Abstract
Analysis of clonotypic immunoglobulin M (IgM) from 15 patients with Waldenstrom macroglobulinemia (WM) showed a strong preferential use of the VH3/JH4 gene families. Identification of the WM IgM V/D/J was validated using single-cell analysis, confirming its presence in most B cells. Despite the extensive hypermutated VH genes in 13 of 15 patients, statistical analysis of framework/complementary-determining region (FR/CDR) mutation patterns suggests that they might have escaped antigenic selection. Neither intraclonal diversity nor isotype switching was detectable. Membranous and secreted forms of clonotypic IgM transcripts were present in bone marrow and blood. Single-cell analysis showed that clonotypic B cells coexpress CD20, surface IgM (sIgM), and sIgD but that they lack CD138. Most B cells lacked memory marker CD27 despite their hypermutated variable regions otherwise suggestive of memory status. At diagnosis, circulating B cells in WM are largely clonotypic. However, when monoclonal IgM levels are decreased, clonotypic frequencies are substantially reduced despite elevated CD20+ cells, shown to be polyclonal by DNA sequencing and CDR3 fragment analysis. Thus, WM includes the expansion of circulating, polyclonal B cells. Overall, this work suggests that WM may originate from a largely VH3-restricted, somatically mutated, predominantly CD27-IgM+IgD+ population that cannot undergo class switching, suggestive of B cells that might have bypassed the germinal center. (Blood. 2004;104:2134-2142)
Introduction
Waldenstrom macroglobulinemia (WM) is characterized by lymphoplasmacytic lymphoma and immunoglobulin M (IgM) monoclonal gammopathy.1 It accounts for 2% of hematologic malignancies, with a median survival of 5 years.2 Approximately 50% of patients have predominant, well-differentiated lymphocytes, whereas 40% of them have more plasmacytic cells.3
WM B cells appear to express somatically mutated IgM.4-6 In a single case, Ciric et al7 report the development of biclonal IgMλ in WM tumor cells. The pattern of mutations within the 2 dominant clones was suggestive of prolonged antigen selection after the malignant WM transformation event. A more recent study of 3 patients with WM shows a complete lack of intraclonal variation.5 Unlike IgM-secreting myeloma, this study shows no evidence for isotype switching in WM.5 Shiokawa et al8 report the existence of clonotypic μ and δ transcripts in bone marrow (BM) and in blood samples of 1 of 3 patients studied. These findings, along with the high rate of somatic mutations, suggest that WM may arise from somatically mutated IgM+IgD+ or IgM-only B cells, or possibly from both.
Phenotypic and immunologic studies show abnormal B lymphocytes in WM patients. WM B cells secrete monoclonal immunoglobulin, express surface immunoglobulin (sIg) receptors, and reside in the BM. Studies of 111 patients with WM showed that 90% of cases were characterized by a CD19+CD20+CD5-CD10-CD23- immunophenotype.9 A higher incidence of CD5 and CD23 expression has, however, been demonstrated in another report of 126 patients with immunocytoma.10 Our previous studies reported increased circulating B cells in WM blood that carried CD19, CD20, CD5, CD10, CD11b, and CD9 markers11 and that over time underwent clonal evolution as defined by shifts in the CD45 isoform expression patterns.12 Moreover, a subset of apparently normal B cells in WM blood have normal B-cell repertoires, as do those of patients with IgM monoclonal gammopathies.13
Recent studies suggest CD27 may be a memory B-cell marker.14 CD27 is a member of the tumor necrosis factor receptor family, which interacts with the protein ligand CD70.15 Although naive B cells carry unmutated VH genes and express IgM+IgD+CD27- phenotypes, memory B cells carry mutated VH genes and express D27 along with sIg markers. Analysis of non-Hodgkin lymphomas showed that CD27 is detectable in naive and memory B cell-derived lymphomas,16 suggesting that in neoplastic B cells, CD27 is not always a faithful memory B-cell antigen. CD27 expression has also been observed in multiple myeloma and has been shown to correlate with disease activity.17 Identification of WM B cells in relation to CD27 expression has not yet been addressed.
Here, we have analyzed clonotypic IgM sequences from 15 patients with WM to characterize the stage of tumor B-cell arrest. Using the IgM signature sequence and single-cell reverse transcription-polymerase chain reaction (RT-PCR), we performed clonotypic transcript analysis of individual WM cells in phenotypically defined B-cell subpopulations to understand tumor distribution and to monitor clonal frequency in WM.
Patients and methods
Patients
After informed consent, BM aspirates and blood samples were obtained from 15 patients with histopathologic diagnoses of lymphoplasmacytic lymphoma and the clinicopathologic diagnosis of WM. BM aspirate and peripheral blood were collected at primary diagnosis for use in identifying clonotypic IgM transcripts. During the follow-up of patients, blood was collected for monitoring of circulating WM B cells.
Identifying IgM V/D/J from WM B cells
Mononuclear cells were isolated from BM aspirate or peripheral blood by gradient-step centrifugation (Ficoll-Hypaque Plus; Amersham Pharmacia Biotech, Uppsala, Sweden). Total RNA was purified from 5 × 106 mononuclear cells using Trizol reagent (Invitrogen, Carlsbad, CA). First-strand cDNA was reverse transcribed from 1 μg total RNA using dT15 primer and Superscript RNaseH- reverse transcriptase (Invitrogen) in a total volume of 20 μL. DNA fragment encoding for the clonotypic variable region was generated from cDNA by polymerase chain reaction (PCR) using VH1 to VH6 leader sequences and the CH1 sequence of IgM (Cμ) as primers (Table 1). Amplification was run for 30 cycles at 94°C for 30 seconds, 60°C or 30 seconds, and 68°C for 30 seconds using standard buffer and Hi Fi Taq DNA polymerase (Invitrogen). VH-Cμ PCR product (450 bp) was further treated with Exo-sapit reagent (Applied Biosystems, Foster City, CA) to remove excess primers and deoxyribonucleotides. Direct sequencing of the treated DNA fragment was made using M13 primer and Big Dye III reagent following the manufacturer's instructions (Applied Biosystems) and run on an ABI Prism 310 Genetic Analyzer (Applied Biosystems) using ABI Prism Sequencing Analysis 3.7 software for data analysis. In some experiments, the PCR product was cloned into pCR4 vector using the TOPO TA cloning system (Invitrogen). Individual clones were analyzed and sequenced using M13 primer. The clonotypic sequence was identified as the consensus sequence expressed by the majority of subclones. For approximately 50% of patients, this was confirmed using single-cell RT-PCR analysis of sorted CD20+ or sIgM+ B cells from BM. Single-cell analysis has been validated in this laboratory as specific, sensitive, and reproducible. Spiking of clonotypic cDNA derived from as little as one sixth of a cell into a PCR reaction of unrelated B cells is detectable by heminested PCR using CDR2/CDR3 patient-specific primers. Sequencing of PCR products confirms the correctness of the amplification. On the other hand, amplification using nonrelevant CDR2/CDR3 primers did not yield a PCR product.
Summary of primer sequences
Primer . | Sequence . |
|---|---|
| VH1 leader | 5′CCATGGACTGGACCTGGAG (G/C/A) A3′ |
| VH2 leader | 5′ATGGACA (T/C) ACTTTG (T/C) TCCAC3′ |
| VH3 leader | 5′CCATGGA (G/A) CT (T/G) GGGCTC3′ |
| VH4 leader | 5′ATGAA (A/G) CACCTGTGGTTCTTC3′ |
| VH5 leader | 5′GGTCAACCGCCATCCT3′ |
| VH6 leader | 5′ATGTCTGTCTCCTTCCTCATCTTC3′ |
| Cμ | 5′GGAATTCTCACAGGAGAC3′ |
| Cμs | 5′AGCTGTGTCGGACATGACC3′ |
| Cμm | 5′CACAGGTTCTCAAAGCCCTC3′ |
| Cδ | 5′GTGTCTGCACCCTGATAT3′ |
| Cγ | 5′GGGGAAGACCGATGGGCCCT3′ |
| Cα | 5′GAGGCTCAGCGGGAAGACCTT3′ |
| FR1c | 5′GGTGCAGCTG (G/C) (A/T) G (G/C) AGTC (G/A/T) GG3′ |
| FR2 | 5′TATGAATTCGGAAAGGGCCTGGAGTGG3′ |
| FR3 | 5′CCGAGGACACGGC (T/C) (C/G) TGTATTACTG3′ |
| JHc | 5′ACCTGAGGAGACGGTGACC (A/G) (G/T) (G/T) GT3′ |
| β2m 5′ | 5′CCAGCAGAGAATGGAAAGTC3′ |
| β2m 5′ int | 5′TGTCTTTCAGCAAGGACTGG3′ |
| β2m 3′ | 5′GATGCTGCTTACATGTCTCG3′ |
Primer . | Sequence . |
|---|---|
| VH1 leader | 5′CCATGGACTGGACCTGGAG (G/C/A) A3′ |
| VH2 leader | 5′ATGGACA (T/C) ACTTTG (T/C) TCCAC3′ |
| VH3 leader | 5′CCATGGA (G/A) CT (T/G) GGGCTC3′ |
| VH4 leader | 5′ATGAA (A/G) CACCTGTGGTTCTTC3′ |
| VH5 leader | 5′GGTCAACCGCCATCCT3′ |
| VH6 leader | 5′ATGTCTGTCTCCTTCCTCATCTTC3′ |
| Cμ | 5′GGAATTCTCACAGGAGAC3′ |
| Cμs | 5′AGCTGTGTCGGACATGACC3′ |
| Cμm | 5′CACAGGTTCTCAAAGCCCTC3′ |
| Cδ | 5′GTGTCTGCACCCTGATAT3′ |
| Cγ | 5′GGGGAAGACCGATGGGCCCT3′ |
| Cα | 5′GAGGCTCAGCGGGAAGACCTT3′ |
| FR1c | 5′GGTGCAGCTG (G/C) (A/T) G (G/C) AGTC (G/A/T) GG3′ |
| FR2 | 5′TATGAATTCGGAAAGGGCCTGGAGTGG3′ |
| FR3 | 5′CCGAGGACACGGC (T/C) (C/G) TGTATTACTG3′ |
| JHc | 5′ACCTGAGGAGACGGTGACC (A/G) (G/T) (G/T) GT3′ |
| β2m 5′ | 5′CCAGCAGAGAATGGAAAGTC3′ |
| β2m 5′ int | 5′TGTCTTTCAGCAAGGACTGG3′ |
| β2m 3′ | 5′GATGCTGCTTACATGTCTCG3′ |
DNA sequence analysis
Clonotypic sequence of the variable region was compared to the closest germline sequence using the International ImMunoGeneTics database (http://imgt.cines.fr/cgi-bin/IMGTdnap.jv?livret=0&Option=humanIg). 18 Base differences between the immunoglobulin genes sequenced and the corresponding germline genes were scored as mutations. CDR and FR locations and the numbering system are according to Kabat et al.19 The multinomial distribution model of Lossos et al20 (www-stat.stanford.edu/immunoglobulin) was used to evaluate the probability of antigenic selection pressure.
Polymerase chain reaction
All PCRs were performed in a total volume of 50 μL, using standard buffer and Taq DNA polymerase (Invitrogen). Amplification was run for 30 cycles at 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds unless otherwise stated. PCR primer sequences and combinations of primer sets for heminested PCR are summarized in Tables 1 and 2.
Combinations of primer sets for heminested PCR
. | PCR primers, sense/antisense . | . | |
|---|---|---|---|
| Transcript type . | First-stage PCR . | Second-stage PCR . | |
| μ, clonotypic | FR1c/Cμ | CDR2/CDR3 or CDR3R/Cμ | |
| μ, secreted, clonotypic | FR1c/CμS.1 | CDR3R/CμS | |
| μ, membrane bound, clonotypic | FR1c/CμM.1 | CDR3R/CμM | |
| σ, clonotypic | FR1c/Cσ | CDR3R/Cσ | |
| β2m, control | β2m 5′/β2m 3′ | β2m 5′int/β2m 3′ | |
| Universal immunoglobulin, control | FR1c/JHc | FR2/JHc | |
. | PCR primers, sense/antisense . | . | |
|---|---|---|---|
| Transcript type . | First-stage PCR . | Second-stage PCR . | |
| μ, clonotypic | FR1c/Cμ | CDR2/CDR3 or CDR3R/Cμ | |
| μ, secreted, clonotypic | FR1c/CμS.1 | CDR3R/CμS | |
| μ, membrane bound, clonotypic | FR1c/CμM.1 | CDR3R/CμM | |
| σ, clonotypic | FR1c/Cσ | CDR3R/Cσ | |
| β2m, control | β2m 5′/β2m 3′ | β2m 5′int/β2m 3′ | |
| Universal immunoglobulin, control | FR1c/JHc | FR2/JHc | |
Isotypic analysis of clonotypic transcripts
cDNA was reverse transcribed from total RNA as described. Clonal transcripts of various isotypes were amplified by CDR3R (patient specific; sense) and CH1 primer (Cα, Cδ, Cγ, or Cμ; antisense), as described, and were analyzed using 2% agarose gel electrophoresis.
Antibodies and cell sorting
Surface antigen expression was assessed by 2-color and 3-color phenotypic analysis using custom-conjugated, anti-CD20 fluorescein isothiocyanate (FITC) (mouse IgG2a; Rituximab21 ), phycoerythrin (PE)-conjugated anti-CD138 (mouse IgG1; Serotec, Raleigh, NC), PE-conjugated anti-IgM (goat Fab'2; Southern Biotechnology, Mississauga, ON, Canada), biotinylated anti-IgD (goat Fab'2; Southern Biotechnology) with SpectralRed (SPRD)-conjugated streptavidin (Southern Biotechnology) and FITC- or PE-conjugated anti-CD27 (mouse IgG1; BD PharMingen, Mississauga, ON, Canada). Isotype-matched antibodies were used as negative controls. Using the Epic Altra high-speed flow cytometer with an Autoclone Cell Deposition Unit (Beckman Coulter, Mississauga, ON, Canada), subpopulations of cells were identified, and single cells were sorted into 0.2-mL PCR tubes. Cells were also sorted at 100 or 1000 cells for analysis of transcript-expressing cells that were present at low cell frequencies. cDNA was prepared by direct lysis, as previously described,22 and was stored at -80°C.
Single-cell RT-PCR and analysis of clonotypic B-cell frequency
Heminested RT-PCR of 1 to 1000 cells was described previously.23 Briefly, 2 μL cDNA generated from single cells (VT = 10 μL) or of 100 to 1000 cells (VT = 40 μL) was used as a template in first-stage PCR. After the first amplification, 1 μL reaction mix was transferred to the second PCR containing heminested primers. PCR product was analyzed on 2% agarose gel electrophoresis. Primer sequences and primer sets are shown in Tables 1 and 2. Amplification of β2-microglobulin or universal immunoglobulin gene product was run as an internal control to verify the integrity of cDNA in each test reaction. Generally, 96 cells were tested for single-cell analysis. Clonal frequency was calculated as the percentage of cells positive to test reaction over the total number of cells positive to internal control.
Analysis of CDR3 by DNA fragment analysis
RT-PCR of the CDR3 region was performed using FR3 and fluorescence-labeled JHc primers following the standard PCR protocol. The PCR product was mixed with formamide and size standard and was analyzed on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems) according to the manufacturer's instructions. Data collection and analysis for the length of PCR product were performed using ABI Prism GeneScan software version 3.7.
Results
Most WM IgM V/D/J sequences exhibit somatic hypermutation, intraclonal homogeneity, and V/J gene bias
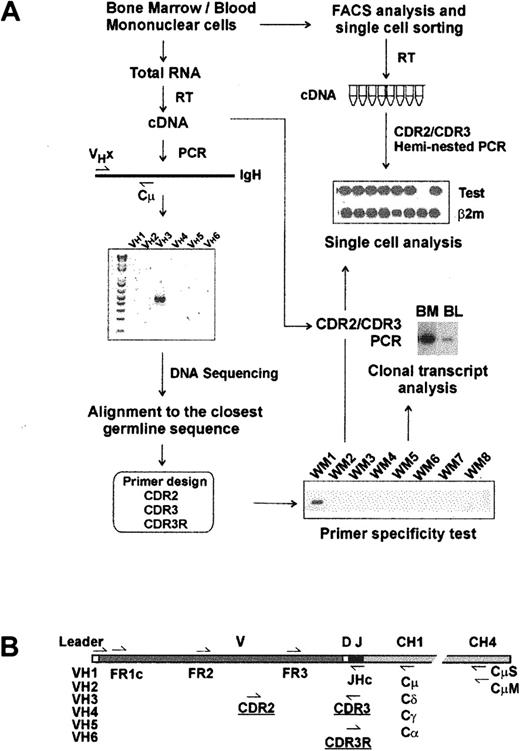
Molecular analysis of IgM V/D/J regions has been studied in WM BM samples, as summarized in Figure 1A. RT-PCR amplification of clonotypic IgM transcripts using a panel of VH leader sequence primers (VH1-VH6) and a Cμ-specific primer was successful in all 15 patients studied. DNA and deduced protein sequence analysis confirmed a functional V/D/J rearrangement for each clonotypic sequence. Compared with the closest VH germline gene, somatic mutations of each sequence were also identified. Our data strongly indicate biased V/J gene usage in WM. Of 15 WM IgH genes, all but one used the VH3 gene family, and 11 of 15 used the JH4 gene family. Most WM IgM involves extensive somatic mutation (3%-10%; n = 13); unexpectedly, 2 patients had unmutated VH genes (0%-1% mutations). PCR products from 4 patients with mutated V/D/J were cloned, sequenced, and compared to determine whether somatic hypermutation was activated in WM cells. For each patient, 8 subclones were sequenced and shown to be identical. For 6 patients, the clonotypic IgM V/D/J sequence was validated by analysis of individual sorted B cells from BM (Figure 2).
Strategy for molecular analysis of VH genes in WM. (A) Identification of clonotypic IgM VDJ sequence and design of primers. (B) Location of PCR primers on VH transcript.
Strategy for molecular analysis of VH genes in WM. (A) Identification of clonotypic IgM VDJ sequence and design of primers. (B) Location of PCR primers on VH transcript.
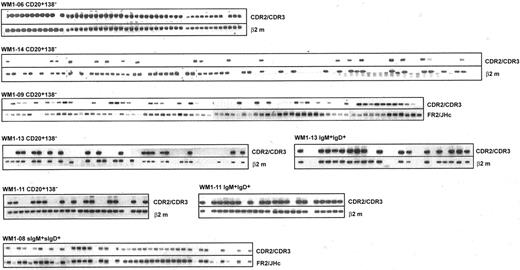
V/D/J sequence designated as clonotypic characterizes phenotypically defined subsets of WM B cells analyzed by single-cell RT-PCR. Clonotypic transcripts were determined by heminested PCR using FR1c/Cμ primers followed by CDR2/CDR3 primers. PCR of β2-microglobulin or FR2/JHc was used as internal control.
V/D/J sequence designated as clonotypic characterizes phenotypically defined subsets of WM B cells analyzed by single-cell RT-PCR. Clonotypic transcripts were determined by heminested PCR using FR1c/Cμ primers followed by CDR2/CDR3 primers. PCR of β2-microglobulin or FR2/JHc was used as internal control.
Most mutated WM clonotypic sequences lack features suggestive of antigenic selection
Analysis of mutated WM V/D/J sequences revealed a high frequency of replacement mutations scattered throughout the FR and CDR regions (Table 3). To further define whether the somatic mutation pattern in WM is characteristic of antigenic selection, each VH sequence was analyzed by a multinomial distribution model, as described by Lossos et al.20 In this analysis, conservation of the FR sequence and excess of replacement mutations in CDRs are determined. The probability that an excess (for CDR, PCDR) or a scarcity (for FR, PFR) of mutations occurred by chance is calculated as a function of the number of replacement and silent mutations in the CDR and FR regions. Concomitant statistical significance of PFR and PCDR indicates antigenic selection. Analysis of mutated VH genes (n = 13) in WM showed that there are more R mutations in CDRs than expected (PCDR < .05) in 11 of 13 sequences. Within these 11 sequences, 5 have fewer replacement mutations than expected because of chance. Therefore, multinomial analysis suggests that at least 5 of 13 mutated VH gene sequences have undergone antigenic selection. However, for 8 of 13 analyzed sequences, evidence for antigen-driven selection is lacking, perhaps reflecting other diversification mechanisms.
VH and JH gene use, rates of somatic hypermutation, multinomial analyses for antigenic selection of immunoglobulins, and lengths of CDR3 in WM B cells
VH . | Patient . | VH . | JH . | Mutation, % . | FR R, aa† . | FR S, aa† . | CDR R, aa† . | CDR S, aa† . | PFR* . | PCDR* . | Antigen selection . | CDR3, nt . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unmutated | ||||||||||||
| WM1-01 | 3 | 6 | 0 | 0 | 0 | 0 | 0 | .00000 | 1.00000 | No | 63 | |
| WM1-02 | 3 | 4 | 1.01 | 1 (1) | 0 | 2(2) | 0 | .23007 | .04402 | No | 45 | |
| Mutated | ||||||||||||
| WM1-03 | 3 | 3 | 3.06 | 3(3) | 2(2) | 4(4) | 0 | .08245 | .03590 | No | 42 | |
| WM1-04 | 3 | 4 | 5.10 | 6(6) | 2(2) | 7(7) | 0 | .09518 | .00756 | No | 27 | |
| WM1-05 | 3 | 4 | 6.0 | 9(9) | 2(2) | 5(7) | 0 | .29486 | .02279 | No | 54 | |
| WM1-06 | 3 | 3 | 6.0 | 7(8) | 4(4) | 5(6) | 0 | .14422 | .06369 | No | 39 | |
| WM1-07 | 5 | 4 | 6.06 | 10(11) | 2(2) | 3(4) | 1(1) | .54927 | .28717 | No | 45 | |
| WM1-08 | 3 | 6 | 6.06 | 4(4) | 3(3) | 10(11) | 0 | .00192 | .00004 | Yes | 60 | |
| WM1-09 | 3 | 4 | 6.27 | 8(10) | 4(4) | 5(5) | 0 | .35667 | .19231 | No | 39 | |
| WM1-10 | 3 | 4 | 7.14 | 8(9) | 3(3) | 9(9) | 0 | .09907 | .00371 | No | 48 | |
| WM1-11 | 3 | 4 | 7.49 | 8(9) | 5(5) | 4(5) | 3(3) | .06757 | .28079 | No | 42 | |
| WM1-12 | 3 | 4 | 8.42 | 9(9) | 5(5) | 7(9) | 2(2) | .02340 | .02137 | Yes | 33 | |
| WM1-13 | 3 | 4 | 8.84 | 6(8) | 3(3) | 10(13) | 2(2) | .00395 | .00005 | Yes | 39 | |
| WM1-14 | 3 | 4 | 9.76 | 9(11) | 4(4) | 7(11) | 3(3) | .01657 | .00481 | Yes | 45 | |
| WM1-15 | 3 | 4 | 9.76 | 6(6) | 8(8) | 9(13) | 2(2) | .00004 | .00041 | Yes | 27 |
VH . | Patient . | VH . | JH . | Mutation, % . | FR R, aa† . | FR S, aa† . | CDR R, aa† . | CDR S, aa† . | PFR* . | PCDR* . | Antigen selection . | CDR3, nt . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unmutated | ||||||||||||
| WM1-01 | 3 | 6 | 0 | 0 | 0 | 0 | 0 | .00000 | 1.00000 | No | 63 | |
| WM1-02 | 3 | 4 | 1.01 | 1 (1) | 0 | 2(2) | 0 | .23007 | .04402 | No | 45 | |
| Mutated | ||||||||||||
| WM1-03 | 3 | 3 | 3.06 | 3(3) | 2(2) | 4(4) | 0 | .08245 | .03590 | No | 42 | |
| WM1-04 | 3 | 4 | 5.10 | 6(6) | 2(2) | 7(7) | 0 | .09518 | .00756 | No | 27 | |
| WM1-05 | 3 | 4 | 6.0 | 9(9) | 2(2) | 5(7) | 0 | .29486 | .02279 | No | 54 | |
| WM1-06 | 3 | 3 | 6.0 | 7(8) | 4(4) | 5(6) | 0 | .14422 | .06369 | No | 39 | |
| WM1-07 | 5 | 4 | 6.06 | 10(11) | 2(2) | 3(4) | 1(1) | .54927 | .28717 | No | 45 | |
| WM1-08 | 3 | 6 | 6.06 | 4(4) | 3(3) | 10(11) | 0 | .00192 | .00004 | Yes | 60 | |
| WM1-09 | 3 | 4 | 6.27 | 8(10) | 4(4) | 5(5) | 0 | .35667 | .19231 | No | 39 | |
| WM1-10 | 3 | 4 | 7.14 | 8(9) | 3(3) | 9(9) | 0 | .09907 | .00371 | No | 48 | |
| WM1-11 | 3 | 4 | 7.49 | 8(9) | 5(5) | 4(5) | 3(3) | .06757 | .28079 | No | 42 | |
| WM1-12 | 3 | 4 | 8.42 | 9(9) | 5(5) | 7(9) | 2(2) | .02340 | .02137 | Yes | 33 | |
| WM1-13 | 3 | 4 | 8.84 | 6(8) | 3(3) | 10(13) | 2(2) | .00395 | .00005 | Yes | 39 | |
| WM1-14 | 3 | 4 | 9.76 | 9(11) | 4(4) | 7(11) | 3(3) | .01657 | .00481 | Yes | 45 | |
| WM1-15 | 3 | 4 | 9.76 | 6(6) | 8(8) | 9(13) | 2(2) | .00004 | .00041 | Yes | 27 |
nt indicates nucleotides,
PFR and PCDR are calculated using the applet available at www-stat.stanford.edu/immunoglobulin. Italics indicates P ≤ .05.
Values indicate the number of amino acids changed; parenthetical values indicate the number of nucleotides mutated.
Analysis of CDR3 indicates N-region diversification at the V/D/J junction, with the total length ranging from 27 to 63 nucleotides (43 ± 10.4 bp; median, 42 bp). The length of CDR3 in 2 unmutated VH clones was 63 and 45 bp, whereas the mutated VH clones varied from 27 to 60 bp.
Clonotypic transcripts encoding secreted and membranous IgM are detectable in WM BM and blood samples
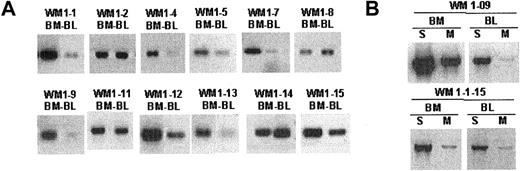
The unique clonotypic V/D/J sequence identified in the CDR regions of each WM clone was used to design patient-specific primers (CDR2, CDR3, CDR3R), as summarized in Figure 1B. Each patient-specific primer was extensively tested against other tumor B cells of the same gene family to ensure its specificity. We found that the combination of CDR2 (sense) and CDR3 (antisense) primers yielded the highest sensitivity in a PCR reaction. Alternatively, the combination of one patient-specific primer (CDR3R, sense) and an immunoglobulin constant region-specific primer (CH1, antisense) permitted clonotypic isotype identification without compromising specificity. Using patient-specific primers in a single-stage RT-PCR reaction, we could routinely detect clonotypic transcripts in tumor B cells. By amplifying cDNA prepared from total RNA of unfractionated mononuclear cells with CDR2/CDR3 or CDR3R/Cμ primers, we were able to detect clonotypic transcripts in BM and peripheral blood for 12 of 12 patients whose matching samples were collected at diagnosis (Figure 3A). In general, we observed that the level of clonotypic transcripts in BM was higher than in blood. Expression of membrane-bound (CDR3R/CμM) and secreted (CDR3R/CμS) clonotypic IgM was also determined in 2 WM patients. Results clearly indicate that both forms of transcripts are present in BM and blood (Figure 3B).
Clonotypic IgM transcripts are detectable in bone marrow and blood of WM patients. Single-stage RT-PCR of unfractionated mononuclear cells prepared from BM and peripheral blood as described in “Patients and methods.” An equal amount of cDNA was used in each sample. Negative control PCR reactions without cDNA or with cDNA from BM or blood of healthy controls (n = 2) or unrelated WM patients (n = 8) lacked bands. Water controls were negative. Amplification was performed using (A) CDR2 (patient specific) and CDR3 (patient specific) primers or (B) CDR3R (patient specific) and CμS (S) or CμM (M) primers.
Clonotypic IgM transcripts are detectable in bone marrow and blood of WM patients. Single-stage RT-PCR of unfractionated mononuclear cells prepared from BM and peripheral blood as described in “Patients and methods.” An equal amount of cDNA was used in each sample. Negative control PCR reactions without cDNA or with cDNA from BM or blood of healthy controls (n = 2) or unrelated WM patients (n = 8) lacked bands. Water controls were negative. Amplification was performed using (A) CDR2 (patient specific) and CDR3 (patient specific) primers or (B) CDR3R (patient specific) and CμS (S) or CμM (M) primers.
WM BM and blood are characterized by elevated levels of CD20+IgM+IgD+ cells, most of which are clonotypic
To better understand tumor cell infiltration of WM BM and blood samples, B-cell populations were phenotypically identified using CD20, IgM, and IgD as cell surface markers and were sorted as single cells into individual tubes for RT-PCR analysis. Tumor B cells were identified by heminested RT-PCR using FR1c/Cμ primers for the first-stage reaction, followed by CDR2/CDR3 or CDR3R/Cμ primers for the second-stage reaction (Tables 1 and 2). We observed elevated levels of CD20+ cells during the active stages of disease, defined by high serum levels of monoclonal IgM, most of which were tumor B cells (Figure 2; Table 4). In 5 patients studied, tumor distributions ranged from 39% to 96% of CD20+ cells (Table 4). In 2 patients (WM1-11 and WM1-13), clonal frequencies of IgM+IgD+ cells and CD20+ cells were directly compared. As predicted, the clonal frequency among IgM+IgD+ cells was slightly higher than that among CD20+ cells (Table 4). This is not surprising because CD20+ cells are likely to include the persisting polyclonal IgM+ B cell population and polyclonal postswitch B cells. IgM+IgD- cells were not detectable in the BM samples studied. CD138, a plasma cell marker, was occasionally detected on a small subset (1%-4%) of WM B cells (CD20+CD138+). Although rare, these cells are exclusively tumor B cells that secrete monoclonal IgM and, in some instances, express membrane-bound IgM (Figure 4).
Clonotypic B cells in WM are characterized as CD20+CD138−IgM+IgD+ and CD20+CD138+ cells
. | . | . | . | Clonotypic B cells with different clonal isotype expression, % . | . | |
|---|---|---|---|---|---|---|
| Phenotypic marker and patient . | Source . | No. cells analyzed . | Clonal frequencies, % . | μ+δ+ . | μ+δ− . | |
| CD20+CD138− | ||||||
| WM1-06.1* | BM | 48 | 96 | NA | NA | |
| WM1-06.2* | BM | 96 | 58 | NA | NA | |
| WM1-09 | BM | 96 | 61 | 30 | 70 | |
| WM1-14 | BM | 96 | 39 | NA | NA | |
| WM1-11 | BM | 96 | 57 | NA | NA | |
| WM1-11 | Blood | 144 | 2 | NA | NA | |
| WM1-13 | BM | 72 | 65 | 35 | 65 | |
| WM1-13 | Blood | 72 | 69 | 39 | 61 | |
| IgM+IgD+ | ||||||
| WM1-08 | BM | 72 | 77 | 64 | 36 | |
| WM1-11 | BM | 72 | 86 | NA | NA | |
| WM1-13 | BM | 72 | 89 | 31 | 69 | |
| WM1-13 | Blood | 72 | 81 | 42 | 58 | |
| CD20+CD138+ | ||||||
| WM1-09 | BM | 48 | 92 | 4 | 96 | |
. | . | . | . | Clonotypic B cells with different clonal isotype expression, % . | . | |
|---|---|---|---|---|---|---|
| Phenotypic marker and patient . | Source . | No. cells analyzed . | Clonal frequencies, % . | μ+δ+ . | μ+δ− . | |
| CD20+CD138− | ||||||
| WM1-06.1* | BM | 48 | 96 | NA | NA | |
| WM1-06.2* | BM | 96 | 58 | NA | NA | |
| WM1-09 | BM | 96 | 61 | 30 | 70 | |
| WM1-14 | BM | 96 | 39 | NA | NA | |
| WM1-11 | BM | 96 | 57 | NA | NA | |
| WM1-11 | Blood | 144 | 2 | NA | NA | |
| WM1-13 | BM | 72 | 65 | 35 | 65 | |
| WM1-13 | Blood | 72 | 69 | 39 | 61 | |
| IgM+IgD+ | ||||||
| WM1-08 | BM | 72 | 77 | 64 | 36 | |
| WM1-11 | BM | 72 | 86 | NA | NA | |
| WM1-13 | BM | 72 | 89 | 31 | 69 | |
| WM1-13 | Blood | 72 | 81 | 42 | 58 | |
| CD20+CD138+ | ||||||
| WM1-09 | BM | 48 | 92 | 4 | 96 | |
One hundred percent of clonotypic B cells expressed clonal μ transcript. Clonal frequency was calculated as the percentage of sorted cells that expressed clonal μ transcript over the total number of cells that showed positive PCR results to β2-microglobulin or universal immunoglobulin control.
NA indicates not applicable.
BM samples from WM1-06 were collected 9 months apart.
WM B cells express transcripts encoding predominantly the secreted form of clonotypic IgM. Heminested RT-PCR of CD20+CD138+ cells in patient WM1-09 was used to detect membrane-bound and secreted clonotypic μ transcripts. The PCR primer combination is as described in Table 2.
WM B cells express transcripts encoding predominantly the secreted form of clonotypic IgM. Heminested RT-PCR of CD20+CD138+ cells in patient WM1-09 was used to detect membrane-bound and secreted clonotypic μ transcripts. The PCR primer combination is as described in Table 2.
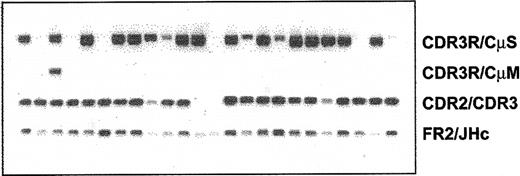
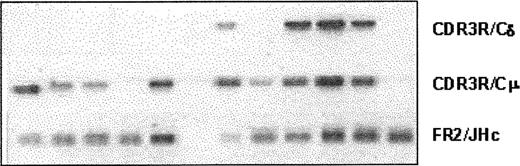
Clonotypic IgM and IgD, but not IgG or IgA, isotype transcripts are detectable in WM B cells
Expression of various clonotypic isotypes in WM was determined by RT-PCR using CDR3R (patient specific, sense) and CH1 (Cμ, Cδ, Cγ, or Cα, antisense) primers in a single-stage PCR reaction. Studies in 8 WM patients indicated the existence of clonotypic μ and δ transcripts in BM and peripheral blood (Figure 5), with relatively larger quantities of μ transcripts observed in most samples tested. By contrast, clonotypic γ or α transcripts were not detected in WM. Coexpression of δ and μ transcripts in WM B cells was further confirmed by single-cell analysis and heminested RT-PCR. A representative result shown in Figure 6 demonstrates the coexpression of clonotypic μ and δ transcripts in a subset of tumor B cells. Table 4 summarizes the distribution of clonotypic μ and δ transcripts in CD20+CD138- BM cells in 4 patients studied. Based on heminested RT-PCR, clonotypic μ transcripts were detected in all, and δ transcripts were detected in approximately 1 of 3 clonotypic B cells in the BM.
Lack of class switching in B cells from bone marrow and peripheral blood of WM patients. RT-PCR of unfractionated mononuclear cells prepared from BM and peripheral blood of WM patients as described in “Patients and methods.” An equal amount of cDNA was used in each sample. Amplification was performed using CDR3R (patient specific) and respective CH1 primers. Negative controls of PCR reaction without cDNA or with cDNA from BM or blood of healthy controls (n = 2), or other WM patients (n = 8) did not yield any bands.
Lack of class switching in B cells from bone marrow and peripheral blood of WM patients. RT-PCR of unfractionated mononuclear cells prepared from BM and peripheral blood of WM patients as described in “Patients and methods.” An equal amount of cDNA was used in each sample. Amplification was performed using CDR3R (patient specific) and respective CH1 primers. Negative controls of PCR reaction without cDNA or with cDNA from BM or blood of healthy controls (n = 2), or other WM patients (n = 8) did not yield any bands.
WM B cells coexpress clonotypic IgM and IgD transcripts. Representative analysis of clonal isotypes in multiple individual cells from the same patient is shown. cDNA from individual CD20+ cells of patient WM1-09 was split into separate aliquots, and heminested PCR was run in parallel experiments using the combination of primer sets described in Table 2. Percentages of cells expressing single or double clonal isotypes of each patient were calculated and summarized in Table 4.
WM B cells coexpress clonotypic IgM and IgD transcripts. Representative analysis of clonal isotypes in multiple individual cells from the same patient is shown. cDNA from individual CD20+ cells of patient WM1-09 was split into separate aliquots, and heminested PCR was run in parallel experiments using the combination of primer sets described in Table 2. Percentages of cells expressing single or double clonal isotypes of each patient were calculated and summarized in Table 4.
CD27- tumor cells predominate over CD27+ tumor cells in WM BM and blood B-cell subpopulations
The observation that WM V/D/J sequences are somatically hypermutated and clonally homogenous but still associated with preswitch constant regions suggests that the point of transformation is at the memory B-cell stage. Because CD27expression on normal B cells correlates with memory status,14 we evaluated the expression of CD27 in B cells from WM blood and BM samples. For 2 WM patients, CD20+CD27+ and CD20+CD27- cells were sorted for clonotypic analysis. Heminested RT-PCR using patient-specific primers was performed to identify tumor B cells in each subpopulation (Table 5). For both patients, the percentage of total CD20+ cells was elevated; as many as one third of CD20+ cells were CD27+. Single-cell analysis clearly demonstrated that clonotypic B cells were present in both subpopulations. Overall, CD27- tumor cells outnumbered CD27+ tumor cells in the 2 BM samples and corresponding blood samples tested (1.5- to 11.2-fold greater numbers). A similar conclusion was reached for a third patient (WM1-13) comparing frequencies of CD27+ B cells by flow cytometry and frequencies of clonotypic transcripts in CD20+ B cells by RT-PCR. In this patient, CD27+ and CD27- frequencies were, respectively, 4% and 55% of the total mononuclear population in BM (Table 5), and clonotypic frequencies were shown to be 65% of CD20+ cells, indicating that most clonotypic cells must have been CD27-. A comparable pattern was observed with these populations in the corresponding blood sample for WM1-13. Taken together with the more detailed study in the first 2 patients, the results of Table 5 show that most WM tumor cells are CD27-, with a smaller proportion of tumor cells in the CD27+ B-cell subpopulation.
WM B cells are not restricted to CD27 expression
Patient and source . | CD20+ MNC, % . | Clonotypic B cells, CD20+, % . | CD20+ CD27− MNC, % . | Clonotypic B cells within CD20+ CD27−, % . | CD20+ CD27+ MNC, % . | Clonotypic B cells within CD20+ CD27+, % . | Ratio of absolute no. clonotypic B cells, CD20+ CD27−/ CD20+ CD27+ . |
|---|---|---|---|---|---|---|---|
| WM1-01 | |||||||
| BM | 23 | NA | 17 | 87 | 6 | 22 | 11.20 |
| Blood | 39 | NA | 37 | 4 | 2 | 50 | 1.48 |
| WM1-11 | |||||||
| BM | 20 | 57 | 14 | 25 | 5 | 17 | 4.12 |
| Blood | 21 | 2 | 13 | 2 | 7 | 2 | 1.86 |
| WM1-13 | |||||||
| BM | 59 | 65 | 55 | NA | 4 | NA | > 1* |
| Blood | 32 | 69 | 24 | NA | 8 | NA | > 1* |
Patient and source . | CD20+ MNC, % . | Clonotypic B cells, CD20+, % . | CD20+ CD27− MNC, % . | Clonotypic B cells within CD20+ CD27−, % . | CD20+ CD27+ MNC, % . | Clonotypic B cells within CD20+ CD27+, % . | Ratio of absolute no. clonotypic B cells, CD20+ CD27−/ CD20+ CD27+ . |
|---|---|---|---|---|---|---|---|
| WM1-01 | |||||||
| BM | 23 | NA | 17 | 87 | 6 | 22 | 11.20 |
| Blood | 39 | NA | 37 | 4 | 2 | 50 | 1.48 |
| WM1-11 | |||||||
| BM | 20 | 57 | 14 | 25 | 5 | 17 | 4.12 |
| Blood | 21 | 2 | 13 | 2 | 7 | 2 | 1.86 |
| WM1-13 | |||||||
| BM | 59 | 65 | 55 | NA | 4 | NA | > 1* |
| Blood | 32 | 69 | 24 | NA | 8 | NA | > 1* |
MNC indicates mononuclear cell.
Results were extrapolated by calculating the absolute number of clonotypic B cells in CD20+ cells and the absolute number of CD20+ CD27+ cells.
Persistently elevated CD20+ cells detected at stages of WM characterized by low serum IgM are polyclonal cells that do not reflect tumor burden
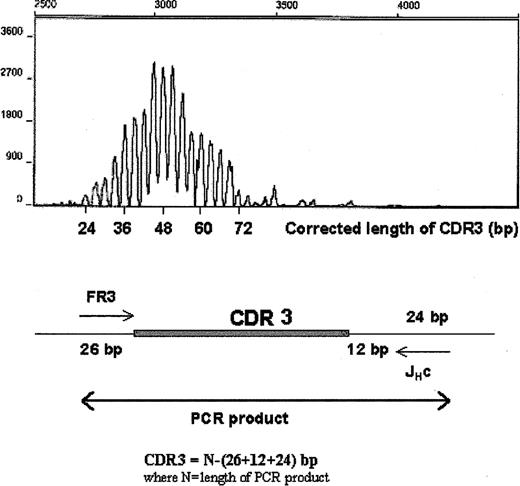
We have monitored WM B cells over time in the peripheral blood of WM patients. In 4 patients studied, we were able to detect circulating tumor B cells at low clonotypic frequencies during periods of disease when serum monoclonal IgM levels were low after effective fludarabine therapy (Table 6). Interestingly, these same patients often had persistently elevated numbers of CD20+ cells, as measured by phenotypic analysis. To determine whether these numerically increased CD20+ populations were tumor cells that had escaped detection (eg, through mutation of primer target regions), 2 methods were used to evaluate the CDR3 regions expressed. First, CDR3 regions were amplified with FR3 and fluorescence-labeled JH primers, and the length of the PCR product was measured by DNA fragment analysis using capillary electrophoresis. A single peak in the expected size range on the electro-pherogram suggested that the elevated CD20+ population consisted of cells with mutated clonotypic CDR3 that had escaped detection, whereas polyclonal populations were characterized by multiple peaks. Figure 7 shows the presence of multiple CDR3 peaks ranging from 24 to 72 bp (an increment of 3 bp/peak), which suggested that these cells were polyclonal. Second, CDR1, CDR2, CDR3, and J regions from this B-cell population were amplified using universal FR1c and JHc primers. After subcloning and sequencing the PCR product, we observed that in each patient tested, 24 of 24 subclones had unique V/D/J sequences, providing direct evidence that these B-cell populations were polyclonal. In these 2 experiments, we used 2 different sets of consensus primers to reduce the possibility that the amplification would fail because of a mutation in primer target sequences. Our studies showed that these elevated numbers of circulating B cells largely comprised polyclonal B cells. It is not yet clear whether these polyclonal B cells bore a functional relationship to the tumor itself.
Clonotypic frequencies at follow-up of WM patients
. | Follow-up sample . | . | . | Date sample collected for clonotypic VH sequence identification . | ||
|---|---|---|---|---|---|---|
| Selected subset and patient . | Date . | CD20+ cells, % . | Clonotypic frequency . | . | ||
| CD20+ CD138− | ||||||
| WM1-07 | September 2002 | 27 | 1/4 000* | October 1997 | ||
| WM1-14 | August 2003 | 36 | < 1/12 000* | December 2002 | ||
| WM1-15 | January 2003 | 20 | 1/4 000* | June 2002 | ||
| WM1-11 | July 2003 | 21 | 1/4 000* | June 2003 | ||
| IgM+ IgD+ | ||||||
| WM1-1 | March 2003 | 30 | 11/100† | December 2002 | ||
. | Follow-up sample . | . | . | Date sample collected for clonotypic VH sequence identification . | ||
|---|---|---|---|---|---|---|
| Selected subset and patient . | Date . | CD20+ cells, % . | Clonotypic frequency . | . | ||
| CD20+ CD138− | ||||||
| WM1-07 | September 2002 | 27 | 1/4 000* | October 1997 | ||
| WM1-14 | August 2003 | 36 | < 1/12 000* | December 2002 | ||
| WM1-15 | January 2003 | 20 | 1/4 000* | June 2002 | ||
| WM1-11 | July 2003 | 21 | 1/4 000* | June 2003 | ||
| IgM+ IgD+ | ||||||
| WM1-1 | March 2003 | 30 | 11/100† | December 2002 | ||
Clonotypic B cells were tested in sorted cell populations collected at 100 and 1000 cells/tube. Clonotypic frequency was determined as the number of sample tubes with clonal μ transcript divided by the total number of cells in all sample tubes analyzed.
Clonotypic frequency was determined by single-cell analysis. Clonal frequency was calculated as described in Table 4.
Elevated CD20+ cell numbers in the peripheral blood of WM patients during the stable phase of disease are polyclonal B cells. CDR3 of unfractionated peripheral blood B cells was analyzed by RT-PCR using FR3 and fluorescence-labeled JHc primer (Table 2) and DNA fragment analysis using capillary electrophoresis. The PCR product includes 26 bp upstream and 36 bp downstream of CDR3. Corrected length of CDR3 is as indicated.
Elevated CD20+ cell numbers in the peripheral blood of WM patients during the stable phase of disease are polyclonal B cells. CDR3 of unfractionated peripheral blood B cells was analyzed by RT-PCR using FR3 and fluorescence-labeled JHc primer (Table 2) and DNA fragment analysis using capillary electrophoresis. The PCR product includes 26 bp upstream and 36 bp downstream of CDR3. Corrected length of CDR3 is as indicated.
Discussion
Analysis of WM IgM sequences reveals a pronounced bias toward use of the VH3 family (14 of 15 patients) and a prevalent use of JH4 (11 of 15 patients). This is the first report, in 2 patients, of WM clones having unmutated VH segments, a phenomenon not reported for multiple myeloma, a related B-lineage malignancy. Clonotypic WM sequences were validated as accurate using single-cell RT-PCR analysis of clonotypic IgM in sorted BM B cells. This study provides the first quantitative frequency analysis of WM tumor cells in phenotypically identified B-cell subpopulations. We have demonstrated the colocalization of clonotypic μ and δ transcripts within 30% and 64% of clonotypic tumor B cells and the predominance of CD20+CD27- over CD20+CD27+ in the WM clone. WM cells may arise from an unusual VH3-expressing B-cell population with an often somatically mutated VH segment that in some patients lacks evidence of antigenic selection. WM includes IgM+IgD+ B cells that are not restricted to CD27 expression. At diagnosis and in relapse, the frequency of clonotypic B cells is high in BM and blood. In periods of disease characterized by low serum monoclonal IgM, the frequency of circulating clonotypic B cells is substantially decreased, even though the aggregate number of CD20+ B cells, shown here to be polyclonal, is often significantly elevated.
The biased VH3 and JH4 gene use detected in WM is consistent with previous reports by Sahota5 and Aoki.24 Although VH3 is frequent in the normal B-cell repertoire, the incidence of VH3 gene use in patients with WM (93%) exceeds that in healthy persons (50%).25 Restricted VH and JH segment use has been reported in patients with various types of lymphoma,26-28 but the significance of this observation remains to be determined. We have also observed a lack of intraclonal diversity in WM V/D/J sequences. VH genes were analyzed from 4 WM samples by sequencing of individual subclones, with complete homogeneity among subclones derived from the same patient. Longitudinal analysis indicated that the VH sequence derived from WM B cells is highly stable over the course of disease. In 2 WM patients (WM1-01 and WM1-06), one of whom expressed a germline VH sequence (WM1-01), an identical clonotypic IgM sequence was independently derived from BM samples collected at different time points (3 years or 9 months apart; data not shown).
Detailed analysis of the distribution of mutations by the multinomial model of Lossos et al20 reveals that only 5 of 15 WM VH sequences show evidence of having undergone antigenic selection. It has been suggested that mutations in the FR and CDR regions, the key variables in calculating antigenic selection by the multinomial method, may not accurately reflect in vivo circumstances. FR regions with relatively high R/S ratios, which are believed to have a deleterious effect on antibody structure, are nevertheless observed in some WM VH genes that make stable IgM.5 CDR mutations, attributed to the selection of amino acid replacements that increase affinity for target antigen, may arise independently of the selection process at mutational hotspots.29,30 Furthermore, it is unclear whether a direct correlation exists between VH gene variables, such as CDR3 length, and antigenic selection. In our studies, the apparently antigen-selected clones have variable lengths of CDR3; some are as short as 27 bp, others are as long as 60 bp. Long and short CDR3 sequences have been associated with hypermutated antigen-selected cells.31-34 Thus, statistical analysis of mutations and CDR3 lengths in WM clones only partially describes the complex, clone-specific, in vivo conditions of antigenic selection, if present.
IgM paraproteinemia in WM suggests preswitch transformation coupled with an inability or an impairment of class-switch recombination. Our studies show that only clonotypic μ and δ, not γ or α, transcripts were present in 12 of 12 WM patients studied, suggesting an absence of isotype switching. Sahota et al5 reported similar results using a smaller patient cohort. Absence of isotype switching in the hyper-IgM syndrome may reflect a defect in the CD40-CD40L signaling pathway,35-38 mutation of activation-induced cytidine deaminase (AID),39,40 or uracil DNA glycosylate deficiency.41 Recent studies in IgM chronic lymphocytic leukemia indicate that defective class-switch recombination may result from a partial deletion of switch μ sequences.42 It is possible that the exclusive expression of IgM in WM results from the transformation of preswitch IgM cells lacking a class-switch mechanism. Although this interpretation is consistent with our VH gene mutation analysis, it is more difficult to reconcile with our detection of tumor cells in some patients with the properties of late-stage B cells, such as CD138 expression and IgM secretion. In contrast, for IgM multiple myeloma, thought to involve postgerminal center IgM+ B cells, somewhat decreased but nevertheless detectable isotype switching has been described.43
Our use of single-cell sorting and direct lysis RT-PCR specific for WM V/D/J transcripts permits the estimation of tumor cell frequency within sorted B-cell subpopulations. Clonal frequencies in the circulation vary among WM patients and stages of disease but are comparable to or lower than those in the BM. Based on analysis of clonotypic IgM transcripts, WM B cells reside in the CD20+CD138- IgM+IgD+ compartment, a definitive demonstration consistent with the phenotypic studies reported by others.1,9,44 We demonstrated the coexpression of clonotypic μ and δ transcripts in approximately one third of tumor cells. A large compartment of WM B cells exclusively expresses IgM transcripts, but detecting clonotypic δ in 64% of sorted sIgM+sIgD+ tumor cells suggests subthreshold levels of the δ transcript in some WM cells. Although it remains possible that some tumor cells may belong to IgM+IgD- B cells, our phenotypic analysis indicated few, if any, IgM+IgD- B cells in the samples studied. Thus, we conclude that these clonotypic B cells are most likely IgM+IgD+ cells.
Infrequently in WM, we detected small numbers (1%-4%) of CD20+CD138+ cells that lack sIg. These were sorted and confirmed to be clonotypic. The secretion of monoclonal IgM and the expression of CD138, a late-stage B- and plasma-cell marker, suggest that clonotypic B-lineage differentiation may persist after transformation. This is supported by our previous observations of a transition in the WM clone over a 3-year period from the expression of CD45RA to the predominant expression of CD45R0, characteristic of late-stage B or early-stage plasma cells.12
We have studied clonal frequencies of WM B cells in the selected CD20+CD27+ and CD20+CD27- cell subpopulations and have shown that tumor B cells are scattered throughout both B-cell compartments, with most residing in the CD20+CD27- subset. Our molecular analysis is supported by immunophenotypic studies reported by San Miguel et al,44 showing that CD27 is heterogeneously expressed in 100% of patients with WM. Our study is the first to show that clonotypic WM B cells are not restricted to CD27 surface antigen expression. Reduced CD27 expression in plasma cells correlates with poor prognosis in multiple myeloma.17 In systemic lupus erythematosus, patients with high disease activity had increased frequencies of CD19+ B cells and CD27high plasma cells.45 In some lymphomas, on the other hand, strong CD27 expression was detected regardless of memory or naive B-cell origin.16 Furthermore, patients with unmutated and mutated B-cell chronic lymphocytic leukemia (B-CLL) had comparable levels of CD27 surface antigen,46 indicating that CD27 expression does not distinguish between neoplastic cells of naive and memory B-cell origin. Our studies also support this idea and suggested that CD27 should not be used as a marker for identifying clonotypic WM B cells.
Our previous report6 and present studies showed that elevated CD20+ cells were consistently observed in the peripheral blood of patients with WM, even in the stable phases of disease. Monitoring of tumor B cells by single-cell RT-PCR revealed decreased clonotypic frequencies in the peripheral blood, even though the number of total B cells considerably exceeded normal levels. This relative absence of clonotypic WM B cells is consistent with the low serum monoclonal protein concentrations in these samples (data not shown). Analysis of CDR3 and sequencing of VH genes in the B-cell population showed the persistently elevated numbers of CD20+ B cells to be of polyclonal origin. Such abnormally high polyclonal B-cell populations in the blood suggested a disturbance of B-cell homeostasis by WM tumor B cells. Thus, neither CD27 nor CD20 is a reliable indicator of circulating malignant B cells. These observations highlight the problems inherent in phenotypic analysis of lymphocyte populations as a means of assessing tumor burden, and they emphasize the importance of molecular analysis to confirm the extent of the malignant WM clone.
In conclusion, this analysis of clonotypic B cells in 15 patients with WM showed that in most patients, WM B cells expressing somatically mutated clonotypic transcripts were IgM+IgD+ cells not restricted to CD27 expression and that clonotypic frequency in the blood may be independent of the total B-cell number, especially during stable disease when most circulating B cells may be polyclonal. VH gene characteristics of WM B cells encouraged speculation that the transformation event that gave rise to WM might have targeted an unusual B-cell subset. The unmutated VH genes in 2 patients, the dominant use of VH3, the apparent lack of antigenic selection in CDR regions, and the failure to undergo class switching (a germinal center event) are consistent with the transformation of a rare, potentially extra-germinal center B cell in WM.
Prepublished online as Blood First Edition Paper, February 5, 2004; DOI 10.1182/blood-2003-11-4024.
Supported by a Smokler Research Grant from the Research Fund for Waldenstrom's, Ltd, and the Research Fund for Waldenstrom's at the Dana-Farber Cancer Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal