Abstract
Novel agents to treat acute myeloid leukemia (AML) are needed with increased efficacy and specificity. We have synthesized a dual-specificity fusion toxin DTU2GMCSF composed of the catalytic and translocation domains of diphtheria toxin (DT) fused to the granulocyte-macrophage colony-stimulating factor (GM-CSF) in which the DT furin cleavage site 163RVRRSV170 is modified to a urokinase plasminogen activator (uPA) cleavage site 163GSGRSA170, termed U2. DTU2GMCSF was highly toxic to the TF1-vRaf AML cell line (proliferation inhibition assay; IC50 = 3.14 pM), and this toxicity was greatly inhibited following pretreatment with anti-uPA and anti-GM-CSF antibodies. The activity of this toxin was then tested on a larger group of 13 human AML cell lines; 5 of the 13 cell lines were sensitive to DTU2GMCSF. An additional 5 of the 13 cell lines became sensitive when exogenous pro-uPA was added. Sensitivity to DTU2GMCSF strongly correlated with the expression levels of uPA receptors (uPARs) and GM-CSF receptors (GM-CSFRs) as well as with total uPA levels. DTU2GMCSF was less toxic to normal cells expressing uPAR or GMCSFR alone, that is, human umbilical vein endothelial cells and peripheral macrophages, respectively. These results indicate that DTU2GMCSF may be a selective and potent agent for the treatment of patients with AML. (Blood. 2004;104:2143-2148)
Introduction
Combination induction and consolidation chemotherapy yields complete remissions in a high proportion of patients with acute myeloid leukemia (AML). However, the majority of patients eventually have a relapse due to the persistence of chemotherapy-resistant blasts.1 Because most chemotherapy drugs cause DNA damage or block cell proliferation, we sought to target refractory AML blasts with agents that use different molecular cytotoxicity mechanisms. One such approach consists of targeting the protein synthesis inhibitor diphtheria toxin (DT) to the AML cells.
DT is composed of a cell-binding domain, a translocation domain, a furin-sensitive loop, and a catalytic domain.2 Recombinant DT molecules have been prepared by replacing the cell-binding domain of DT with tumor-specific peptide ligands.3 Our laboratory has focused on the development of recombinant DT molecules for the treatment of AML.
We and others first synthesized DT388GMCSF consisting of the catalytic and translocation domains of diphtheria toxin (DT388) fused to human granulocyte-macrophage colony-stimulating factor (GM-CSF).4,5 DT388GMCSF was selectively toxic to leukemic blasts in vitro, efficacious in animal models, and active in patients.6-8 Remissions were observed in 10% of patients, but liver injury secondary to toxicity to normal GM-CSF receptor (GM-CSFR)-expressing cells (eg, liver macrophages) limited dose escalation.
To enhance the specificity of DT388GMCSF, we sought to find additional cell surface AML-specific molecules absent from normal tissues expressing GM-CSFR. Liu et al9 pioneered redirecting toxins by modifying the furin cleavage site in the toxin to a tumor-selective urokinase plasminogen activator (uPA) cleavage sequence. uPA is a serine protease that cleaves plasminogen into its active form, plasmin. uPA is synthesized and released as pro-uPA, a single-chain inactive form, which is then cleaved into the double-chain active uPA by plasmin and other proteases. Active uPA binds to its receptor (uPAR), forming a potent uPA/uPAR extracellular serine protease system. The uPA/uPAR system is overexpressed in a variety of tumors including leukemias.10,11 AML blasts overexpress uPA and uPAR,12 whereas most normal tissues have no or little uPA and uPAR expression, except when they are up-regulated during certain physiologic processes such as wound healing and extracellular matrix remodeling.10,11 Consequently, we chose to enhance DT388GMCSF specificity to AML by changing its furin cleavage region (163RVRRSV170) to a uPA cleavage sequence (163GSGRSA170), yielding DTU2GMCSF.
In this study, we describe the yield and the biologic properties of DTU2GMCSF. In particular, we studied the specificity, range, and potency of this new AML recombinant toxin.
Materials and methods
DTU2GMCSF construction
To construct the expression plasmid coding for uPA-activated DTGMCSF, a mutagenic DNA fragment was amplified using 5′ T7 promoter primer (TAATACGACTCACTATAG), 3′ mutagenic primer U2 (GATTTATGCATGACAATGAGCTACCTGCTGATCTTCCACTTCCATTTCCTGCACAGGCTTG; NsiI site is in boldface, the antisense sequence coding for uPA substrate peptide GSGRSA is underlined), and the DT388GMCSF expression plasmid pRKDTGM as a template.4 The PCR product was digested with XbaI and NsiI, cloned into the XbaI-NsiI sites of pRKDTGM, yielding DTU2GMCSF expression plasmid pRKDTGM-U2.
DTU2GMCSF expression and purification
Expression and purification of DTU2GMCSF was done according to the same strategy we used for the expression and purification of DT388GMCSF.4 Briefly, pRKDTGM-U2 was used to transform BL21 (DE3) Escherichia coli harboring the pUBS500 plasmid. Transformants were grown in Superbroth and induced with isopropylthiolgalactoside (IPTG). Inclusion bodies were isolated, washed, and denatured in guanidine hydrochloride with dithioerythritol. Recombinant protein was refolded by diluting 100-fold in cold buffer with arginine and oxidized glutathione. After dialysis, purified protein was obtained after anion-exchange, size exclusion on fast protein liquid chromatography (FPLC), and polymixin B affinity chromatography. After refolding, the production yield was 16 mg/L bacterial culture, and the purity was more than 95% as determined by Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Other toxins used
Cells and cell lines
Human AML cell lines HL60, U937, ML1, ML2, ML17, Monomac 1, Monomac 6, CTV-1, KG-1, Sig M5, and TF1v-Src were grown as we previously described.11 The 2 other human AML cell lines used in this study, TF1-vRaf and TF1-vSrc, were cultured as previously described.13 Human umbilical vein endothelial cells (HUVECs) were purchased from American Type Culture Collection (Manassas, VA) and cultured according to the directions provided. Peripheral monocytes were obtained and isolated from healthy adult individuals with informed consent under a protocol approved by the Wake Forest University Institutional Review Board.14
Cell line sensitivity to DTU2GMCSF
Aliquots of 104 cells were incubated in 100 μL medium (same as that used to grow the cells) in Costar (Corning, NY) 96-well flat-bottomed plates in duplicate. Exogenous human pro-uPA (American Diagnostica, Stamford, CT) was added to 36 wells of each duplicate plate (100 ng/mL in each well). Then, 50 μL DTU2GMCSF in medium was added to each column to yield concentrations ranging from 0.1 to 10 000 pM, and the cells were incubated at 37°C/5% CO2 for 50 hours. Then, 1 μCi (0.037 MBq) 3H-thymidine (NEN DuPont, Boston, MA) in 50 μL medium was added to each well, and incubation continued for an additional 18 hours at 37°C/5% CO2. Cells were then harvested using a Skatron Cell Harvester (Skatron Instruments, Lier, Norway) onto glass fiber mats and counts per minute (cpm) of incorporated radiolabel were counted using an LKB-Wallac 1205 Betaplate liquid scintillation counter (Perkin-Elmer, Gaithersburg, MD) gated for 3H. The concentration of toxin that inhibited thymidine incorporation by 50% compared to control wells (IC50). The percent maximal 3H-thymidine incorporation was plotted versus the log of the toxin concentration, and nonlinear regression with a variable slope sigmoidal dose-response curve was generated along with IC50 using GraphPad Prism software (GraphPad Software, San Diego, CA). All assays were performed at least twice with an interassay range of 30% or less for IC50.
Monocyte and HUVEC sensitivity to DTU2GMCSF
Normal cell sensitivity to DTU2GMCSF was determined using the same cytotoxicity assay described (see “Cell line sensitivity to DTU2GMCSF”) with the following differences. Both normal monocytes and HUVECs were plated 24 hours prior to incubation with DTGMCSF, DTU2GMCSF, and DTAT (only for HUVECs). Exogenous pro-uPA was not added in these assays. Cells were incubated with the toxins for 5 hours in the case of peripheral monocytes and 48 hours for HUVECs. The efficacy of DTU2GMCSF, DTGMCSF, and DTAT on HUVECs was determined by 3H-thymidine incorporation inhibition assay; 50 μL of a 10 μCi (0.37 MBq)/mL 3H-thymidine solution was added to each well, and the cells were harvested 24 hours after the addition of 3H-thymidine. The efficacy of DTU2GMCSF and DTGMCSF was determined by 3H-leucine incorporation inhibition for normal monocytes; 100 μL of a 20 μCi (0.74 MBq)/mL 3H-leucine solution (NEN DuPont) was added to each well, and the cells were harvested 6 hours following the addition of 3H-leucine.
GM-CSFR expression
GM-CSFR expression levels were determined in all cell lines as described previously.15 In short, aliquots of 3 to 5 × 106 cells were mixed with varying amounts of 125I Bolton-Hunter-labeled human GM-CSF (80-120 μCi [2.96-4.44 MBq]/μg, NEX249; NEN DuPont, Boston, MA) with or without excess (1800g [1500 ng]) cold GM-CSF (Immunex, Seattle, WA). Cells were incubated at 37°C for 1 hour and then layered over a 200 μL oil phthalate mixture. After centrifugation at 12 000 rpm for 1 minute in a microfuge at room temperature, both pellets and supernatants were saved and counted in an LKB-Wallac 1260 multigamma counter (Turku, Finland) gated for 125I with 50% counting efficiency. Background counts per minute were calculated by linear extrapolation from incubations with excess cold GM-CSF. Experiments were performed in duplicate. Receptor number/cell (Bmax) as well as dissociation constant (Kd) were calculated using the GraphPad Prism software.
uPAR expression
The uPAR expression levels were determined following the same protocol used for the determination of GMCSFR levels.14 The amino-terminal fragment (ATF) of uPA (amino acids 1-133) was a generous gift from Dr Michael Ploug. 125I labeling of the ATF was done in Iodogen-coated tubes; 8 μg ATF was incubated with 0.5 mCi (18.5 MBq) 125I and 10 μL binding buffer for 15 minutes. The reaction was then stopped with the addition of excess amounts of binding buffer and 10 μL of the reaction mixture was added to 900 μL radioimmunoassay (RIA) buffer and 100 μL tricarboxylic acid (TCA). The mixture was then centrifuged at 2000 rpm for 10 minutes, and the percentage of labeled ATF as well as the specific activity of labeled ATF was calculated by determining the amount of radioactivity in the pellet versus the amount of radioactivity in the supernatant. 125I-labeled ATF was separated from free 125I on a desalting column. Calculations and analysis were done using the GraphPad Prism software.
uPA and PAI-1 levels
The uPA and plasminogen activator inhibitor 1 (PAI-1) levels were determined using enzyme-linked immunosorbent assay (ELISA) kits (American Diagnostica), and the assays were done according to the description provided in each assay kit. For each cell line 100 μL cell culture supernatant (cell density > 106 cells/mL) was used, in duplicate, in each ELISA. Both these kits detect total levels of uPA and PAI-1.
Blocking assays
Blocking assays were performed to test the ability of specific anti-GM-CSF and anti-uPA antibodies to block the killing of the sensitive cell lines by DTU2GMCSF. The same thymidine incorporation inhibition assays were performed as described (see “Cell line sensitivity to DTU2GMCSF”) but without the addition of pro-uPA. In the blocking assays, 10 μg/mL monoclonal anti-uPA antibody (American Diagnostica) or 2 μg/mL monoclonal anti-GM-CSF antibody (Oncogene Research, San Diego, CA) was added to the cells 2 hours before the addition of DTU2GMCSF. The rest of the assay proceeded as described.
Results
DTU2GMCSF expression and purification
Recombinant protein was expressed in E coli, purified from inclusion bodies with a yield of 16 mg/L bacterial culture and a purity of more than 95% by Coomassie-stained SDS-PAGE. DTU2GMCSF is refolded as described previously for the refolding of DT388GMCSF.4 The percent of properly folded protein recovered from the inclusion bodies is about 25%.
DTU2GMCSF cytotoxicity
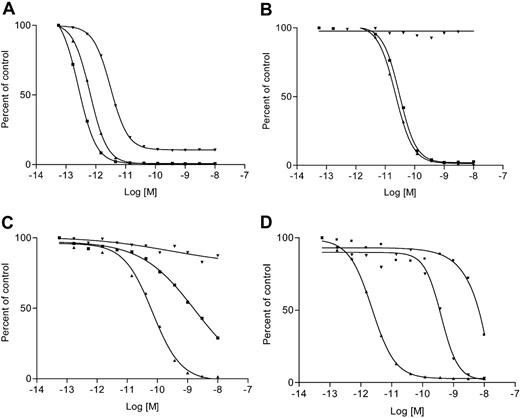
To determine the biologic activity of DTU2GMCSF, we tested the cytotoxicity of this fusion protein on a panel of 13 AML cell lines (Table 1). Tritiated thymidine incorporation inhibition assays were done using DTU2GMCSF with or without the addition of exogenous pro-uPA as well as using DT388GMCSF. All cell lines tested had a high sensitivity to DT388GMCSF except CTV-1, which had an IC50 for DT388GMCSF more than 4000 pM. This cell line was also not sensitive to DTU2GMCSF with or without the addition of exogenous pro-uPA. Five of 13 cell lines, namely, U937, TF1-vRaf, KG-1, HL60, and ML2, were highly sensitive to DTU2GMCSF without the addition of exogenous pro-uPA (IC50 = 3.14-34.7 pM; Table 1; Figure 1A). When these cells where coincubated with DTU2GMCSF and exogenous pro-uPA, the cytotoxicity of DTU2GMCSF significantly increased and became similar to that of DT388GMCSF (IC50 = 2.6-3.5 pM). Another 5 of 13 cell lines, namely, TF1-vSrc, TF1-HaRas, Sig M5, ML1, and ML17, were not sensitive to DTU2GMCSF. These cell lines, however, when coincubated with both DTU2GMCSF and exogenous pro-uPA had significantly increased sensitivity to DTU2GMCSF (IC50 = 0.55-42 pM), becoming similar to the sensitivity of these cells to DT388GMCSF (Table 1; Figure 1B). The remaining 3 cell lines, Monomac 1, Monomac 6, and CTV-1, were not sensitive to DTU2GMCSF even when exogenous pro-uPA was added. Monomac 1 and Monomac 6 were sensitive to DT388GMCSF (IC50 = 3.2 and 68.3 pM, respectively), whereas CTV-1 was not sensitive even to DT388GMCSF (Table 1; Figure 1C).
AML cell line sensitivity to DTU2GMCSF, DTU2GMCSF plus pro-uPA, and DT388GMCSF as determined by 3H-thymidine incorporation inhibition assays
Cell line . | DTU2GMCSF, IC50, pM . | DTU2GMCSF + pro-uPA, IC50, pM . | DT388GMCSF, IC50, pM . |
|---|---|---|---|
| U937 | 34.7 | 0.48 | 0.47 |
| TF1-vRaf | 3.14 | 0.26 | 0.64 |
| KG-1 | 9.8 | 3.5 | 2.5 |
| HL60 | 7.8 | 1.5 | 2.7 |
| ML2 | 5.8 | 0.76 | 1.9 |
| TF1-vSrc | >4000 | 0.55 | 0.84 |
| TF1-HaRas | >4000 | 0.67 | 0.27 |
| Sig M5 | >4000 | 42 | 21.1 |
| ML1 | >4000 | 30 | 22 |
| ML17 | >4000 | 4.5 | 3.1 |
| Monomac 6 | >4000 | 1730 | 68.3 |
| Monomac 1 | >4000 | >4000 | 3.2 |
| CTV-1 | >4000 | >4000 | >4000 |
Cell line . | DTU2GMCSF, IC50, pM . | DTU2GMCSF + pro-uPA, IC50, pM . | DT388GMCSF, IC50, pM . |
|---|---|---|---|
| U937 | 34.7 | 0.48 | 0.47 |
| TF1-vRaf | 3.14 | 0.26 | 0.64 |
| KG-1 | 9.8 | 3.5 | 2.5 |
| HL60 | 7.8 | 1.5 | 2.7 |
| ML2 | 5.8 | 0.76 | 1.9 |
| TF1-vSrc | >4000 | 0.55 | 0.84 |
| TF1-HaRas | >4000 | 0.67 | 0.27 |
| Sig M5 | >4000 | 42 | 21.1 |
| ML1 | >4000 | 30 | 22 |
| ML17 | >4000 | 4.5 | 3.1 |
| Monomac 6 | >4000 | 1730 | 68.3 |
| Monomac 1 | >4000 | >4000 | 3.2 |
| CTV-1 | >4000 | >4000 | >4000 |
Proliferation inhibition assays on 3 AML cell lines. Results of inhibition assays are shown for TF1-vRaf (A), ML1 (B), and Monomac 6 (C) using DT388GMCSF (▴), DTU2GMCSF (▾), and DTU2GMCSF plus exogenous pro-uPA (▪). The x-axis represents the log of the molar drug concentration and the y-axis represents cell viability expressed as percent control of 3H-thymidine incorporation in counts per minute. TF1-vRaf w as sensitive to DTU2GMCSF (IC50 = 3.14 pM); the sensitivity was enhanced by the addition of exogenous pro-uPA (IC50 = 0.26 pM) and became similar to that of DT388GMCSF (IC50 = 0.64 pM) (A). ML1 was not sensitive to DTU2GMCSF unless exogenous pro-uPA was added (IC50 = 30 pM). The IC50 for DT388GMCSF was 22 pM (B). Monomac 6 was not sensitive to DTU2GMCSF even when exogenous pro-uPA was added (C). (D) Blocking assay. The proliferation inhibition assay on TF1-vRaf with DTU2GMCSF (▴), DTU2GMCSF plus anti-uPA (▪), and DTU2GMCF plus anti-GM-CSF (▾) is shown. On the x-axis is the log molar drug concentration, on the y-axis is the percentage of control 3H-thymidine incorporation. Both anti-GM-CSF and anti-uPA greatly decreased DTU2GMCSF efficacy (IC50 = 400 pM and 0.67 μM, respectively) compared to an IC50 = 2.3 pM for DTU2GMCSF alone, thus demonstrating the dual specificity of DTU2GMCSF, which requires the expression of both GM-CSFR and the uPA/uPAR protease system.
Proliferation inhibition assays on 3 AML cell lines. Results of inhibition assays are shown for TF1-vRaf (A), ML1 (B), and Monomac 6 (C) using DT388GMCSF (▴), DTU2GMCSF (▾), and DTU2GMCSF plus exogenous pro-uPA (▪). The x-axis represents the log of the molar drug concentration and the y-axis represents cell viability expressed as percent control of 3H-thymidine incorporation in counts per minute. TF1-vRaf w as sensitive to DTU2GMCSF (IC50 = 3.14 pM); the sensitivity was enhanced by the addition of exogenous pro-uPA (IC50 = 0.26 pM) and became similar to that of DT388GMCSF (IC50 = 0.64 pM) (A). ML1 was not sensitive to DTU2GMCSF unless exogenous pro-uPA was added (IC50 = 30 pM). The IC50 for DT388GMCSF was 22 pM (B). Monomac 6 was not sensitive to DTU2GMCSF even when exogenous pro-uPA was added (C). (D) Blocking assay. The proliferation inhibition assay on TF1-vRaf with DTU2GMCSF (▴), DTU2GMCSF plus anti-uPA (▪), and DTU2GMCF plus anti-GM-CSF (▾) is shown. On the x-axis is the log molar drug concentration, on the y-axis is the percentage of control 3H-thymidine incorporation. Both anti-GM-CSF and anti-uPA greatly decreased DTU2GMCSF efficacy (IC50 = 400 pM and 0.67 μM, respectively) compared to an IC50 = 2.3 pM for DTU2GMCSF alone, thus demonstrating the dual specificity of DTU2GMCSF, which requires the expression of both GM-CSFR and the uPA/uPAR protease system.
Inhibition assays
To show the absolute requirement for the presence of GM-CSFR and an active uPA/uPAR protease system for DTU2GMCSF toxicity, we inhibited each of these components separately and looked at the effects of these inhibitions on the efficiency of DTU2GMCSF. We inhibited the uPA/uPAR system by preincubating TF1-vRaf cells with a specific monoclonal antibody directed against an epitope close to the active site of double-chain uPA. This antibody binds and inactivates all forms of uPA (single-chain pro-uPA and double-chain uPA), and it inhibits the proteolytic activity of active double-chain uPA even in its uPAR-bound form. Incubation of the cells with 10 μg/mL anti-uPA antibody increased the IC50 of DTU2GMCSF by 106-fold (IC50 = 0.62 μM) as compared to DTU2GMCSF alone (IC50 = 16.6 pM; Figure 1D). We separately inhibited the binding of DTU2GMCSF to its GM-CSFR by cotreating cells with an anti-GM-CSF monoclonal antibody that targets an epitope near the GM-CSFR-binding site on GM-CSF and subsequently blocks binding of GM-CSF to its receptor. Incubation of the cells with 2 μg/mL anti-GM-CSF increased the IC50 of DTU2GMCSF by 200-fold (IC50 = 400 pM) as compared to the IC50 of DTU2GMCSF alone (2.3 pM; Figure 1D).
Receptor levels
To determine the minimal expression levels of GM-CSFR and uPAR needed for DTU2GMCSF to be active, we determined the expression levels of these 2 receptors in the 13 AML cell lines tested for cytotoxicity and correlated these expression levels to the sensitivity of each cell line for DTU2GMCSF. GM-CSFR levels varied between 108 receptors/cell for CTV-1 and 11 430 receptors/cell for TF1-HaRas (Figure 2A). The low GM-CSFR-expressing CTV-1 cells were resistant to both DT388GMCSF and DTU2GMCSF. GM-CSFR density correlated with the sensitivity of each cell line to DT388GMCSF (Table 2). Among the 12 cell lines that were sensitive to DT388GMCSF, Sig M5 had the lowest number of GM-CSFRs (236 receptors/cell). Therefore, the minimum number of GM-CSFRs required for a cell to be sensitive to DT388GMCSF in our survey was 236 receptors/cell. The 2 cell lines that were sensitive to DT388GMCSF but not sensitive to DTU2GMCSF had very low levels of uPAR expression. uPAR levels in these cell lines were 33 receptors/cell for Monomac 1 and 45 receptors/cell for Monomac 6. On the other hand, the 10 cell lines that were sensitive to DTU2GMCSF with or without the addition of pro-uPA had high levels of uPAR expression. uPAR levels varied between 387 and 1848 receptors/cell for the 5 cell lines that were sensitive to DTU2GMCSF alone (Table 3). Receptor levels varied between 54 and 991 receptors/cell for the 5 cell lines that were sensitive to DTU2GMCSF only when pro-uPA was added (Figure 2B-C; Table 3). The sensitive cell line with the lowest uPAR expression levels (ML17) had 54 receptors/cell and was sensitive to DTU2GMCSF when excess uPA was added. Therefore, the minimum expression levels of uPAR required for the cells to be sensitive to DTU2GMCSF in our survey was 54 receptors/cell. uPAR expression levels in the DTU2GMCSF-resistant cell lines were significantly lower than expression levels in all other cells lines. On the other hand, uPAR expression levels in cell lines sensitive to DTU2GMCSF did not distinguish cell lines requiring the addition of exogenous pro-uPA from those not requiring exogenous pro-uPA
uPAR levels as determined by 125I-ATF receptor-binding assay. (A) HL60 GM-CSFR levels were determined by 125I-GM-CSF receptor-binding assay. (B) uPAR expression levels of Sig M5. The x-axis represents the concentration of 125I-ATF (pg/mL); the y-axis represents the amount of cell-bound 125I-ATF (cpm). (C) uPAR levels expressed as number of receptors/cell in the 13 AML cell lines. DTU2GMCSF-resistant cells (black) have lower uPAR expression levels than DTU2GMCSF-sensitive cells (white, without the addition of pro-uPA; gray, requiring the addition of pro-uPA).
uPAR levels as determined by 125I-ATF receptor-binding assay. (A) HL60 GM-CSFR levels were determined by 125I-GM-CSF receptor-binding assay. (B) uPAR expression levels of Sig M5. The x-axis represents the concentration of 125I-ATF (pg/mL); the y-axis represents the amount of cell-bound 125I-ATF (cpm). (C) uPAR levels expressed as number of receptors/cell in the 13 AML cell lines. DTU2GMCSF-resistant cells (black) have lower uPAR expression levels than DTU2GMCSF-sensitive cells (white, without the addition of pro-uPA; gray, requiring the addition of pro-uPA).
Comparison of DT388GMCSF efficacy (IC50) and GMCSFR expression levels
Cell line . | GMCSFR levels, receptors/cell . | DT388GMCSF, IC50, pM . |
|---|---|---|
| TF1-HaRas | 11 430 | 0.27 |
| U937 | 2 329 | 0.47 |
| TF1-vRaf | 7 624 | 0.64 |
| TF1-vSrc | 5 846 | 0.84 |
| ML2 | 1 209 | 1.9 |
| KG-1 | 869 | 2.5 |
| HL60 | 3 262 | 2.7 |
| ML 17 | 456 | 3.1 |
| Monomac 1 | 304 | 3.2 |
| Sig M5 | 236 | 21 |
| ML-1 | 358 | 22 |
| Monomac 6 | 712 | 68.3 |
| CTV-1 | 108 | >4000 |
Cell line . | GMCSFR levels, receptors/cell . | DT388GMCSF, IC50, pM . |
|---|---|---|
| TF1-HaRas | 11 430 | 0.27 |
| U937 | 2 329 | 0.47 |
| TF1-vRaf | 7 624 | 0.64 |
| TF1-vSrc | 5 846 | 0.84 |
| ML2 | 1 209 | 1.9 |
| KG-1 | 869 | 2.5 |
| HL60 | 3 262 | 2.7 |
| ML 17 | 456 | 3.1 |
| Monomac 1 | 304 | 3.2 |
| Sig M5 | 236 | 21 |
| ML-1 | 358 | 22 |
| Monomac 6 | 712 | 68.3 |
| CTV-1 | 108 | >4000 |
uPAR expression levels, total uPA levels, and DTU2GMCSF efficacy, with or without pro-uPA, on the 12 AML cell lines that were sensitive to DT388GMCSF
Cell line . | uPAR levels, no. receptors/ cell . | Total uPA levels, ng/mL . | DTU2GMCSF, IC50, pM . | DTU2GMCSF + pro-uPA, IC50, pM . |
|---|---|---|---|---|
| TF1-vRaf | 1848 | 0.116 | 3.14 | 0.26 |
| ML2 | 784 | 0.283 | 5.8 | 0.76 |
| U937 | 480 | 0.135 | 34.7 | 0.48 |
| KG-1 | 399 | 0.326 | 9.8 | 3.5 |
| HL60 | 387 | 0.489 | 7.8 | 1.5 |
| ML1 | 991 | 0.094 | >4000 | 30 |
| TF1-HaRas | 581 | 0.092 | >4000 | 0.67 |
| TF1-vSrc | 491 | 0.054 | >4000 | 0.55 |
| Sig M5 | 241 | 0.090 | >4000 | 42 |
| ML17 | 54 | 0.294 | >4000 | 4.5 |
| Monomac 6 | 45 | 0.559 | >4000 | 1730 |
| Monomac 1 | 33 | 0.931 | >4000 | >4000 |
| CTV-1* | 110 | 0.32 | >4000 | >4000 |
Cell line . | uPAR levels, no. receptors/ cell . | Total uPA levels, ng/mL . | DTU2GMCSF, IC50, pM . | DTU2GMCSF + pro-uPA, IC50, pM . |
|---|---|---|---|---|
| TF1-vRaf | 1848 | 0.116 | 3.14 | 0.26 |
| ML2 | 784 | 0.283 | 5.8 | 0.76 |
| U937 | 480 | 0.135 | 34.7 | 0.48 |
| KG-1 | 399 | 0.326 | 9.8 | 3.5 |
| HL60 | 387 | 0.489 | 7.8 | 1.5 |
| ML1 | 991 | 0.094 | >4000 | 30 |
| TF1-HaRas | 581 | 0.092 | >4000 | 0.67 |
| TF1-vSrc | 491 | 0.054 | >4000 | 0.55 |
| Sig M5 | 241 | 0.090 | >4000 | 42 |
| ML17 | 54 | 0.294 | >4000 | 4.5 |
| Monomac 6 | 45 | 0.559 | >4000 | 1730 |
| Monomac 1 | 33 | 0.931 | >4000 | >4000 |
| CTV-1* | 110 | 0.32 | >4000 | >4000 |
This cell line was not sensitive to DT388GMCSF because of low GM-CSFR expression levels and was not included in the correlation of DTU2GMCSF efficacy with uPAR expression levels and total uPA levels.
uPA and PAI-1 levels
We determined total uPA levels in cultures with densities comprised between 1.5 and 2 million cells/mL, using an ELISA kit that detects all forms of uPA, that is, inactive single-chain pro-uPA as well as double-chain active uPA. Total uPA levels in the 13 cell lines tested varied between 0.1 and 0.93 ng/mL (Table 3). The highest levels of total uPA were observed in the cell lines that are resistant to DTU2GMCSF; these levels were 0.93 ng/mL for Monomac 1, 0.55 ng/mL for Monomac 6, and 0.32 ng/mL for CTV-1. We do not have any explanation for why the 3 cell lines that were not sensitive to DTU2GMCSF had the highest total uPA levels. However, this result indicates that in the absence of adequate uPAR expression, cells were resistant to DTU2GMCSF even in the presence of relatively high levels of total uPA. Further, cells that were sensitive to DTU2GMCSF without the addition of pro-uPA had significantly higher total uPA levels than cell lines that needed the addition of exogenous pro-uPA to be sensitive (Figure 3A). PAI-1 levels varied between 0.018 and 0.903 ng/mL and did not correlate with the sensitivity of each cell line to DTU2GMCSF. Three cell lines, namely, HL60, Sig M5, and Monomac 1, did not have any detectable levels of PAI-1 (Figure 3B). However, this finding does not rule out the possibility of PAI-1 activity being a major regulator of DTU2GMCSF efficacy in vivo. Therefore, we were unable in this study to determine the role played by PAI-1 in the efficacy of DTU2GMCSF.
uPA and PAI-1 levels. (A) Average uPA levels (ng/mL) in the supernatant of each of the 3 categories of AML cell lines as determined by ELISA are shown. Nonsensitive cell lines (black) had the highest concentrations of total uPA in their supernatants, which indicates that DTU2GMCSF efficacy is determined by uPAR expression levels rather than uPA concentration. Cell lines that were sensitive to DTU2GMCSF only when exogenous pro-uPA was added (gray) had significantly lower total uPA levels in their supernatants as compared to cells that did not require the addition of exogenous pro-uPA (white; P = .04). (B) Total PAI-1 concentrations in the supernatants of the AML cell lines as determined by ELISA are shown. PAI-1 levels did not correlate with DTU2GMCSF efficacy.
uPA and PAI-1 levels. (A) Average uPA levels (ng/mL) in the supernatant of each of the 3 categories of AML cell lines as determined by ELISA are shown. Nonsensitive cell lines (black) had the highest concentrations of total uPA in their supernatants, which indicates that DTU2GMCSF efficacy is determined by uPAR expression levels rather than uPA concentration. Cell lines that were sensitive to DTU2GMCSF only when exogenous pro-uPA was added (gray) had significantly lower total uPA levels in their supernatants as compared to cells that did not require the addition of exogenous pro-uPA (white; P = .04). (B) Total PAI-1 concentrations in the supernatants of the AML cell lines as determined by ELISA are shown. PAI-1 levels did not correlate with DTU2GMCSF efficacy.
DTU2GMCSF toxicity to normal cells
To determine the toxicity of DTU2GMCSF to normal cells we tested the sensitivity of both peripheral monocytes and HUVECs to DTU2GMCSF and DTGMCSF. Normal peripheral monocytes, with GM-CSFR but low uPA/uPAR protease activity, were 103-fold less sensitive to DTU2GMCSF than to DT388GMCSF. HUVECs, cells that express uPAR but low or absent GM-CSFR, were resistant to both DTGMCSF and DTU2GMCSF but sensitive to DTAT (IC50 = 26.8 pM). Thus, DTU2GMCSF is a dual-specificity cytotoxin requiring the presence of GM-CSFR and an active uPA/uPAR protease system (Table 4).
DTU2GMCSF toxicity on normal cells
Cell type . | DTU2GMCSF, IC50, pM . | DT388GMCSF, IC50, pM . | DTAT, IC50, pM . |
|---|---|---|---|
| HUVECs | > 4000 | > 4000 | 26.8 |
| Human monocytes | 124 | 0.16 | — |
Cell type . | DTU2GMCSF, IC50, pM . | DT388GMCSF, IC50, pM . | DTAT, IC50, pM . |
|---|---|---|---|
| HUVECs | > 4000 | > 4000 | 26.8 |
| Human monocytes | 124 | 0.16 | — |
— indicates not tested.
Discussion
Our laboratory has focused on the development of DT fusion toxins for therapy of chemotherapy-resistant AML. The first generation of DT fusion toxins for AML have only a single targeting moiety—the ligand. Thus, our first AML fusion toxins (DT388GMCSF, DT388IL3, and DTAT) were potentially toxic to normal tissues bearing their respective receptors—GM-CSFR, interleukin 3 receptor (IL-3R), or uPAR.4,12,16 Clinical testing of the first AML fusion toxin DT388GMCSF confirmed clinical efficacy but with associated damage to GM-CSFR-containing normal cells. This led to significant liver injury. We chose to re-engineer fusion toxins to enhance specificity and reduce normal tissue toxicity. Based on the studies of Liu and colleagues9 with anthrax protective antigen, we reasoned that the AML specificity of DT388GMCSF could be increased by modifying the DT furin site to require an active AML-selective protease system. We and others have shown uPA and uPAR in patient AML blasts.12,17 Thus, we reasoned that by replacing DT 163RVRRSV170 with the uPA cleavage sequence 163GSGRSA170 in DT388GMCSF, the new AML fusion toxin DTU2GMCSF would retain potency but have enhanced AML specificity. The new dual-specificity fusion toxin DTU2GMCSF was produced in the same yields and purity as DT388GMCSF. The protein remained stable and biologically active after storage at -80°C for over 1 month (R.J.A.-H. and A.F., unpublished observations, February 2003). Further, DTU2GMCSF remained fairly stable after incubation with serum at 37°C for 48 hours (IC50 = 12.8 pM as compared to an IC50 = 6.3 pM for freshly thawed DTU2GMCSF in a tritiated thymidine incorporation inhibition assay on HL60 cells; data not shown).
Remarkably, DTU2GMCSF showed potency similar to DT388GMCSF when exogenous single-chain pro-uPA was added. This suggests that uPAR is expressed in adequate levels on most AML cell lines. The deficiency of uPA, in some cases, may be an artifact of in vitro culture or may occur during cell line generation. In vivo, there may be higher concentrations of uPA in the tumor environment.18 Even without exogenous pro-uPA, 5 (39%) of 13 cell lines were killed by DTU2GMCSF. Thus, a significant fraction of patients may have disease sensitive to this fusion toxin.
DTU2GMCSF is a dual-specificity fusion toxin. Blocking assays, normal tissue toxicities, and cell line receptor studies show that both uPA/uPAR and GM-CSFR are required for DTU2GMCSF toxicity. Several groups have made toxins targeting more than one tumor cell receptor.19,20 In most cases, the toxins had 2 ligands and thus were able to intoxicate cells with either ligand, for example, IL-13R and uPAR,19 and epidermal growth factor (EGF) receptor and erbB-2.20 Hence, although the new molecules were able to bind a larger range of tumor cells, they had less rather than more specificity.
We provide a proof of principle for the efficacy and increased specificity of dual-specificity fusion toxins. Further studies are needed to determine the in vivo efficacy and safety of DTU2GMCSF. Hopefully, this dual-specificity AML fusion toxin will be clinically beneficial to patients with chemotherapy-refractory AML. Similar approaches with other fusion toxins may provide opportunities to treat metastatic solid tumors. Many of the receptor targets for fusion toxins have limited specificity; hence, addition of a requisite toxin-processing step with tumor-selective proteases may yield safer active fusion toxins for cancer treatment.
Prepublished online as Blood First Edition Paper, May 25, 2004; DOI 10.1182/blood-2004-01-0339.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal