Abstract
We examined the immunoglobulin (Ig) heavy chain variable region genes (VH genes) used by leukemia cells of 1220 unrelated patients with chronic lymphocytic leukemia (CLL). We found 1188 (97%) expressed Ig encoded by a single Ig VH subgroup, the most common of which was VH3 (571 or 48.1%), followed by VH1 (319 or 26.8%) and VH4 (241 or 20.2%). Using allele-specific primers, we found 13.8% of all samples (n = 164) used one major VH1-69 allele, designated 51p1, 163 of which were not somatically mutated. For these cases, there was marked restriction in the structure of the Ig third complementarity determining regions (CDR3s), which were encoded by a small number of unmutated D and JH gene segments. Strikingly, 15 of the 163 cases had virtually identical CDR3s encoded by the second reading frame of D3-16 and JH3. Further analysis revealed that each of these 15 samples used the same unmutated Ig kappa light-chain gene, namely A27. These data reveal that approximately 1.3% (15/1220) of all patients had leukemia cells that expressed virtually identical Ig. This finding provides compelling evidence that the Ig expressed by CLL B cells are highly selected and not representative of the Ig expressed by naive B cells.
Introduction
The mutational status of the immunoglobulin (Ig) genes expressed in B-cell chronic lymphocytic leukemia (CLL) can be used to segregate patients into 2 subsets that have significantly different tendencies for disease progression. Those patients with leukemia cells that express unmutated Ig heavy chain variable region genes (VH genes) have a greater tendency for disease progression and shorter survival than those who have leukemia cells that express Ig VH genes with less than 98% nucleic acid sequence homology with their germ-line counterparts.1-7 Generally, the Ig VH genes expressed by any leukemia cell population do not display significant intraclonal diversity or tendency to accumulate additional somatic mutations over time.8 As such, the leukemia cells that express mutated Ig genes apparently do not evolve from cases that originally expressed unmutated Ig genes. Because of this, the presence or absence of Ig somatic mutations was thought by some to reflect 2 subtypes of CLL, each with distinctive cytogenesis.1,2,9 The subtype that expressed unmutated Ig VH genes was considered to be derived from naive or pre–germinal center B cells, whereas the CLL cells expressing mutated receptors were thought likely to be derived from a post–germinal center or “memory-type” B cells.10
However, molecular analyses of the Ig VH genes expressed by CLL B cells suggest that neither subtype is derived from a naive, nonselected B cell. CLL B cells express an Ig repertoire that appears more restricted than that of adult blood B cells. Several genes, such as the 51p1 allele of VH1-69, are expressed at high frequency,11 are rarely mutated,12 and constitute a large proportion (eg, ≈ 20%) of the cases that lack Ig somatic mutation. In addition, prior studies found that 51p1-expressing CLL B cells preferentially use certain diversity (D) and JH gene segments with restricted reading frames (RFs), encoding relatively long third complementarity determining regions (CDR3s) with conserved amino acid motifs.12 The CDR3s of 51p1-expressing CLL B cells contrasted with those of the Ig heavy chains expressed by CLL B cells that used other Ig VH1 genes or by nonneoplastic tonsillar or blood B cells that used 51p1.12-14 As the CDR3 is the most variable region of the heavy chain and is directly involved in antigen binding, these results suggest that CLL B cells that express unmutated Ig might have been selected by virtue of their capacity to react with an undefined, yet specific, antigen(s). As such, these leukemia cells are not likely to be derived from naive B cells.
To exclude the possibility that the noted restriction in 51p1 used by CLL reflected a small sample bias or a skewed collection of samples obtained at a single institution, we investigated the Ig VH gene used by CLL B cells of nonselected patients monitored by the CLL Research Consortium (CRC). In this large study, we confirm the high level of expression of 51p1 by the CLL B cells and extend these studies to report that a high proportion of patients expressed virtually identical Ig heavy and light chains. This study provides compelling evidence that the Ig expressed by CLL B cells are highly selected and not representative of the Ig used by naive B cells.
Patients, materials, and methods
Patient samples
Blood was collected from consenting patients who satisfied diagnostic and immunophenotypic criteria for B-cell CLL15 and who presented for evaluation at the referral centers of the CLL Research Consortium (CRC). Institutional review board approval and informed consent were obtained for the procurement of the samples in all cases, in accordance with the Declaration of Helsinki. Peripheral blood mononuclear cells were isolated by density gradient centrifugation using Ficoll-Hypaque 1077 (Sigma, St Louis, MO), washed twice, and analyzed directly or suspended in fetal calf serum containing 10% dimethylsulfoxide (DMSO; Sigma) for storage in liquid nitrogen. All samples contained more than 90% CLL B cells as assessed by flow cytometric analyses.
Flow cytometry analyses
CLL B cells (5 × 105) were stained with optimized amounts of monoclonal antibodies (mAbs) conjugated with fluorescein isothiocyanate, phycoerythrin, or allophycocyanin (BD PharMingen, La Jolla, CA) specific for human CD19, CD3, CD5, CD38, Igκ, and Igλ, or with fluorochrome-conjugated isotype control antibodies of irrelevant specificity. Cells were examined by 4-color, multiparameter flow cytometry using a dual laser FACScalibur (Becton Dickinson, San Jose, CA). Data were analyzed using FlowJo analysis software (Treestar, San Carlos, CA). Viable lymphocytes were defined by exclusion of propidium iodide and light scatter characteristics.
VH and VL gene analyses
Total cellular RNA was isolated from 5 × 106 CLL B cells using RNeasy reagents (Qiagen, Valencia, CA), per the manufacturer's instructions. First-strand cDNA was synthesized from one third of the total purified RNA using an oligo-dT primer and Superscript II RT (Life Technologies, Grand Island, NY). The remaining RNA was removed with RNase H and the cDNA purified using QIAquik purification columns (Qiagen). The purified cDNA was poly-dG–tailed using deoxyguanosine triphosphate and terminal deoxytransferase (Roche, Indianapolis, IN). The VH gene expressed by the CLL B cells was determined by reverse transcription–polymerase chain reaction (RT-PCR) enzyme-linked immunosorbent assay (ELISA) technique.16,17 The cDNA from each sample was amplified using VH or Vκ family-specific primers for the sense strand of the gene of interest and antisense Cμ or Cκ consensus primers. The PCR products were size selected by electrophoresis in 2% agarose containing 0.5 μg/mL ethidium bromide (Life Technologies). The expected products were excised and purified using Geneclean III (BIO 101, Carlsbad, CA). Most PCR products were sequenced directly, although in several cases, amplified products were cloned into pGEM-T (Promega, Madison, WI) and analyzed, as described.18 Nucleic acid sequence analyses were conducted using the fluorescence-dideoxy-chain-termination method and an Applied Biosystems 377 automated nucleic acid sequence analyzer (ABI, Foster City, CA). Nucleotide sequences were analyzed using DNASTAR (DNASTAR, Madison, WI) and compared with the sequences deposited in the V BASE and GenBank sequence databases. Somatic mutations were identified by comparison with the most homologous germ-line VH gene. The percent of homology was calculated by counting the number of nucleotide differences between the 5′ end of framework 1 (FW1) and the 3′ end of FW3. Cloned Ig VH genes with less than 98% homology with the corresponding germ-line Ig VH sequence were considered mutated. The method of Corbett et al was used to assign D genes of the longer gene families (D2 and D3), and 7 consecutive nucleotides were used for the shorter D gene families.19 Heavy chain CDR3 (HCDR3) length was determined by the method of Kabat et al20 and defined by the number of amino acids between codon 94 at the end of FW3 and the conserved Trp of position 102 at the beginning of FW4. Light chain CDR3 (LCDR3) length was defined by the number of amino acids between codon 89 at the end of FW3 and the conserved Phe of position 97 at the beginning of FW4. Cluster analysis of all sequences was performed using MegAlign (DNASTAR).
Results
We performed RT-PCR ELISA on 1220 CLL samples from unrelated patients to determine the expressed Ig VH gene subgroups. We found that 1188 (97%) of the CLL samples expressed Ig heavy chains (IgH) encoded by VH genes of a single VH family. This analysis also revealed that the most common VH subgroup used was VH3 (seen in 571 cases or 48.1% of the total), followed by VH1 (319 or 26.8%) and VH4 (241 or 20.2%) (Table 1). The VH2, VH5, VH6, and VH7 subgroups were used less frequently, accounting for 29 (2.4%), 16 (1.3%), 8 (0.7%), and 4 (0.3%) samples, respectively. In addition, we performed immunophenotypic analysis of all samples by flow cytometry. Of the samples, 64% (730 of 1138) and 36% (408 of 1138) expressed kappa and lambda light chains, respectively, reflecting the 3:2 ratio of kappa to lambda light chains noted for B cells of healthy adults.
Relative expression of Ig VH and VL gene subgroups
. | . | Associated light chain . | . | |
|---|---|---|---|---|
| VH subgroup . | No. (%) . | Kappa (%*) . | Lambda (%*) . | |
| Total | 1188 (100) | 730 (64) | 408 (36) | |
| VH1 | 319 (27) | 224 (72) | 87 (28) | |
| VH2 | 29 (2) | 14 (48) | 15 (52) | |
| VH3 | 571 (48) | 336 (62) | 206 (38) | |
| VH4 | 241 (20) | 138 (60) | 92 (40) | |
| VH5 | 16 (1) | 10 (67) | 5 (33) | |
| VH6 | 8 (0.7) | 6 (75) | 2 (25) | |
| VH7 | 4 (0.3) | 2 (67) | 1 (33) | |
. | . | Associated light chain . | . | |
|---|---|---|---|---|
| VH subgroup . | No. (%) . | Kappa (%*) . | Lambda (%*) . | |
| Total | 1188 (100) | 730 (64) | 408 (36) | |
| VH1 | 319 (27) | 224 (72) | 87 (28) | |
| VH2 | 29 (2) | 14 (48) | 15 (52) | |
| VH3 | 571 (48) | 336 (62) | 206 (38) | |
| VH4 | 241 (20) | 138 (60) | 92 (40) | |
| VH5 | 16 (1) | 10 (67) | 5 (33) | |
| VH6 | 8 (0.7) | 6 (75) | 2 (25) | |
| VH7 | 4 (0.3) | 2 (67) | 1 (33) | |
The percent of the VH subgroup.
We focused on the 319 CLL samples found to express the VH1 subgroup, in particular those that expressed IgH encoded by VH1-69. There are several alleles at the VH1-69 locus that can be grouped into 2 major alleles (eg, 51p1 versus 1263) based upon nonconservative differences in the second complementarity determining region (CDR2) that can be recognized by anti-idiotypic mAb, G6.13,21 We performed RT-PCR ELISA using allele-specific primers to detect CLL samples that expressed 51p1-like alleles of VH1-69. All samples typed as expressing 51p1-like alleles of VH1-69 also were typed as expressing VH genes of the VH1 subgroup. We found that 164 cases (13.8% of all samples and 51.4% of VH1-expressing CLL samples) used 51p1-like alleles of VH1-69 using this method. Immunophenotypic analysis of these samples by flow cytometry revealed that 72% (118 of 164) and 26.8% (44 of 164) of the samples expressed kappa and lambda light chains, respectively.
To analyze the mutational status and molecular characteristics of the CDR3, we sequenced all VH1 genes identified as VH1-69 by RT-PCR ELISA. All samples were found to have a single functional IgH rearrangement. Of the 164 patient samples, 163 expressed IgH encoded by unmutated 51p1-related alleles. Of the 5 51p1-related alleles, 4 differed only by single conservative nucleotide changes. Variants 1, 2, 5, and 11 therefore have identical coding regions and were present in 141 (86.5%) of 163 of the unmutated samples. Variant 7 differs from the other 4 alleles by a Glu to Lys substitution in FW3 and was present in 22 (13.5%) samples.
In addition to being predominantly expressed without somatic mutation, the rearrangements of the 51p1-like VH1-69 genes expressed by CLL cells were highly restricted (Figure 1A). The 2 major D gene families, D2 and D3, accounted for the vast majority of the D segments used. Of the D2 family, 4 D segments were used, namely D2-2, D2-8, D2-15, and D2-21, accounting for 25.2%, 2.5%, 5.5%, and 2.5% of all the VH1-69 rearrangements, respectively. Of the D3 family, 5 D segments were used, namely D3-3, D3-9, D3-10, D3-16, and D3-22, accounting for 24.5%, 3.1%, 8.0%, 12.9%, and 4.3% of all the VH1-69 rearrangements, respectively. In contrast, the VH1-69–expressing B cells of healthy adults only seldom use the 3 most frequent D genes identified in this group, namely D2-2, D3-3, and D3-16.14,22,23 In addition, we noted that JH6 was the most frequently used JH gene segment (103 of 163, 63.1%), followed by JH3, then JH4 and JH5 (Figure 1B). This is in contrast to the B cells of healthy adults, which most frequently use the JH4 gene segment, including normal B cells that use the VH1-69 gene.14
Restricted D segment and JH gene use by the 163 CLL B-cell cases that express unmutated Ig heavy chains encoded by 51p1. (A) Relative expression of the most common D segments used by the 51p1-encoded heavy chains expressed in this cohort. Values are expressed as a percentage of the 163 samples found to use each of the different D segments. Some of the D segments found to be frequently used by the heavy chains analyzed in this cohort also display a restricted use of certain reading frames (RF), as indicated by the distribution of RF used for D2-2. (B) Relative expression of the JH genes used by the 51p1-encoded heavy chains expressed in this cohort. Values are expressed as a percentage of the 163 samples found to use genes from each of the JH families.
Restricted D segment and JH gene use by the 163 CLL B-cell cases that express unmutated Ig heavy chains encoded by 51p1. (A) Relative expression of the most common D segments used by the 51p1-encoded heavy chains expressed in this cohort. Values are expressed as a percentage of the 163 samples found to use each of the different D segments. Some of the D segments found to be frequently used by the heavy chains analyzed in this cohort also display a restricted use of certain reading frames (RF), as indicated by the distribution of RF used for D2-2. (B) Relative expression of the JH genes used by the 51p1-encoded heavy chains expressed in this cohort. Values are expressed as a percentage of the 163 samples found to use genes from each of the JH families.
The restricted use of D and JH genes leads to CDR3s with conserved molecular features. As noted in prior studies, the average length of the CDR3 for the VH1-69–expressing CLL cells (19.6 ± 3.0 codons) was significantly longer than that of most Ig gene rearrangements of normal blood B cells, including those that use 51p1-like VH1-69 genes in young (14.6 ± 4.3 codons, n = 39) or aged (16.6 ± 3.4 codons, n = 38) adults (P < .001).14,22,23 Furthermore, there was restricted use of certain reading frames of unmutated D gene segments, resulting in CDR3 that shared amino acid sequence motifs (Table 2). The motif DIVVVPAA(I/M), for example, was found in the CDR3 of 20 (74%) of the 27 VH1-69–encoded heavy chains that used the D2-2 gene segment and JH6, resulting from restricted use of this D gene segment's third reading frame. Of these 20 Ig heavy chains, 16 also had long CDR3s of 19 or 20 amino acids. Similarly, 15 (83%) of the 18 51p1-encoded heavy chains that used D3-3 and JH6 shared the CDR3 motif GGYDFWSGYY, owing to the restricted use of the second reading frame of D3-3.
Common CDR3 amino acid motifs found in 51p1-encoded heavy chains
. | . | . | . | . | Associated light chain . | . | Specific Vκ genes . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| CDR3 amino acid sequence motif . | D gene . | RF . | JH gene . | Sample no. . | Lambda (%*) . | Kappa (%*) . | A27 (%†) . | O2 (%†) . | ||
| DIVVVPAA(I/M) | D2-2 | 3 | JH6 | 20 | 4 (20) | 16 (75) | 0 (0) | 10 (63) | ||
| YDFWSGY | D3-3 | 2 | JH3/4 | 6 | 0 (0) | 6 (100) | 1 (17) | ND | ||
| GGYDFWSGYY | D3-3 | 2 | JH6 | 15 | 10 (67) | 5 (33) | 1 (20) | 0 (0) | ||
| TIFGVVI | D3-3 | 3 | JH4/6 | 10 | 2 (20) | 8 (80) | 1 (13) | ND | ||
| GGXYDY(I/V)WGSYRPNDAFDI | D3-16 | 2 | JH3 | 15 | 0 (0) | 15 (100) | 15 (100) | 0 (0) | ||
. | . | . | . | . | Associated light chain . | . | Specific Vκ genes . | . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| CDR3 amino acid sequence motif . | D gene . | RF . | JH gene . | Sample no. . | Lambda (%*) . | Kappa (%*) . | A27 (%†) . | O2 (%†) . | ||
| DIVVVPAA(I/M) | D2-2 | 3 | JH6 | 20 | 4 (20) | 16 (75) | 0 (0) | 10 (63) | ||
| YDFWSGY | D3-3 | 2 | JH3/4 | 6 | 0 (0) | 6 (100) | 1 (17) | ND | ||
| GGYDFWSGYY | D3-3 | 2 | JH6 | 15 | 10 (67) | 5 (33) | 1 (20) | 0 (0) | ||
| TIFGVVI | D3-3 | 3 | JH4/6 | 10 | 2 (20) | 8 (80) | 1 (13) | ND | ||
| GGXYDY(I/V)WGSYRPNDAFDI | D3-16 | 2 | JH3 | 15 | 0 (0) | 15 (100) | 15 (100) | 0 (0) | ||
Underlined letters (X) indicate the presence of that amino acid in more than 67% of the samples.
ND indicates not determined.
The percent of the VH subgroup.
The percent of the Vκ subgroup.
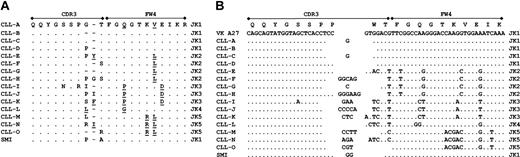
The most striking sequence homology, though, was noted for VH1-69–expressing CLL cell samples that used the D3-16 gene segment along with JH3. By cluster analysis of the amino acid sequence of all 51p1-expressing samples, we identified 15 distinct CLL samples from unrelated patients that used virtually identical heavy chains. Each heavy chain rearrangement has a CDR3 that is 19 amino acids in length and is encoded by D3-16 and JH3 genes. They all used the second reading frame (RF2) and had an amino acid sequence that was highly conserved in the CDR3, namely GG(X)YDY(I/V)WGSYR(P/S)NDAFDI (Figure 2A). Although they have virtually identical amino acid sequences, they were each derived from distinct genomic rearrangements (Figure 2B). For the CDR3 of each sample, 8 and 5 amino acids are encoded by D3-16 and JH3, respectively. Remarkably, although the other 6 amino acids are encoded by nontemplated nucleotides (N segments) that were added during the process of Ig gene rearrangement, 3 amino acids at the VH-D junction of each of these 15 samples are encoded by N segments, each encoding the same 2 amino acids (Gly-Gly) in the first 2 positions of CDR3, and likely a result of insertion of guanosine and cytidine by terminal deoxynucleotidyl transferase. CLL-H has a Tyr in the third position that is encoded by the D3-16 gene. The others have various amino acids in the third position that are the result of exonuclease activity and nucleotide addition at the 5′ end of the D gene, and account for the primary difference between the 15 heavy chains. N segments also encode 3 amino acids at the D-JH junction, and encode the same 3 amino acids (Arg-Pro-Asn) for 13 of 15 CLL samples, and (Arg-Ser-Asn) for the other 2. The processing of the D segment by exonuclease activity for all samples is remarkably similar, as the last 2 codons encoded by D3-16 are removed, and each results in the Arg in position 12. The Pro in position 13 is encoded only by N segments, whereas the 2 samples with Ser likely use the thymidine from the D3-16 gene as the first nucleotide in this codon, and nontemplated nucleotides in the other 2. The Asn in position 14 of all rearrangements is likely encoded by a combination of N segment addition and the thymidine from the 5′ end of the JH3 gene. Although 15 of the 21 amino acids can be encoded by a codon with a thymidine in the third position, all but one have Asn. These Ig heavy chains were virtually identical to those expressed by one other CLL sample noted in our prior studies, designated SMI.24 This previously identified CLL sample also expressed a particular Vκ that frequently is expressed in CLL, namely A27, which encodes the cross-reactive idiotype (CRI) recognized by the mAb 17.109.25
Amino acid and nucleotide sequences of the CDR3 of virtually identical Ig heavy chains expressed by unrelated CLL patients. Alignments of the amino acid (A) and nucleotide (B) sequences of the CDR3 of the fifteen 51p1-encoded heavy chains are shown, compared with CLL sample A (CLL-A). Dots indicate homology at that position. Listed on the left margin is the designation for each CLL sample. Labels above indicate the relative positions of FW3, CDR3, and FW4. The sequences of the CDR3 of the SMI heavy chain and the germ-line sequences of the D3-16 and JH3 genes are included at the bottom for reference.
Amino acid and nucleotide sequences of the CDR3 of virtually identical Ig heavy chains expressed by unrelated CLL patients. Alignments of the amino acid (A) and nucleotide (B) sequences of the CDR3 of the fifteen 51p1-encoded heavy chains are shown, compared with CLL sample A (CLL-A). Dots indicate homology at that position. Listed on the left margin is the designation for each CLL sample. Labels above indicate the relative positions of FW3, CDR3, and FW4. The sequences of the CDR3 of the SMI heavy chain and the germ-line sequences of the D3-16 and JH3 genes are included at the bottom for reference.
We noted that each of the CLL samples identified in this study that expressed virtually the same Ig heavy chains also expressed kappa light chains (Table 2). Because of this we isolated the kappa light chain cDNA from each for nucleic acid sequence analyses. This analysis revealed that each of the CLL samples expressed the same Vκ gene, namely A27. This Vκ gene also was the same one used by the prototype CLL IgM protein, SMI. All samples were derived from distinct genomic rearrangements (Figure 3B), and, similar to SMI, all A27 encoded Vκ light chains did not have any Ig somatic mutations. The restriction of Jκ genes was less than for the heavy chains, as A27 was rearranged 4 times each with Jκ1 or Jκ2 gene segments, 3 times each with Jκ3 and Jκ5, and once with Jκ4. However, because these Jκ gene segments encode highly homologous amino acid sequences, the CDR3 segments of the rearranged A27 light chains used by these CLL samples also were highly similar in both length and amino acid sequence composition (Figure 3). In contrast, only 13 (13%) of the other 102 samples that expressed unmutated 51p1-encoded heavy chains expressed A27-encoded light chains (Table 2). Instead, the other 51p1-encoded heavy chains that had common CDR3 motifs other than GG(X)YDY(I/V)WGSYR(P/S)NDAFDI appeared to have restricted pairing with light chains encoded by V genes other than A27. For example, 10 (50%) of the 20 51p1-encoded Ig heavy chains with the CDR3 motif of DIVVPAA(I/M) were paired with kappa light chains with variable regions encoded by the unmutated VκO2 gene, a Vκ gene of the VκI gene subgroup that is quite distinct from VκA27. Moreover, 10 (56%) of the 18 samples that express 51p1-encoded heavy chains with the CDR3 motif of GGYDFWSGYY are paired with lambda light chains. As such, the skewed association of VκA27 with the 51p1-encoded heavy chains with the highly conserved CDR3 of GG(X)YDY(I/V)WGSYR(P/S)NDAFDI is not a general property of all 51p1-encoded heavy chains.
Amino acid and nucleotide sequences of the CDR3 of virtually identical Ig kappa light chains expressed by unrelated CLL patients. Alignments of the amino acid (A) and nucleotide (B) sequences of the CDR3 of the 15 Vκ A27-encoded light chains are shown, compared with CLL sample A (CLL-A). Dots indicate homology and dashes indicate no sequence at that position. Listed on the left margin is the designation for each CLL sample. Indicated on the right is the most homologous JK gene. Underlined letters (X) designate amino acid differences between the germ-line JK genes, relative to JK1. Labels above indicate the relative positions of FW3, CDR3, and FW4. The sequences of the CDR3 of the SMI light chain and the germ-line sequences of Vκ A27 are included for reference.
Amino acid and nucleotide sequences of the CDR3 of virtually identical Ig kappa light chains expressed by unrelated CLL patients. Alignments of the amino acid (A) and nucleotide (B) sequences of the CDR3 of the 15 Vκ A27-encoded light chains are shown, compared with CLL sample A (CLL-A). Dots indicate homology and dashes indicate no sequence at that position. Listed on the left margin is the designation for each CLL sample. Indicated on the right is the most homologous JK gene. Underlined letters (X) designate amino acid differences between the germ-line JK genes, relative to JK1. Labels above indicate the relative positions of FW3, CDR3, and FW4. The sequences of the CDR3 of the SMI light chain and the germ-line sequences of Vκ A27 are included for reference.
Discussion
Previous studies have identified restriction in the VH genes used in CLL, particularly those that express unmutated heavy chains.15,26,27 These studies have defined that not only is Ig VH gene use in CLL not random, but that CLL cells that express unmutated Ig VH genes preferentially use certain Ig VH genes,11,28 whereas CLL B cells belonging to the mutated subset frequently express a different set of Ig VH genes.28,29 However, Ig VH gene use in CLL must be compared with Ig VH gene use by normal B cells or the normal counterpart to the CLL B cell. It is clear that VH gene use in normal B cells is also nonrandom. However, the frequent use of certain genes might vary between the 2 groups.
Distinction between normal B cells and CLL is made clearer when there is frequent expression of one Ig VH coupled with critical evaluation of junctional diversity in CDR3. Our previous studies have shown that VH1-69 is frequently expressed in CLL,11 and that 51p1-encoded heavy chains preferentially use certain D and JH gene segments with restricted reading frames, encoding relatively long CDR3 regions with distinct amino acid motifs.12 These features are unique but not exclusive to CLL, as they are infrequently present in normal tonsillar and blood B cells of young14 and aged22,23 healthy adults. To exclude the possibility that the noted restriction reflected a small sample bias, we investigated the Ig VH gene used by the CLL B cells of a larger cohort of nonselected patients monitored by the CLL Research Consortium. In this large cohort of CLL samples, not only do we find similar restricted expression of 51p1 as noted in previous reports, we found that approximately 1.3% of all cases expressed virtually identical Ig.
The expression of these conserved heavy chains is not a consequence of chance, as the frequency of coexpression of these 3 distinct heavy chain genes alone is 1 in 7650 (1/51 × 1/25 × 1/6), representing the probability of using each particular gene relative to the total number of functional genes in each group. This probability is further diminished by similar consideration of 1 in 200 for rearrangement of a specific Vκ and Jκ gene (1/40 × 1/5). Therefore, one would expect that only 1 of every 1 530 000 B cells would randomly express the heavy and light chains encoded by the same 5 genes. This estimate is also without regard to generation of junctional diversity, which would decrease the probability to less than 10–12.
Expression of these Ig heavy chains also is not associated with aging, as similar 51p1-encoded heavy chains are not prevalent in a recent study of healthy aged individuals.22,23 Also, these samples are not a reflection of 51p1 expression in CLL, as neither D3-16 nor JH3 is highly expressed in CLL,12 and these samples represent 15 of the 21 51p1-encoded heavy chains that use D3-16 and 15 of the 26 that use JH3. It is also not a reflection of preferential rearrangement of certain genes, as D3-16 and JH3 are not frequently expressed in the normal B-cell repertoire19,30,31 or in CLL overall. Rather it is likely a result of selection, as preferential rearrangement would not provide for the conservation of junctional diversity noted in these heavy chain rearrangements.
The highly restricted and virtually identical structure of the B-cell receptors (BCRs) expressed by this set of CLL B cells strongly suggests selection for a particular reactivity. As CLL B cells frequently express IgM antibodies that display reactivity to self-proteins,24,32,33 including several 51p1-encoded Ig expressed in CLL that are polyreactive,24,34-37 we and others have previously suggested that expression of Ig with restricted CDR3 structures may influence the development of CLL. As noted previously, the heavy and light chains presented here are virtually identical to those of SMI, a previously characterized CLL that expresses a polyreactive IgM/κ autoantibody with low affinity for a number of self-antigens, including human IgG.24 The SMI heavy chain also was encoded by the 51p1 variant 7 that has a Glu to Lys replacement in FW3, which distinguishes this allele from the other 4 51p1-like variants. Variant 7 is also used by 10 of the 15 samples that express 51p1-encoded heavy chains that are identified here, but this number represents almost half of those that express this allele, which is only 22 of the 164 samples analyzed. In addition, the CDR3 of the SMI heavy chain is homologous with the 15 cases identified in this study to express homologous Ig.
Previous Ig structure-function studies that used the SMI IgM/κ, and other 51p1 and A27-encoded heavy and light chains produced as transfectoma proteins, demonstrated that polyreactivity did not merely result from the use of certain combinations of Ig heavy and light chains encoded by particular germ-line V genes.24 Binding reactivity also was dependent upon the sequences of the CDR3 that were generated during Ig rearrangement. Moreover, mutational analysis of selected amino acid residues also demonstrated that certain CDR3 residues were critical for polyreactivity, as several single amino acid substitutions could dramatically alter the specificity of the SMI autoantibody.38,39 As such, such polyreactive self-reactivity is a selected specificity.
Engagement of the SMI-like BCR with self-antigen or autoantigen might influence the fate and/or differentiation of B cells in lymphoid tissues. Evidence for this comes from transgenic mice that express the SMI IgM/κ antibody.40 Transgenic expression of the SMI IgM/κ induced mouse B cells to differentiate into nonnaive, memory-type B cells that were hyperresponsive to nonspecific T-cell help. SMI mice, but not control mice that express a human Ig in the absence of specific antigen, had increased numbers of human Ig transgene–expressing B1 and marginal zone (MZ) B cells. Such cells were found in the periarteriolar sheath, MZ, and interfollicular areas of the spleen that typically are populated by memory or antigen-experienced B cells. SMI B cells could secrete human IgM/κ when provided with nonspecific T-cell help alone, further demonstrating that these cells are antigen experienced, rather than naive B cells.
In this regard, it is noteworthy that microarray studies demonstrated that both subtypes of CLL share a common gene expression profile that is distinct from that of other B-cell lymphomas or normal blood B cells.41,42 This CLL gene expression profile appeared similar to that of MZ B cells of secondary lymphoid tissues. In addition, several genes found expressed in CLL B cells are typically up-regulated as a consequence of BCR ligation, suggesting that CLL B cells might be stimulated via the Ig receptor in vivo. Although expression of Ig such as SMI IgM/κ might not in itself result in leukemia in transgenic mice, the continuous low-level stimulation afforded by expression of such self-reactive Ig receptors still appears to enhance B-cell turnover and/or survival,43 potentially increasing the risk for neoplastic transformation.
The restricted repertoire of the Ig expressed in CLL potentially could be exploited for development of idiotype-directed immune therapy. CLL cells of unrelated patients frequently express Ig with common CRI that can be recognized by mAbs.44,45 Studies were performed testing the biologic activity of such mAbs in the SMI/κ transgenic mouse. Although the B cells expressing the SMI IgM/κ transgenes expressed relatively low levels of surface human Ig, these transgene-expressing B cells still could be selectively deleted in vivo by treatment with mAbs specific for the CRI of the SMI IgM/κ.40 Conceivably, anti-idiotypic mAbs could be used during early pathogenesis to target cells that express CRI of Ig associated with an increased risk for neoplastic transformation. In addition, vaccine strategies also could be developed to induce cellular immune responses against peptide epitopes of common motifs found within the CDR3 of Ig frequently expressed in CLL. Conceivably, batteries of mAb anti-CRI reagents and/or peptide vaccines could be assembled for immune therapy of a relatively large proportion of patients with this disease.
Prepublished online as Blood First Edition Paper, June 24, 2004; DOI 10.1182/blood-2004-03-0818.
Supported in part by National Institutes of Health grant PO1-CA81534 for the CLL Research Consortium, and R37-CA49870 (T.J.K.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the investigators who have contributed samples to the CRC Tissue Core, Andrew Greaves for management of the Tissue Core Management System, and Esther Avery for excellent technical assistance.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal