temperature for 20 minutes before the probe was added. The antibodies for USF1 and USF2 referred to in “Western blotting” and the control immunoglobulin G (IgG, anti-TFIID [SI-1]; Santa Cruz Biotechnology) were used for supershift assays.
In vitro transcription/translation
In vitro translated USF1 and 2 were prepared with the TNT coupled transcription-translation reticulocyte lysate (T7 polymerase version) (Promega, Madison, WI) according to the manufacturer's instructions. The obtained USF1 and 2 were analyzed by Western blotting (Figure 2), and 3/50 of the total product of each protein was used for the EMSA.
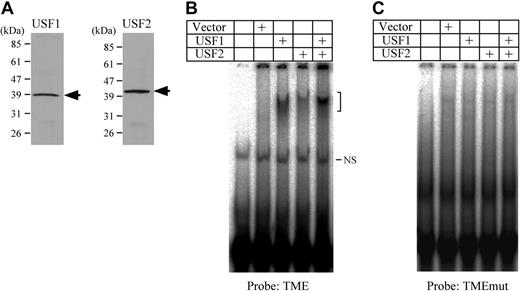
Binding activities of USF1 and USF2 to the TME. (A) Western blotting detection of USF1 and USF2 prepared by in vitro translation. Arrows indicate the band of either USF1 or USF2. (B) EMSA using the TME probe and USF1 and USF2 prepared by in vitro translation. The bracket indicates the specific shifted bands. NS indicates a nonspecific band. (C) EMSA was performed using the TMEmut probe. This probe includes the mutations in the E-box motifs.
Binding activities of USF1 and USF2 to the TME. (A) Western blotting detection of USF1 and USF2 prepared by in vitro translation. Arrows indicate the band of either USF1 or USF2. (B) EMSA using the TME probe and USF1 and USF2 prepared by in vitro translation. The bracket indicates the specific shifted bands. NS indicates a nonspecific band. (C) EMSA was performed using the TMEmut probe. This probe includes the mutations in the E-box motifs.
Transient transfection assay
Transient transfection assays were performed using reporter plasmid, PF4luc or hPF4luc. For the reporter gene assays using HepG2 cells, 0.5 μg of each reporter plasmid was transfected, together with 0.5 μg pβactin-lacZ for the internal control, using Lipofectamine2000 reagent (GIBCO BRL, Gaithersburg, MD). For the overexpression experiments, 0.5 or 1 μg of the expression vector was used with PF4luc or hPF4luc, respectively. In the case of the reporter gene assay using HEL cells, 7 μg of each reporter plasmid was transfected with 7 μg pβactin-lacZ by the electroporation method described previously.8 Transfected cells were then incubated with RPMI 1640 or IMDM medium supplemented with 10 nM phorbol 12-myristate 13-acetate (PMA; SIGMA), 10% FBS or FCS, 100 IU/mL penicillin, and 100 μg/mL streptomycin. In all assays, cells were harvested approximately 48 hours after transfection. Each assay was performed in duplicate more than 3 times.
ChIP assay
The method of purification of rat megakaryocytes has been described previously.11 Purified megakaryocytes were incubated for 2 hours in IMDM containing 5% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 0.2 ng/mL thrombopoietin (TPO; donated by Kirin Brewery). After the incubation, the ChIP assay was performed using a Chromatin Immunoprecipitation Assay Kit (Upstate, Lake Placid, NY) according to the manufacturer's instructions. For the assay, 10 μg of antibodies for USF1 and 2 described above or subtype-matched normal rabbit IgG (Santa Cruz Biotechnology) was used. The PCR amplification for the fragment including the TME was performed by 35 cycles of 3 steps (94° C for 30 seconds, 55° C for 30 seconds, and 72° C for 30 seconds) using the primers, forward 5′-CATACAGCATACCTTCTGCG-3′ and reverse 5′-AAGCAGCTGAGGCCAGCTGTCAGCA-3′. The PCR amplification for hypoxanthine phosphoribosyltransferase (Hprt) was performed by 40 cycles of 3 steps (94° C for 30 seconds, 60° C for 30 seconds, and 72° C for 30 seconds) using the primers, forward 5′-GGAGATTGAGGGAGAGCAAACTCAG-3′ and reverse 5′-CTCTTCCCACTGATGTCCTACAAGG-3′. PCR products were separated on a 2% agarose gel and stained by ethidium bromide.
Reverse transcriptase (RT)–PCR using human megakaryocytes
Cord blood (CB) was collected following normal pregnancies and deliveries, and with the informed consent of the parents. Mononuclear cells were isolated from CB and were subjected to immunomagnetic separation using a magnetic-activated cell separation (MACS) CD34 Progenitor Cell Isolation Kit (Miltenyi Biotech, Auburn, CA), as described previously.35 Purified CD34+ cells were cultured in IMDM containing 20% human serum, 50 IU/mL penicillin, 50 μg/mL streptomycin, and 10 ng/mL TPO (PeproTech, Rocky Hill, NJ). The same amount of TPO was added to the medium every 2 days, and half of the medium was replaced with new medium after 6 days of incubation. Megakaryocytes were collected after 0, 4, or 10 days of incubation. Total RNA was isolated using ISOGEN (Nippon Gene, Tokyo, Japan), and each cDNA was synthesized from 0.5 μg total RNA using ReveTra Ace (TOYOBO, Osaka, Japan) according to the manufacturers' instructions. The obtained cDNAtemplates and a set of primers (PF4: forward, 5′-AGCATGAGCTCCGCAGCCGGGTTCT-3′ and reverse, 5′-GTAGGCAGCTAGTAGCTAACTCTCC-3′; glycoprotein IIb [GPIIb]: forward, 5′-CAAGAACAGCCAGAATCCAAACAG-3′ and reverse, 5′-TACGAGAACTGGATCCTGAAGCCT-3′; USF1: forward, 5′-GATGAGAAACGCAGGGCTCAGCATA-3′ and reverse, 5′-TTAGTTGCTGTCATTCTTGATGACG-3′; USF2: forward, 5′-CCGGACACACCCTTACTCTCCAAAA-3′ and reverse, 5′-TCACTGCCGGGTGCCCTCGCCCACC-3′; and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]: forward, 5′-ACTGCTTAGCACCCCTGGCCAAGGT-3′ and reverse, 5′-GAGTGGGTGTCGCTGTTGAAGTCAG-3′) was used for PCR amplification. The following describes the temperature and time of each step of the PCR (denaturing, annealing, and extension), the number of PCR cycles, and the PCR product length obtained, respectively: PF4 and GPIIb: 94°C 30 seconds, 55°C 30 seconds, 72°C 30 seconds, 35 cycles, 323 and 459 bp; USF1 or USF2: 94°C 30 seconds, 60°C30 seconds, 72°C 30 seconds, 40 cycles, 345 or 395 bp; and GAPDH: 94°C 30 seconds, 55°C 30 seconds, 72°C 30 seconds, 30 cycles, 438 bp.
Statistical analyses
Data were expressed as mean ± standard error (SE). The statistical significance of differences of the means was determined by Student t test (2 groups comparison) or one-way analysis of variance and multiple comparisons that were performed with Tukey-Kramer multiple range test (more than 3 groups comparison).
Results
Identification of the TME binding proteins
In our previous study, we demonstrated that homeodomain proteins (MEIS1, PREP1, and PBXs) are the TME binding proteins.11,12 Although it was speculated that other proteins might bind to the E-box motif in the TME, we could not identify them from the binding motifs. In order to identify other TME binding proteins, total proteins obtained by TME affinity chromatography were separated by SDS-PAGE and stained by SYPRO Orange and all bands were cut from the gel. After in-gel tryptic digestion, eluted peptides were analyzed using the LC-MS/MS system. From the obtained amino acid sequence data, the TME binding proteins were identified using SEQUEST software (Table 1). Many nuclear proteins and transcription factors were identified. Among them, we focused on the 2 transcription factors, PBXs and USF2. Because they were each identified by more than 2 peptide fragments obtained with tryptic digestion (Table 2), it is highly probable that they are legitimate TME binding proteins. We previously identified PBXs as TME binding proteins. We, therefore, concentrated on USF2 to evaluate it as a new TME binding factor.
Identified proteins by mass spectrometry
kDa . | Search result . |
|---|---|
| 114 | Heterogeneous nuclear ribonucleoprotein U |
| 90 | PTB-associated splicing factor |
| 80 | ATP-dependent DNA helicase, II, 70-kDa subunit (KU70) |
| 80 | Nucleolar protein NOP56 |
| 80 | Probable RNA-dependent helicase p72 (dead box protein p72) |
| 80 | Putative pre-mRNA splicing factor RNA helicase |
| 80 | Proto-oncogene tyrosine-protein kinase ABL (P150) |
| 65 | Probable RNA-dependent helicase p68 (dead box protein p68) |
| 65 | GAP-associated tyrosine phosphoprotein p62 |
| 65 | RNA-binding protein (FUS) |
| 55 | Similar to zinc finger protein (BR140) |
| 55 | 54-kDa nuclear RNA-binding protein |
| 45 | Heterogeneous nuclear ribonucleoprotein G |
| 45 | Transcription factor NF-AT 45k chain |
| 43 | Upstream stimulatory factor |
| 43 | Pre B-cell leukemia transcription factor |
| 43 | Heterogeneous nuclear ribonucleoprotein C-like protein |
| 43 | Repressor ZEB |
| 40 | Heterogeneous nuclear riboproteins A2/B1 |
| 40 | Paired box protein 8 |
| 40 | Heterogeneous nuclear riboprotein A1 |
| 40 | Heterogeneous nuclear ribonucleoprotein A3 |
| 40 | Heterogeneous ribonucleoprotein homolog |
| 40 | Heterogeneous nuclear proteins C1/C2 |
| 28 | Transcription factor CBF alpha 2, splice form 1, AML1a protein |
| 28 | Nonhistone chromosomal protein (HMG-1) |
| 28 | Transcription initiation factor TFIID 30-kDa subunit |
kDa . | Search result . |
|---|---|
| 114 | Heterogeneous nuclear ribonucleoprotein U |
| 90 | PTB-associated splicing factor |
| 80 | ATP-dependent DNA helicase, II, 70-kDa subunit (KU70) |
| 80 | Nucleolar protein NOP56 |
| 80 | Probable RNA-dependent helicase p72 (dead box protein p72) |
| 80 | Putative pre-mRNA splicing factor RNA helicase |
| 80 | Proto-oncogene tyrosine-protein kinase ABL (P150) |
| 65 | Probable RNA-dependent helicase p68 (dead box protein p68) |
| 65 | GAP-associated tyrosine phosphoprotein p62 |
| 65 | RNA-binding protein (FUS) |
| 55 | Similar to zinc finger protein (BR140) |
| 55 | 54-kDa nuclear RNA-binding protein |
| 45 | Heterogeneous nuclear ribonucleoprotein G |
| 45 | Transcription factor NF-AT 45k chain |
| 43 | Upstream stimulatory factor |
| 43 | Pre B-cell leukemia transcription factor |
| 43 | Heterogeneous nuclear ribonucleoprotein C-like protein |
| 43 | Repressor ZEB |
| 40 | Heterogeneous nuclear riboproteins A2/B1 |
| 40 | Paired box protein 8 |
| 40 | Heterogeneous nuclear riboprotein A1 |
| 40 | Heterogeneous nuclear ribonucleoprotein A3 |
| 40 | Heterogeneous ribonucleoprotein homolog |
| 40 | Heterogeneous nuclear proteins C1/C2 |
| 28 | Transcription factor CBF alpha 2, splice form 1, AML1a protein |
| 28 | Nonhistone chromosomal protein (HMG-1) |
| 28 | Transcription initiation factor TFIID 30-kDa subunit |
Identified peptide sequences of USF2 and PBXs
Identified transcription factor . | Identified peptide sequence . |
|---|---|
| Upstream stimulatory factor 2 | AQLQQHNLEMVGEGTRQ |
| QQIEELKNENALLR | |
| TVLSIR | |
| Pre B-cell leukemia transcription factor | GGSAAAAAAAAASGGAGSDNSVEHSDYR |
| GAQEEEPTDPQLMR | |
| NIGKFQEEANLYAAK |
Identified transcription factor . | Identified peptide sequence . |
|---|---|
| Upstream stimulatory factor 2 | AQLQQHNLEMVGEGTRQ |
| QQIEELKNENALLR | |
| TVLSIR | |
| Pre B-cell leukemia transcription factor | GGSAAAAAAAAASGGAGSDNSVEHSDYR |
| GAQEEEPTDPQLMR | |
| NIGKFQEEANLYAAK |
Both USF1 and 2 bind to E-box motifs in the TME and activate the PF4 promoter
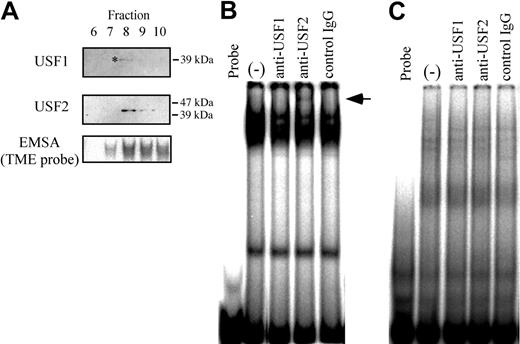
USF1 is a homolog protein of USF2. In order to determine the presence of USF1 and 2 in the purified fractions, Western blotting was performed (Figure 1A). Although USF1 showed a weak band, both USF1 and 2 were detected. In addition, a supershift assay using nuclear extracts from HEL cells indicates that a supershifted band derived from USF2 is present (Figure 1B). This supershifted band as well as the shifted bands derived from TME binding proteins disappeared when using the TMEmut probe containing mutations in the 3 E-box motifs (Figure 1C). These data suggest that USF2 is a major binding protein to the TME, and that the E-box motifs are important for TME binding activity in HEL cells.
Binding of USF1 and USF2 to the TME in HEL cells. (A) Detection of USF1 and USF2 among the TME binding proteins. Western blotting was performed with fractions from a DNA affinity column. Fraction numbers of affinity column elute are indicated above. *Faint USF1 band. TME binding activity of each fraction was measured by EMSA. (B-C) Detection of USF1 and USF2 in the TME binding complex in HEL cells by the supershift assay. A supershift assay was performed with nuclear extracts of HEL cells and factor-specific antibodies. Arrow indicates supershifted band. The TME probe (B) or TMEmut probe (C) that includes the mutations in the E-box motifs was used. (–) indicates no antibody.
Binding of USF1 and USF2 to the TME in HEL cells. (A) Detection of USF1 and USF2 among the TME binding proteins. Western blotting was performed with fractions from a DNA affinity column. Fraction numbers of affinity column elute are indicated above. *Faint USF1 band. TME binding activity of each fraction was measured by EMSA. (B-C) Detection of USF1 and USF2 in the TME binding complex in HEL cells by the supershift assay. A supershift assay was performed with nuclear extracts of HEL cells and factor-specific antibodies. Arrow indicates supershifted band. The TME probe (B) or TMEmut probe (C) that includes the mutations in the E-box motifs was used. (–) indicates no antibody.
To certify the bindings and identify binding sites, USF1 and 2 were prepared by in vitro translation (Figure 2A). EMSA using these proteins indicated that both USF1 and USF2 bound to the TME (Figure 2B). On the other hand, neither USF1 nor USF2 bound to the TMEmut probe (Figure 2C). These results indicate that USF1 and 2 bind to the E-box motifs in the TME.
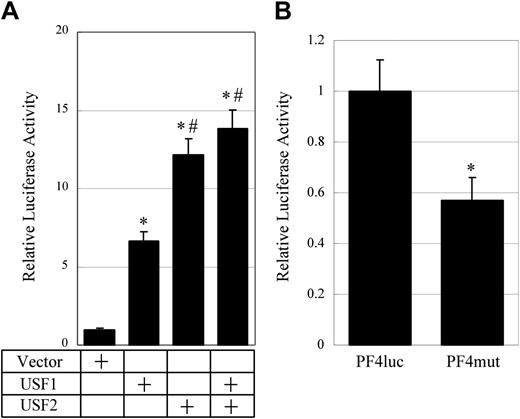
To investigate the effects of USF1 and 2 on the PF4 promoter, transient transfection assays were performed. Reporter plasmid, PF4luc, which contains 1.1 kb of the rat PF4 promoter, and the expression vectors for USF1 or 2 were cotransfected into HepG2 cells (Figure 3A). In order to detect unambiguously the contributions of USF1 and 2 to the PF4 promoter, we use nonmegakaryocytic HepG2 cells in which little endogenous USF2 is expressed. The overexpression of each of USF1 and USF2 strongly activated the PF4 promoter 7- and 12-fold, respectively. The overexpression of both USF1 and 2 activated the PF4 promoter 14-fold. These results indicate that both USF1 and 2, especially USF2, strongly transactivate the PF4 gene expression.
Activation of the rat PF4 promoter by USF1 and USF2 through the E-box motifs. (A) Activation of the rat PF4 promoter by USF1 and USF2. A transient transfection assay was performed with PF4luc and USF expression vectors. PF4luc contains 1.1 kb of PF4 promoter in front of the luciferase gene. pcDNA3 vectors (0.5 μg), expressing either USF1 or USF2 or a mix of both vectors, were cotransfected with PF4luc into HepG2 cells. Relative luciferase activities were measured. The columns and vertical bars represent mean ± SE of 6 replicates. *A significant difference (P < .05) between control and USF1, USF2, and USF1 + USF2. #A significant difference (P < .05) between USF1 and USF2 and USF1 + USF2. (B) Activation of the rat PF4 promoter through E-box motifs. PF4luc or PF4mut, both of which constructed by mutating the E-box motifs in the rat PF4 promoter, was transfected into HEL cells. After the transfection, cells were incubated with medium containing 10 nM PMA, and luciferase activities were measured. The columns and vertical bars represent mean ± SE of 6 replicates. *A significant difference (P < .05).
Activation of the rat PF4 promoter by USF1 and USF2 through the E-box motifs. (A) Activation of the rat PF4 promoter by USF1 and USF2. A transient transfection assay was performed with PF4luc and USF expression vectors. PF4luc contains 1.1 kb of PF4 promoter in front of the luciferase gene. pcDNA3 vectors (0.5 μg), expressing either USF1 or USF2 or a mix of both vectors, were cotransfected with PF4luc into HepG2 cells. Relative luciferase activities were measured. The columns and vertical bars represent mean ± SE of 6 replicates. *A significant difference (P < .05) between control and USF1, USF2, and USF1 + USF2. #A significant difference (P < .05) between USF1 and USF2 and USF1 + USF2. (B) Activation of the rat PF4 promoter through E-box motifs. PF4luc or PF4mut, both of which constructed by mutating the E-box motifs in the rat PF4 promoter, was transfected into HEL cells. After the transfection, cells were incubated with medium containing 10 nM PMA, and luciferase activities were measured. The columns and vertical bars represent mean ± SE of 6 replicates. *A significant difference (P < .05).
To evaluate the function of the E-box motifs in megakaryocytic cells, PF4luc or PF4mut reporter plasmid that contains mutations in the E-box motifs was transfected into HEL cells. After incubation in medium containing 10 nM PMA, the luciferase activities were evaluated (Figure 3B). The mutation of the E-box motifs in the TME decreased the promoter activity by approximately 40%. This result indicated that the E-box motifs are important for the rat PF4 promoter activity. Taken together, these data suggest that the bindings of USF1 and 2 to the E-box motifs in the TME are important for the activation of the rat PF4 promoter.
USF1 and 2 physiologically bind to the TME in megakaryocytes
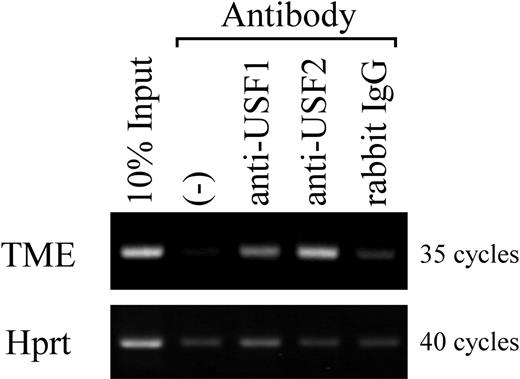
To investigate the bindings of USF1 and 2 to the TME in vivo, a ChIP assay was performed. Rat bone marrow cells were differentiated into megakaryocytes with TPO for 3 days. These differentiated cells were purified with a MACS system using the anti-CD61 antibody. Purified megakaryocytes were used in a ChIP assay. Specific bands were detected only when antibodies for USF1 and 2 were used, but were not detected in the control lane (Figure 4). This result indicates that USF1 and 2 physiologically bind to the TME.
Analysis of USF1 and USF2 bindings to the TME in rat megakaryocytes. ChIP assay was performed using purified rat megakaryocytes. Antibodies for USF1 and USF2 or isotype-matched controls were used. Precipitated DNA fragments and 10% of total DNA samples were amplified by PCR using primers specific for the rat PF4 promoter (–300 to –182), including the TME (–219 to –182), or primers for Hprt as a control. PCR products were separated on a 2% agarose gel and stained by ethidium bromide. (–) indicates no antibody.
Analysis of USF1 and USF2 bindings to the TME in rat megakaryocytes. ChIP assay was performed using purified rat megakaryocytes. Antibodies for USF1 and USF2 or isotype-matched controls were used. Precipitated DNA fragments and 10% of total DNA samples were amplified by PCR using primers specific for the rat PF4 promoter (–300 to –182), including the TME (–219 to –182), or primers for Hprt as a control. PCR products were separated on a 2% agarose gel and stained by ethidium bromide. (–) indicates no antibody.
USF1 and 2 also regulate the human PF4 promoter
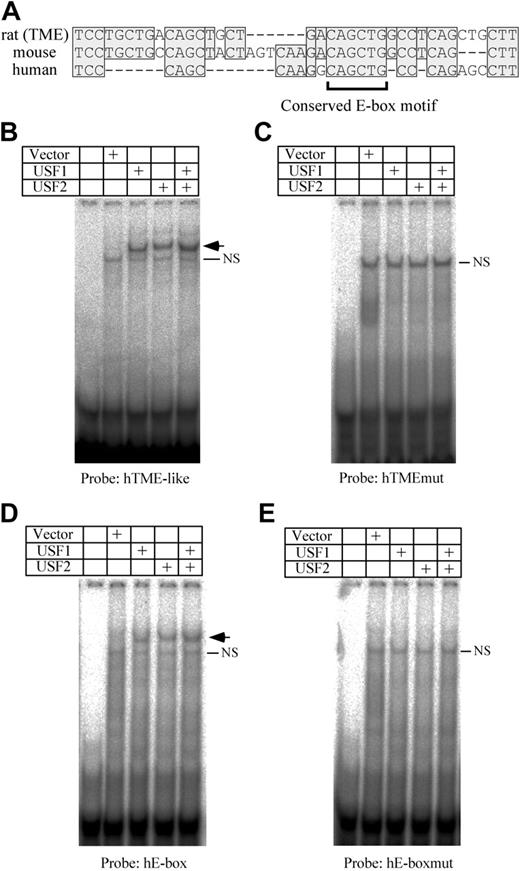
The TME is located immediately upstream of the T-cluster region in the rat PF4 promoter. We previously reported that TME-like sequences exist in both human and mouse promoters, and are located immediately upstream of each T-cluster region.11 By comparing the TME with the TME-like sequences, we found that one E-box motif is conserved among the species (Figure 5A). To investigate the bindings of USF1 and 2 to the human TME-like sequence (hTME-like), an EMSA was performed. USF1 and 2 bind to the hTME-like probe (Figure 5B). On the other hand, neither USF1 nor USF2 bind to the human TME-like mutant (hTMEmut) probe that is mutated in the E-box motif (Figure 5C). These results indicate that USF1 and 2 bind to the E-box motif in the hTME-like. In addition to the E-box in the hTME-like, another CAGCTG type E-box motif exists in the upstream region of the hTME-like (–430 to –425) of the human PF4 promoter. To investigate the bindings of USF1 and 2 to this E-box motif (hE-box), EMSA using the hE-box probe was performed. USF1 and 2 bound to the hE-box, but not to the hE-boxmut probe that is mutated in the E-box motif (Figure 5D-E). These results indicate that USF1 and 2 bind not only to the TME but also to the upstream E-box motif in the human PF4 promoter.
Analysis of USF1 and USF2 bindings to the human PF4 promoter. (A) Comparison of the rat TME promoter with the human and mouse TME-like sequences. Homologous sequences are indicated by shaded boxes. Dashes indicate deletion sequences. The bracket below the sequence indicates the conserved E-box motif. EMSA was performed using hTME-like (B), hTMEmut (C), hE-box (D), or hE-boxmut (E) probes. Arrows indicate the specific shifted bands. NS indicates a nonspecific band. The hTMEmut or hE-boxmut probes were prepared by mutating the E-box motif of the hTME or hE-box probe, respectively.
Analysis of USF1 and USF2 bindings to the human PF4 promoter. (A) Comparison of the rat TME promoter with the human and mouse TME-like sequences. Homologous sequences are indicated by shaded boxes. Dashes indicate deletion sequences. The bracket below the sequence indicates the conserved E-box motif. EMSA was performed using hTME-like (B), hTMEmut (C), hE-box (D), or hE-boxmut (E) probes. Arrows indicate the specific shifted bands. NS indicates a nonspecific band. The hTMEmut or hE-boxmut probes were prepared by mutating the E-box motif of the hTME or hE-box probe, respectively.
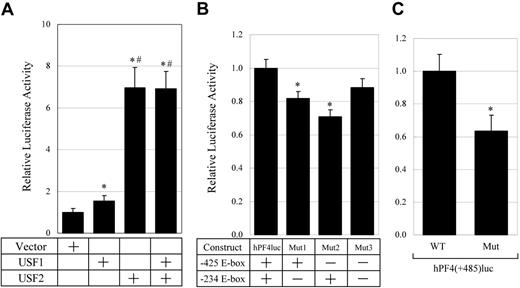
To investigate the effect of USF1 and 2 on the human PF4 promoter, transient transfection assays were performed. hPF4luc that contains 1.7 kb human PF4 promoter and expression vectors for USF1 or 2 were cotransfected into the HepG2 cells. The overexpression of each of USF1 or USF2 activated the human PF4 promoter 1.5- and 7-fold, respectively. In addition, the overexpression of both USF1 and 2 activated the PF4 promoter 7-fold. These data indicate that USF1 and 2 bind to the human PF4 promoter, and both USF1 and 2, especially USF2, strongly activate the human PF4 promoter.
To evaluate the function of the E-box motifs in megakaryocytic cells, hPF4mut1, 2, and 3 plasmids were prepared. These constructs contain mutations in each upstream (–425) or downstream (–234) E-box motif, or both E-box motifs. These plasmids were transfected into HEL cells and luciferase activities were evaluated after incubation with 10 nM PMA (Figure 6B). The mutations in either the upstream or downstream E-box motif significantly decreased the promoter activity by approximately 20% to 30%. This result indicates that both the upstream and downstream E-box motifs play a role in the activation of the human PF4 promoter. Furthermore, hPF4mut3 that was mutated in both E-box motifs decreased transcriptional activity by approximately 10%. To confirm the significance of this result, further transfection assays were performed using hPF4(+485)luc and hPF4(+485)Mut that contain the extra 3′ region of the human PF4 promoter (Figure 6C). This result indicated that the mutations in both the E-box motifs decreased transcriptional activity by approximately 40%. Taken together, these data suggest that the bindings of USF1 and 2 to the E-box motifs play a significant role in the activation of the human PF4 promoter as well as the rat PF4 promoter.
Activation of the human PF4 promoter by USF1 and USF2. (A) Transient transfection assays were performed with hPF4luc and transcription factor expression vectors. hPF4luc containing 1.7 kb of 5′ flanking region of human PF4 gene in front of the luciferase gene, and 1 μg of pcDNA3 vector, either expressing USF1 or USF2 or both, were cotransfected into HepG2 cells. Relative luciferase activities were measured. The columns and vertical bars represent mean ± SE of 6 replicates. *A significant difference (P < .05) between control and USF1, USF2, and USF1 + USF2. #A significant difference (P < .05) between USF1 and USF2 and USF1 + USF2. (B) Contribution of the E-box motifs in the human PF4 promoter in HEL cells. hPF4mut1, hPF4mut2, and hPF4mut3 contain mutations in the downstream (–234), upstream (–425), or both E-box motifs in hPF4luc. After these plasmids and hPF4luc were transfected into HEL cells, cells were incubated with the medium containing 10 nM PMA, and relative luciferase activities were measured. The columns and vertical bars represent mean ± SE of 12 replicates. *A significant difference (P < .05). (C) The function of E-box motifs in the human PF4 promoter. The same procedure was used as shown in panel B using hPF4(+485)luc and hPF4(+485)Mut. Each construct contains the downstream region from the transcriptional start site in the human PF4 gene. hPF4 (+485)Mut contains mutations in 2 E-box motifs. The columns and vertical bars represent mean ± SE of 6 replicates. *A significant difference (P < .05).
Activation of the human PF4 promoter by USF1 and USF2. (A) Transient transfection assays were performed with hPF4luc and transcription factor expression vectors. hPF4luc containing 1.7 kb of 5′ flanking region of human PF4 gene in front of the luciferase gene, and 1 μg of pcDNA3 vector, either expressing USF1 or USF2 or both, were cotransfected into HepG2 cells. Relative luciferase activities were measured. The columns and vertical bars represent mean ± SE of 6 replicates. *A significant difference (P < .05) between control and USF1, USF2, and USF1 + USF2. #A significant difference (P < .05) between USF1 and USF2 and USF1 + USF2. (B) Contribution of the E-box motifs in the human PF4 promoter in HEL cells. hPF4mut1, hPF4mut2, and hPF4mut3 contain mutations in the downstream (–234), upstream (–425), or both E-box motifs in hPF4luc. After these plasmids and hPF4luc were transfected into HEL cells, cells were incubated with the medium containing 10 nM PMA, and relative luciferase activities were measured. The columns and vertical bars represent mean ± SE of 12 replicates. *A significant difference (P < .05). (C) The function of E-box motifs in the human PF4 promoter. The same procedure was used as shown in panel B using hPF4(+485)luc and hPF4(+485)Mut. Each construct contains the downstream region from the transcriptional start site in the human PF4 gene. hPF4 (+485)Mut contains mutations in 2 E-box motifs. The columns and vertical bars represent mean ± SE of 6 replicates. *A significant difference (P < .05).
USF1 and 2 are expressed during human megakaryocytopoiesis
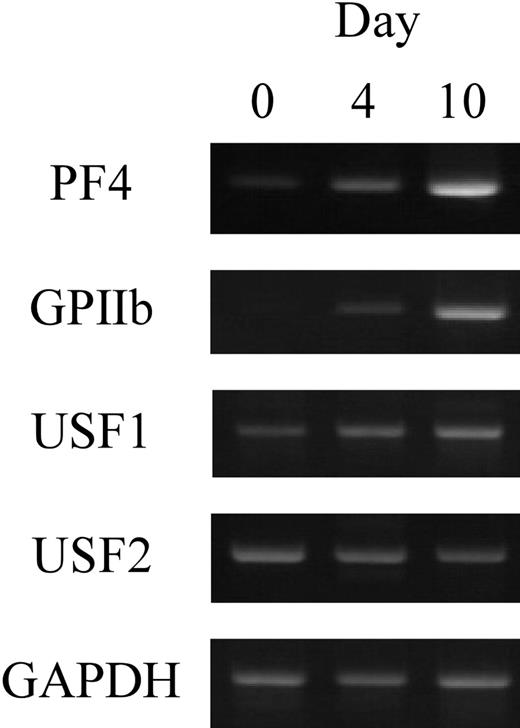
To investigate the expression of USF1 and 2 in human megakaryocytes, RT-PCR was performed. Human CD34+ cells prepared from cord blood were differentiated into megakaryocytes with TPO. After differentiation for 0, 4, or 10 days, the cultured cells were collected and total RNAs were isolated. RT-PCR was performed with the specific primers for PF4, GPIIb, USF1, or USF2 genes (Figure 7). The results show that USF1 and USF2 genes are expressed in the differentiated cells. These data indicate that USF1 and 2 are physiologically expressed in human megakaryocytes.
The expression of USF1 and USF2 during megakaryocytic differentiation. The expression levels of USF1 and USF2 genes in human megakaryocytes were evaluated by RT-PCR. Purified CD34+ cells were differentiated by TPO. The cultured cells were collected after 0, 4, or 10 days of differentiation, and total RNAs were isolated. RT-PCR was performed with specific primers for USF1 and USF2, PF4, and GPIIb.
The expression of USF1 and USF2 during megakaryocytic differentiation. The expression levels of USF1 and USF2 genes in human megakaryocytes were evaluated by RT-PCR. Purified CD34+ cells were differentiated by TPO. The cultured cells were collected after 0, 4, or 10 days of differentiation, and total RNAs were isolated. RT-PCR was performed with specific primers for USF1 and USF2, PF4, and GPIIb.
Discussion
Megakaryocytic gene expression is not well understood because an insufficient number of transcription factors that work in megakaryocytes have been identified. Through the analysis of the rat PF4 promoter, we identified the TME that contains a variety of binding motifs of transcription factors.11 We further studied this element and identified the homeodomain proteins (MEIS1, PREP1, and PBXs) as TME binding proteins.11,12 In addition to the binding motifs of homeodomain proteins, the TME contains 3 E-box motifs. Many E-box binding factors regulate cell differentiation. TAL-1 is an E-box binding transcription factor that forms a heterodimer with E2A and is essential for the early stage of hematopoiesis.13,14,36 TAL-1 is expressed during erythroid/megakaryocytic differentiation.15 Enforced TAL-1 expression stimulates the differentiation of primitive, erythroid, and megakaryocytic progenitors.16 These reports suggest that TAL-1 is important for megakaryocytic differentiation and that TAL-1 might bind to the E-box motifs in the TME. Therefore, in a previous report, we investigated whether TAL-1 was included in the TME affinity-purified fractions. Although TAL-1 is expressed in HEL cells, it is not included in these fractions.11,37 Because other E-box binding proteins have not yet been identified as regulators of megakaryocytic gene expression, we had no other candidate for TME binding proteins. We, therefore, tried to identify the TME-binding proteins using the LC-MS/MS system. At first, we isolated TME binding proteins by performing several steps of DNA affinity column purification. However, the TME binding activity decreased with every purification step. We speculated that a binding complex constructed of several factors was important for effective TME binding. Therefore, we next tried to determine all proteins purified by one-step affinity column chromatography. Although it is difficult to obtain high purity by the one-step DNA affinity column chromatography, it is useful for obtaining proteins in binding complexes, in this case including both direct and indirect binding proteins to the TME. By this method, we successfully identified many proteins in the TME binding complex, including PBXs that we had already identified as TME binding proteins. Furthermore, USF2, which is an E-box binding protein, was included among the identified proteins. Because USF2 regulates gene expression by forming a heterodimer with USF1, itself a homolog protein of USF2, bindings of both USF1 and 2 to the TME were investigated. Unexpectedly, only a tiny amount of USF1 was detected by Western blotting and could hardly be detected in EMSA using the nuclear extracts of HEL cells (Figure 1). However, EMSA (using USF1 prepared by in vitro translation) and reporter assays (dependent on the TME) show that USF1 binds to the TME and activates the rat PF4 promoter (Figures 2, 3). The result of the ChIP assay also indicates that USF1 physiologically binds to the TME in megakaryocytes (Figure 4). From these data, we conclude that USF1 and USF2 regulate rat PF4 gene expression and that USF2 more effectively contributes to PF4 promoter activation than USF1.
Next, we compared the rat PF4 promoter sequence with the human and mouse PF4 promoters (Figure 5A). One E-box motif is conserved in both human and mouse TME-like sequences. It was hypothesized that USF1 and 2 and the conserved E-box motif might be important for the regulation of the PF4 gene expression because USF1 and 2 also activate the erythroid-specific genes, human β-globin and glycophorin B, through the E-box motifs near the transcription start sites.38,39 To evaluate the effects of USF1 and 2 on the human PF4 promoter, an overexpression assay was performed (Figure 6A). The results from this indicated that USF1 and 2 activated the human PF4 promoter in HepG2 cells. In addition to this, we performed a transient transfection assay using the mutated human PF4 promoter (Figure 6B). Interestingly, we found that not only the conserved downstream E-box motif but also the upstream E-box motif contributed to the activation of human PF4 promoter. The significant effect of the E-box motifs was also detected with hPF4(+485)luc (Figure 6C). All of these results suggest that USF1 and 2, especially USF2, strongly activate the human PF4 promoter through the E-box motifs.
In the overexpression assays with HepG2 cells using the rat and human PF4 promoter that are mutated in the E-box motifs, we did not detect complete suppression of the increased activity derived from overexpressed USF1 and 2 (data not shown). We speculate that a protein complex that contains USF1 and 2 can be recruited to the E-box motif and activate the PF4 promoter without binding strongly to the individual E-box motifs. It has been demonstrated that USF1 and 2 interact with basal transcription factors.40-43 These reports support a part of our hypothesis and suggest the necessity to investigate the binding and function of a more complete complex on the PF4 promoter. Through the investigation of TME binding proteins, we have identified many candidates that may regulate PF4 gene expression. Further investigation of the interaction and synergistic functions of these proteins will contribute to understanding of the whole mechanism of PF4 and possibly other megakaryocytic gene expression.
It was recently reported that USF1 and 2 regulate HOXB4 gene expression, and TPO enhances USF1 and 2 bindings to the HOXB4 promoter in a p38 mitogen-activated protein kinase–dependent manner in a human megakaryocytic cell line.44,45 In addition, these reports suggest that USF1 and 2 are downstream factors of TPO signaling. Our study demonstrates that the PF4 gene is a downstream target gene of TPO signaling (Figure 7) and that USF1 and 2 regulate PF4 gene expression. Furthermore, we confirmed that USF1 and 2 bind to the TME in megakaryocytes differentiated by TPO (Figure 4). From these data, we speculate that TPO signaling enhances USF1 and 2 bindings to the TME and activates the PF4 promoter without changing the mRNA expression levels of USF1 and 2 (Figure 7). Therefore, USF1 and 2 may function as downstream transducers of TPO signaling in the regulation of PF4 gene expression. We suggest that this function might be a megakaryocyte-specific function of USF1 and 2, and are now studying this point.
Prepublished online as Blood First Edition Paper, June 8, 2004; DOI 10.1182/blood-2003-09-3107.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Takahiro Sato and Ivo G. Schoots for the useful suggestions. We also thank Kirin Brewery for the gift of thrombopoietin.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal