Abstract
Fms-like tyrosine kinase 3 (FLT3) mutations are associated with unfavorable outcomes in children with acute myeloid leukemia (AML). We used DNA microarrays to identify gene expression profiles related to FLT3 status and outcome in childhood AML. Among 81 diagnostic specimens, 36 had FLT3 mutations (FLT3-MUs), 24 with internal tandem duplications (ITDs) and 12 with activating loop mutations (ALMs). In addition, 8 of 19 specimens from patients with relapses had FLT3-MUs. Predictive analysis of microarrays (PAM) identified genes that differentiated FLT3-ITD from FLT3-ALM and FLT3 wild-type (FLT3-WT) cases. Among the 42 specimens with FLT3-MUs, PAM identified 128 genes that correlated with clinical outcome. Event-free survival (EFS) in FLT3-MU patients with a favorable signature was 45% versus 5% for those with an unfavorable signature (P = .018). Among FLT3-MU specimens, high expression of the RUNX3 gene and low expression of the ATRX gene were associated with inferior outcome. The ratio of RUNX3 to ATRX expression was used to classify FLT3-MU cases into 3 EFS groups: 70%, 37%, and 0% for low, intermediate, and high ratios, respectively (P < .0001). Thus, gene expression profiling identified AML patients with divergent prognoses within the FLT3-MU group, and the RUNX3 to ATRX expression ratio should be a useful prognostic indicator in these patients. (Blood. 2004;104:2646-2654)
Introduction
Genomewide analysis of diagnostic bone marrow samples from patients for whom therapy fails may provide insight into particular profiles of gene expression that determine outcome. Understanding the relationships of known biologic markers to outcome using genomic profiles should help develop rational targeted therapies.
In acute myeloid leukemia (AML), mutations of the Fms-like tyrosine kinase 3 (FLT3) receptor are markers of poor outcomes. Several studies in children and adults suggested that FLT3 internal tandem duplication (FLT3-ITD) confers poor prognosis in AML.1-6 The FLT3 receptor is a member of the class III receptor tyrosine kinase (RTK) family.7,8 High FLT3 receptor expression has been reported in more than 60% of precursor B-cell acute lymphocytic leukemia and AML cases,9-11 and it is the most frequently mutated RTK2,4,6 in adult and childhood de novo AML. Most FLT3 mutations result in constitutive activation and autophosphorylation of the FLT3 receptor. This results in downstream activation of the signal transducer and activator of transcription 5 (STAT5), phosphatidylinositol 3 (PI-3) kinase, and Ras pathways.6,12-23 Activation of these pathways leads to enhanced proliferation and resistance to apoptosis.24,25
There are 2 types of FLT3 receptor mutations that have been described: internal tandem duplications (ITDs) in the juxtamembrane region26-31 and activation loop mutations (ALMs) in the tyrosine kinase domain.32-34 The incidence of FLT3-ITD is 11% to 16% in children3,5 and 20% or more in adults2,35 ; whereas, the incidence of point mutations is 7% to 8% in adults and children.6,35 In studies by Whitman et al4 and Liang et al,5 a higher ratio of ITD to wild-type (WT) allele had a strong correlation with poor outcome, whereas a lower ratio correlated with an outcome similar to a FLT3-WT receptor. Meshinchi et al3 reported that FLT3-ITD correlated highly with refractoriness to induction therapy in the Children's Cancer Group study 2891. Zwaan et al36 reported similar results after analyzing a larger set of samples from several Berlin-Frankfurt-Müenster and Dutch Childhood Leukemia Group studies. As with most biologic markers, FLT3-ITD and FLT3-ALM convey a high but not absolute risk of poor outcome.
In this study, we used DNA microarray analysis to identify a pattern of gene expression associated with a high-risk of treatment failure in specimens with FLT3 mutations (FLT3-MUs) in childhood AML patients. There are 2 genes included in this expression profile that are also predictive of outcome when analyzed separately and provide insight into a possible mechanism by which FLT3-MUs confer a poor prognosis.
Patients, materials, and methods
Arrays
DNA microarrays are based on IMAGE clones37 prepared by Research Genetics (Huntsville, AL). Microarrays from several print batches were used, comprising 42 749 elements (42K), representing 32 275 unique Unigene clusters (Build no. 158, released on January 18, 2003). All arrays were printed at the Stanford University School of Medicine according to the Stanford Functional Genomics Facility protocols.
Childhood de novo acute myeloid leukemia specimens
The specimens used in this study were archived at the Children's Oncology Group AML cell bank from Pediatric Oncology Group (POG) study 9421. Institutional review board (IRB) approval was obtained for gene expression profiling and correlative studies. The total number of eligible patients enrolled in POG-9421 was 622, and 353 specimens were archived for further studies. In this study we used specimens from 81 patients with high-quality RNA, selected from among 3 clinical outcome groups: refractory to induction (IR) with daunorubicin, cytarabine, and thioguanine (n = 23); relapse (R) any time after achieving postinduction remission (n = 30); and continued complete remission (CCR) for more than 3 years (n = 28). A disproportionately large number of IR patients were included to have a numerically balanced set to compare with patients who achieved remission (R and CCR groups). An additional 19 paired specimens were collected at relapse from among the 30 patients who had initial remissions. The total number of specimens collected at diagnosis was 81, and the total number of samples was 100 (81 at diagnosis and 19 at relapse).
Analysis of FLT3 status
RNA and genomic DNA were obtained from bone marrow specimens using the Qiagen RNEasy and DNEasy Kits (Valencia, CA) according to the manufacturer's protocol. Analyses methods for FLT3-ITD and FLT3-ALM are published.3
Common reference
A common reference control for each microarray analysis consisted of equal amounts of total RNA from 11 human cancer cell lines (Stratagene, Cedar Creek, TX). Reference amplified RNA (aRNA, 2 μg) was labeled with cyanin 3-deoxyuridine triphosphate (Cy3-dUTP) in each experiment.
Isolation of RNA
Total RNA was isolated using Qiagen RNEasy Midi Kit. The amount of starting material varied from 20 to 100 × 106 cells per sample. The average blast percentage for all diagnostic samples was 86%, with 5 of 100 samples between 51% and 71%, and 2 of 100 samples between 25% and 50%. Poly(A)+ RNA was then amplified using the MessageAmp aRNA Kit from Ambion (Austin, TX). The total and amplified RNA quality was assessed by examining the size distribution of the 28S and 18S ribosomal RNA bands on 1% agarose gel electrophoresis, and by using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Labeling and hybridization
For all specimens, 2 μg amplified total RNA (aRNA) was used for labeling with Cy5-dUTP and hybridization. The aRNA was reverse transcribed with Superscript II (Invitrogen Life Technologies, Carlsbad, CA) using a random hexamer primer (Operon Technologies, Alameda, CA). Each sample was compared with the amplified common reference total RNA labeled with Cy3-dUTP.38-41 Fluorescent dyes were purchased from Amersham Pharmacia Biotech (Piscataway, NJ). Hybridizations were done as published.40 Complete experimental details may be found at http://cmgm.stanford.edu/pbrown/protocols/5_hyb_human.html.42
Data analysis and clustering
Data selection. Data were analyzed using GenePix 3.0 and 4.0 (Axon Instruments, Alameda, CA). Spots with aberrant measurements due to array artifacts or poor quality were manually flagged and removed from further analysis. A filter was applied to omit measurements when fluorescent signal from the DNA spot was less than 20% above the measured background fluorescence surrounding the printed DNA spot in the Cy3 and Cy5 channels. Genes that did not meet these criteria for at least 80% of the measurements across the cases were excluded from further analysis. Data were retrieved as log2(Cy5/Cy3). The (Cy5/Cy3) ratio is defined in the Stanford Microarray Database (SMD) as the normalized ratio of the background-corrected intensities.43
Data selection for the FLT3-MU/WT and clinical outcome analysis. For our set of 100 samples, genes were selected for further analysis if they displayed at least a 2½-fold variation from their mean expression value for all samples, in at least one of the samples. Arrays and genes were clustered by Pearson correlation using a noncentered metric. Only spots with fluorescent signal at least 2-fold greater than the local background were included in the analysis. In addition, genes that did not meet these criteria for at least 80% of measurements across cases were excluded from further analysis. Results were 23 867 genes used for statistical analyses with predictive analysis of microarrays (PAM) and significance analysis of microarrays (SAM).
The data set was adjusted to remove array batch, date of amplification, and date of hybridization bias by 3-way analysis of variance (ANOVA) with a simple additive model, Yijkl = μ + Bi + Hj + Ak + ϵijkl, where Yijkl is the lth observation of the ith batch, the jth date of hybridization, and the kth date of amplification. The errors were assumed to be independent and normally distributed with mean zero and variance σ2. The residuals were taken as the true signals after accounting for biases.
Predictive analysis of microarrays (PAM). PAM analysis is a modified version of the nearest-centroid method. Complete details of this method were published recently.44 We estimated missing values across the 23 867 genes selected for the set of 100 samples, using the K nearest neighbor (KNN) impute engine as published.44,45 PAM was carried out using the statistical package R.
Significance analysis of microarrays (SAM). SAM is a statistical approach to identify genes for which expression patterns are associated significantly with specific characteristics of sample sets.47
Complete details of statistical methods for PAM and SAM may be found in Lacayo et al (http://genome-www5.stanford.edu/cgi-bin/publication/viewPublication.pl?pub_no=351) under the respective subheadings.47 PAM and SAM analyses were applied to subsets from our 100-sample data set to examine differential expression patterns between FLT3-WT and MU (ITD and ALM) samples, as well as in the comparative analysis of gene expression among FLT3-MU cases with death or survival as outcomes.
Statistical methods
Data from POG-9421 through December 2002 were used to compare subgroups with and without FLT3 mutations. Event-free survival (EFS) was defined as the length of time from registration to treatment failure (relapse or failure to enter remission). Among 81 cases, there were 2 patients who died early and 3 with relapses that coincided with death. Overall survival (OS) was defined as the time from registration to death. Except for 9 of 81 cases, all deaths were more than 65 days after relapses. Event-free survival curves were constructed with the Kaplan-Meier method. Findings with respect to EFS were tested for significance with the log-rank test. Reported P values are 2-sided.
Quantitative polymerase chain reaction
Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) was done with the ABI 7700 system with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) as follows: nonamplified total RNA was purified further with a DNA-free Kit (Ambion) that included DNase I digestion to remove any genomic DNA contamination. Human RUNX3, ATRX, and GAPDH primers (Applied Biosystems) were used in 3-step (95° for 10 seconds → 55° for 30 seconds → 72° for 30 seconds) TaqMan 5′→3′ reactions for 40 cycles. A non-RT control was included to rule out genomic DNA contamination. A nontemplate reaction also was included. Transcript quantification was performed in triplicate for every sample and reported relative to GAPDH. The primers used for RUNX3 were as follows: forward, 5′AGCGTGGGAGGCACCTTT-3′; reverse, 5′-CAGGATCAGGAAGGGTTAGTAATCT-3′. For the ATRX, the primers were as follows: forward, 5′-CATCATCTAGTTGAACTTTGGCATTT-3′; reverse, 5′-CAATAAACGGCCAGAATTT-CCA-3′. For GAPDH, the primers were as follows: forward, 5′-TGCACCACCAACTGCTTAGC-3′; reverse, 5′-GGCATG-GACTGTGGTCATGAG-3′.
Gene expression array data were calculated using the following formula: RUNX3 expression on array = log2 (RUNX3 sample [Cy5]/reference [Cy3]); the ATRX expression on array = log2 (ATRX sample [Cy5]/reference [Cy3]). Gene ratios on the array were calculated as follows: log2 (RUNX3 sample [Cy5]/reference [Cy3])/log2 (ATRX sample [Cy5]/reference [Cy3]) = 2[log2 (RUNX3 sample [Cy5]/reference [Cy3])]-[log2 (ATRX sample [Cy5]/reference [Cy3])]. For quantitative RT-PCR with 95% efficiency, Ct values were normalized using CtGAPDH for each sample (ΔCtRUNX3 [or ATRX] = Ct RUNX3 [or ATRX] - Ct GAPDH). This was used to calculate RUNX3-to-ATRX ratio as follows: [1:1.95][(ΔCt RUNX3) - (ΔCt ATRX)].
Results
Study population
Patients were selected based on clinical outcome as defined in “Patients, materials, and methods.” Approximately equal numbers of patients with IR disease (n = 23), postremission R (n = 30), and CCR (n = 28) were selected randomly for FLT3 status determination. Population characteristics are summarized in Table 1. Median age was higher for FLT3-MU than FLT3-WT patients. Diagnostic white blood cell (WBC) counts were higher in FLT3-ITD patients than FLT3-ALM or FLT3-WT patients. Prevalence of FLT3-MU was higher in patients in the refractory/relapsed groups than the CCR group. The 3-year EFS for FLT3-MU was 25% compared with 47% for FLT3-WT (P = .12). Among FLT3-MU patients, there was no significant difference in EFS between patients with FLT3-ITD (n = 24, EFS = 22%) and FLT3-ALM (n = 12, EFS = 23%).
Patient characteristics
. | Clinical outcome, no. = 81 . | . | . | FLT3 status, no. = 72 . | . | . | All patients, POG-9421 . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics . | IR . | R . | CCR . | ITD . | ALM . | WT . | . | ||||
| No. | 23 | 30 | 28 | 24 | 12 | 36 | 622 | ||||
| Median age, y (range) | 12.8 (0.1-18.4) | 10.2 (0.4-19.3) | 8.3 (0.5-18.8) | 12.9 (2.5-18.4) | 10.2 (0.4-15.3) | 7.4 (0.1-19.3) | − | ||||
| Median WBC count (range) | 90.2 (8.5-483.0) | 77 (2.1-634.0) | 75.4 (1.5-492.0) | 98.0 (8.5-492.0) | 46.0 (2.1-483.0) | 65.1 (1.3-633.0) | − | ||||
| FAB | |||||||||||
| MO | 0 | 3 | 1 | 3 | 0 | 1 | 25 | ||||
| M1 | 8 | 7 | 9 | 7 | 3 | 11 | 102 | ||||
| M2 | 7 | 6 | 10 | 10 | 3 | 7 | 148 | ||||
| M4 | 6 | 7 | 2 | 3 | 3 | 7 | 91 | ||||
| M5 | 2 | 5 | 4 | 0 | 2 | 8 | 113 | ||||
| M6 | 0 | 1 | 0 | 0 | 0 | 1 | 8 | ||||
| M7 | 0 | 1 | 1 | 1 | 1 | 0 | 49 | ||||
| MX | 0 | 0 | 0 | 0 | 0 | 0 | 29 | ||||
| DS | 0 | 0 | 1 | 0 | 0 | 1 | 57 | ||||
| Cytogenetics | |||||||||||
| Normal | 7 | 6 | 9 | 7 | 5 | 10 | 156 | ||||
| t (8;21) | 1 | 0 | 1 | 0 | 0 | 2 | 59 | ||||
| inv (16) | 0 | 3 | 7 | 1 | 3 | 2 | 38 | ||||
| 11q2 3 | 1 | 6 | 6 | 2 | 1 | 9 | 85 | ||||
| Miscellaneous | 9 | 10 | 3 | 9 | 2 | 10 | 177 | ||||
| NA | 5 | 5 | 2 | 5 | 1 | 3 | 107 | ||||
| 3-y EFS, % (± SE) | 0 | 0 | 100 | 22 (± 9) | 23 (± 12) | 46 (± 8) | 42 (± 2) | ||||
. | Clinical outcome, no. = 81 . | . | . | FLT3 status, no. = 72 . | . | . | All patients, POG-9421 . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics . | IR . | R . | CCR . | ITD . | ALM . | WT . | . | ||||
| No. | 23 | 30 | 28 | 24 | 12 | 36 | 622 | ||||
| Median age, y (range) | 12.8 (0.1-18.4) | 10.2 (0.4-19.3) | 8.3 (0.5-18.8) | 12.9 (2.5-18.4) | 10.2 (0.4-15.3) | 7.4 (0.1-19.3) | − | ||||
| Median WBC count (range) | 90.2 (8.5-483.0) | 77 (2.1-634.0) | 75.4 (1.5-492.0) | 98.0 (8.5-492.0) | 46.0 (2.1-483.0) | 65.1 (1.3-633.0) | − | ||||
| FAB | |||||||||||
| MO | 0 | 3 | 1 | 3 | 0 | 1 | 25 | ||||
| M1 | 8 | 7 | 9 | 7 | 3 | 11 | 102 | ||||
| M2 | 7 | 6 | 10 | 10 | 3 | 7 | 148 | ||||
| M4 | 6 | 7 | 2 | 3 | 3 | 7 | 91 | ||||
| M5 | 2 | 5 | 4 | 0 | 2 | 8 | 113 | ||||
| M6 | 0 | 1 | 0 | 0 | 0 | 1 | 8 | ||||
| M7 | 0 | 1 | 1 | 1 | 1 | 0 | 49 | ||||
| MX | 0 | 0 | 0 | 0 | 0 | 0 | 29 | ||||
| DS | 0 | 0 | 1 | 0 | 0 | 1 | 57 | ||||
| Cytogenetics | |||||||||||
| Normal | 7 | 6 | 9 | 7 | 5 | 10 | 156 | ||||
| t (8;21) | 1 | 0 | 1 | 0 | 0 | 2 | 59 | ||||
| inv (16) | 0 | 3 | 7 | 1 | 3 | 2 | 38 | ||||
| 11q2 3 | 1 | 6 | 6 | 2 | 1 | 9 | 85 | ||||
| Miscellaneous | 9 | 10 | 3 | 9 | 2 | 10 | 177 | ||||
| NA | 5 | 5 | 2 | 5 | 1 | 3 | 107 | ||||
| 3-y EFS, % (± SE) | 0 | 0 | 100 | 22 (± 9) | 23 (± 12) | 46 (± 8) | 42 (± 2) | ||||
DS indicates Down syndrome; NA, not available; -, not applicable.
FLT3 mutations
Among 81 diagnostic samples, there were 24 with ITD and 12 with ALM of codon 835/836 in the FLT3 gene. Of 81 cases, 9 were not evaluated for FLT3-ALM and 2 of 81 were not evaluated for FLT3-ITD status due to sample availability. These samples were censored from analysis. Among 19 relapse samples (data not shown), we identified 2 with de novo FLT3-ITDs and none with de novo FLT3-ALMs. Of 5 patients with FLT3-ALM at diagnosis, 3 did not have FLT3-ALM at relapse, suggesting a loss of the leukemic clone that harbored the FLT3-ALM.
Among 23 IR patients, 11 specimens had FLT3-ITDs (48%) and 3 had FLT3-ALMs (13%). Among 58 patients who achieved remission (R + CCR), there were 7 FLT3-ITDs (25%) and 5 FLT3-ALMs (19%) among R patients (n = 30, 4 not tested for ALM and 2 not tested for ITD and ALM), and 6 FLT3-ITDs (21%), 3 FLT3-ALMs (12%), and 1 case that coexpressed an ITD and ALM among CCR patients (n = 28, 3 not tested for ALM). The sample that coexpressed ITD and ALM was coded as an ALM case.
The total of FLT3-ITD (30%) and ALM (15%) cases at diagnosis in this cohort (72 of 81 patients) analyzed for FLT3 status was larger than expected given the relative incidence of FLT3-ITDs of 11% to 16% and FLT3-ALMs of 8% described in larger cohorts of children with de novo AML. This was the result of the larger number of IR samples selected for our analysis.
Cytogenetics and FLT3 mutations
As expected, there were differences in prevalence of cytogenetic abnormalities among response groups (IR, R, and CCR), which are summarized in Table 1. As reported, there was a strong correlation between IR patients and normal cytogenetics with expression of a FLT3-ITD (57%). Most IR and R patients had miscellaneous cytogenetic findings (41% and 45%, respectively). Among CCR patients, 27% had inv(16). Patients in the R or CCR groups had similar incidence of 11q23 cytogenetic abnormalities (46% each).
Samples with FLT3-ITD had predominantly normal cytogenetics (37%) or miscellaneous cytogenetics (47%). Samples with FLT3-ALM had predominantly normal cytogenetic findings (45%), followed by inv(16) cases (27%) and miscellaneous cytogenetics (18%). Finally, cases with FLT3-WT had an equal incidence of normal, 11q23, and miscellaneous cytogenetic abnormalities (29% each), and lower incidence of inv(16) and t(8;21) (6% each).
FLT3 mutations define a gene expression profile
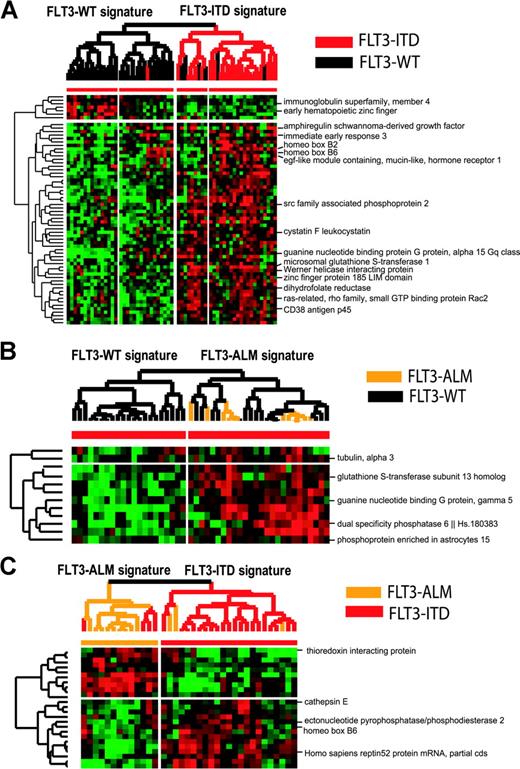
A set of 72 diagnostic samples was analyzed using PAM to identify differences in gene expression among patients with and without FLT3-MU (Figures 1, 2). A gene expression signature was readily identified for FLT3-ITD samples. Only 1 of 24 FLT3-ITD samples was misclassified (misclassification error, 4%). In contrast, the FLT3-WT samples used for comparison were more heterogeneous (misclassification error, 16%). A group of 6 samples with FLT3-WT clustered with FLT3-ITD samples (Figure 1A). A comparison of FLT3-WT samples with FLT3-ALM cases had a higher overall misclassification error (32%). This analysis revealed 2 types of FLT3-WT signatures, 1 more related to FLT3-ALM samples than other FLT3-WT samples (Figure 1B). Next, we used PAM to differentiate FLT3-ITD from FLT3-ALM cases (Figure 1C). ITD and ALM patients had a different mean age and WBC. None of the genes differentially expressed were related directly to the FLT3 receptor signal transduction pathway. A full list of genes differentially expressed is included in the web supplement.
Hierarchic clustering supervised by PAM analysis of FLT3-WT versus FLT3-ITD and FLT3-ALM, and FLT3-ITD versus FLT3-ALM samples. (A) The hierarchic clustering defined by PAM analysis of FLT3-WT versus FLT3-ITD samples; overall misclassification error is 11%. The difference in EFS between these 2 branches is 48% versus 24% (P = 0.1). (B) The differential gene expression between FLT3-WT and FLT3-ALM samples had a higher overall classification error rate, and was unable to differentiate FLT3-ALM samples from all FLT3-WT samples. (C) We show hierarchic clustering of FLT3-ITD versus FLT3-ALM samples. Overall misclassification rate is 19% and differences in gene expression may be related to their different clinical behavior, and to differences in mean age and mean WBC at presentation.
Hierarchic clustering supervised by PAM analysis of FLT3-WT versus FLT3-ITD and FLT3-ALM, and FLT3-ITD versus FLT3-ALM samples. (A) The hierarchic clustering defined by PAM analysis of FLT3-WT versus FLT3-ITD samples; overall misclassification error is 11%. The difference in EFS between these 2 branches is 48% versus 24% (P = 0.1). (B) The differential gene expression between FLT3-WT and FLT3-ALM samples had a higher overall classification error rate, and was unable to differentiate FLT3-ALM samples from all FLT3-WT samples. (C) We show hierarchic clustering of FLT3-ITD versus FLT3-ALM samples. Overall misclassification rate is 19% and differences in gene expression may be related to their different clinical behavior, and to differences in mean age and mean WBC at presentation.
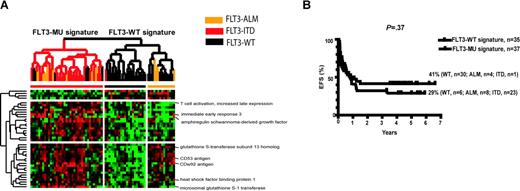
Hierarchic clustering supervised by PAM analysis of FLT3-MUs versus FLT3-WT in 72 diagnostic samples. In this group of patients, FLT3-MU is not a universal prognostic factor of poor outcome because of the IR/R/CCR outcomes in WT and MU FLT3 signature groups. (A) Hierarchic clustering defined by PAM analysis did not include any known genes related to the FLT3 receptor signal transduction pathway. There are 6 samples with the FLT3-WT receptor that have a gene expression profile similar to that of FLT3-MU signature, independent of the FLT3 receptor genotype. One FLT3-ITD also has a gene expression profile similar to FLT3-WT signature, independent of the FLT3 receptor genotype. (B) Pairwise comparison of FLT3-MU versus FLT3-WT shows a trend for different EFS, but it does not achieve statistical significance defined by mutation status or gene expression signatures.
Hierarchic clustering supervised by PAM analysis of FLT3-MUs versus FLT3-WT in 72 diagnostic samples. In this group of patients, FLT3-MU is not a universal prognostic factor of poor outcome because of the IR/R/CCR outcomes in WT and MU FLT3 signature groups. (A) Hierarchic clustering defined by PAM analysis did not include any known genes related to the FLT3 receptor signal transduction pathway. There are 6 samples with the FLT3-WT receptor that have a gene expression profile similar to that of FLT3-MU signature, independent of the FLT3 receptor genotype. One FLT3-ITD also has a gene expression profile similar to FLT3-WT signature, independent of the FLT3 receptor genotype. (B) Pairwise comparison of FLT3-MU versus FLT3-WT shows a trend for different EFS, but it does not achieve statistical significance defined by mutation status or gene expression signatures.
PAM analysis derived a set of 40 genes that were expressed differentially between FLT3-WT and FLT3-MU specimens (Figure 2). The differences in gene expression were in the 0.5- to 1-fold range, and no clusters or genes related to an activated FLT3 receptor were identified. When the 40-gene list was applied to hierarchic clustering of the 72 diagnostic AML specimens, 5 of the 36 FLT3-MU specimens clustered with the FLT3-WT signature group, while 6 of the 36 FLT3-WT specimens clustered with the FLT3-MU signature group, yielding misclassification errors of 16% and 13%, respectively, and 15% overall.
Gene expression profiling supervised by clinical outcome identifies patients with FLT3 mutations at high risk of treatment failure in POG-9421
A set of 42 samples with FLT3-MUs, including the 6 samples from relapse patients with FLT3-MUs, was used to evaluate correlations of gene expression profile with clinical outcome (death versus survival). A signature consisting of 129 genes and expressed sequence tags (ESTs) was derived by PAM analysis from a total of 23 867 genes (Figure 3). The overall misclassification rate for clinical outcome using the 129 genes from PAM was 19% (4% for Group I signature, n = 23 [predominantly IR + R cases]; and 36% for Group II signature, n = 19 [most CCR cases]). SAM analysis also defined a list of 125 genes that discriminated between groups, including 95 that overlapped with the PAM list, with a median false significant number of 5 genes.
Hierarchic clustering FLT3-MU samples supervised by clinical outcome: induction refractory and relapse (Group I) versus continuous complete remission (Group II). (A) Hierarchic clustering based on PAM analysis that differentiates FLT3-MUs with inferior (Group I) versus superior (Group II) clinical outcome. PAM analysis of FLT3-MU samples was supervised by clinical outcome, and the analysis identified several genes involved in transcriptional regulation of gene expression. RUNX3 and ATRX genes, 2 known transcriptional regulators, were selected by PAM and SAM as differentially expressed between FLT3-MU with inferior (Group I) and superior (Group II) clinical outcomes. (B) The statistically different EFS between FLT3-MUs with different gene expression signatures (Group I signature vs Group II signature).
Hierarchic clustering FLT3-MU samples supervised by clinical outcome: induction refractory and relapse (Group I) versus continuous complete remission (Group II). (A) Hierarchic clustering based on PAM analysis that differentiates FLT3-MUs with inferior (Group I) versus superior (Group II) clinical outcome. PAM analysis of FLT3-MU samples was supervised by clinical outcome, and the analysis identified several genes involved in transcriptional regulation of gene expression. RUNX3 and ATRX genes, 2 known transcriptional regulators, were selected by PAM and SAM as differentially expressed between FLT3-MU with inferior (Group I) and superior (Group II) clinical outcomes. (B) The statistically different EFS between FLT3-MUs with different gene expression signatures (Group I signature vs Group II signature).
Table 2 presents a partial list of the genes that were expressed differentially in Group I (predominantly IR + R cases) versus Group II (most CCR cases). Among genes with lower expression in Group II we found the RUNX3, WTAP, and RYBP genes. These genes contribute to transcriptional regulation and cell proliferation. INSIG1 and MAPK62 were 2 genes involved in the cell cycle. The SNRK gene involved in glucose metabolism also was underexpressed. Finally, genes involved in protein kinase activity included PCTK2 and PK1B.
Partial list of genes expressed differentially in specimens from FLT3-MU patients with induction refractoriness or relapse versus CCR as shown inFigure 3
Gene category and name . | Functional class . |
|---|---|
| Lower expression in CCR | |
| RUNX3, runt related transcription factor 3 | Transcriptional regulation, cell proliferation |
| CYLD, cylindromatosis (turban tumor syndrome) | Tumor suppressor gene |
| PCTK2, PCTAIRE protein kinase 2 | ATP binding, protein serine/threonine kinase, transferase activity |
| RAB5A, member RAS oncogene family | Required for the fusion of plasma membranes and early endosomes |
| ETEA, expressed in T cells and eosinophils in atopic dermatitis | Potentiates FAS induced apoptosis |
| SNRK, SNF-1 related kinase | Essential for release from glucose repression, interacts with SNF4, SIP1, SIP2, and GAL83 |
| WTAP, Wilms tumor 1 associated protein | Plays a role in transcriptional and posttranscriptional regulation of genes |
| RYBP, RING1 and YY1 binding protein | Interacts with Polycomb transcriptional repressor complex and with the transcription factor YY1 |
| INSIG1, insulin induced gene 1 | May play a role in tissue growth and differentiation, G0/G1 transition |
| MAPK6, mitogen activated protein kinase 6 | Phosphorylates MAP2, may promote entry into cell cycle |
| RC3, rabconnectin 3 | Calmodulin binding protein |
| PHLDA1, pleckstrin homology-like domain, family A, member 1 | Binds phosphoinositides, involved in platelet cytoskeletal changes |
| PK1B, protein kinase (cAMP-dependent, catalytic) inhibitor beta | May act as a competitive inhibitor of cAMP-dependent protein kinase |
| Higher expression in CCR | |
| ATRX, alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, Saccharomyces cerevisiae) | SWI/SNF chromatin remodeling protein; ATPase/helicase domain; mutations cause changes in pattern of methylation |
| PPP5C, protein phosphatase 5, catalytic subunit | Regulation of RNA biogenesis, or mitosis; dephosphorylates serine residues, and histone H1 |
Gene category and name . | Functional class . |
|---|---|
| Lower expression in CCR | |
| RUNX3, runt related transcription factor 3 | Transcriptional regulation, cell proliferation |
| CYLD, cylindromatosis (turban tumor syndrome) | Tumor suppressor gene |
| PCTK2, PCTAIRE protein kinase 2 | ATP binding, protein serine/threonine kinase, transferase activity |
| RAB5A, member RAS oncogene family | Required for the fusion of plasma membranes and early endosomes |
| ETEA, expressed in T cells and eosinophils in atopic dermatitis | Potentiates FAS induced apoptosis |
| SNRK, SNF-1 related kinase | Essential for release from glucose repression, interacts with SNF4, SIP1, SIP2, and GAL83 |
| WTAP, Wilms tumor 1 associated protein | Plays a role in transcriptional and posttranscriptional regulation of genes |
| RYBP, RING1 and YY1 binding protein | Interacts with Polycomb transcriptional repressor complex and with the transcription factor YY1 |
| INSIG1, insulin induced gene 1 | May play a role in tissue growth and differentiation, G0/G1 transition |
| MAPK6, mitogen activated protein kinase 6 | Phosphorylates MAP2, may promote entry into cell cycle |
| RC3, rabconnectin 3 | Calmodulin binding protein |
| PHLDA1, pleckstrin homology-like domain, family A, member 1 | Binds phosphoinositides, involved in platelet cytoskeletal changes |
| PK1B, protein kinase (cAMP-dependent, catalytic) inhibitor beta | May act as a competitive inhibitor of cAMP-dependent protein kinase |
| Higher expression in CCR | |
| ATRX, alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, Saccharomyces cerevisiae) | SWI/SNF chromatin remodeling protein; ATPase/helicase domain; mutations cause changes in pattern of methylation |
| PPP5C, protein phosphatase 5, catalytic subunit | Regulation of RNA biogenesis, or mitosis; dephosphorylates serine residues, and histone H1 |
ATP indicates adenosine triphosphate; cAMP, cyclic adenosine monophosphate.
Overexpressed genes included ATRX and PPP5C. Both contribute to regulation and modification of chromatin remodeling proteins.
We found a statistically significant difference in EFS and OS between groups defined by hierarchic clustering using the gene expression profile generated by PAM analysis (Figure 3). Group I (22/23 with R [n = 14] or IR [n = 8] cases and 1/23 in CCR) had EFS of 5% compared with 45% for Group II (9/19 in CCR, 5/19 with R, and 5/19 IR; P = .018). For Group I, the OS was 16% versus 63% for Group II (P = .014). The distribution of ALM and ITD mutations in the groups defined by hierarchic clustering was not different.
The PAM- and SAM-generated gene lists were inspected. One gene, RUNX3, had a significant correlation with treatment failure. Therefore, we selected 12 genes from the SAM list that coclustered with RUNX3. This gene list included an EST moderately similar to zinc-finger protein ZBRK1, KIAA0937 protein, zinc finger protein 24 KOX 17, LNK, and GNA12. Only one gene, ATRX, was significantly underexpressed in the resistant group by SAM analysis. Its expression pattern was reciprocal to RUNX3 expression. Therefore, these 13 genes (RUNX3 cluster and ATRX) were used to define a new hierarchic cluster (Figure 4). This analysis resulted in 2 main branches that compared as follows: EFS was 34% (Subgroup II) versus 0% (Subgroup I), P = .009. These differences also were statistically significant for OS (54% vs 7%, P = .02).
Hierarchic clustering FLT3-MU samples supervised by SAM-derived RUNX3 cluster (12 genes) and ATRX gene. SAM-derived RUNX3 cluster identified a significantly different gene expression pattern that correlated with clinical outcome. The ATRX helicase had an inverse correlation with expression level of RUNX3. Cases with poor outcome had elevated RUNX3 expression and low expression of ATRX.
Hierarchic clustering FLT3-MU samples supervised by SAM-derived RUNX3 cluster (12 genes) and ATRX gene. SAM-derived RUNX3 cluster identified a significantly different gene expression pattern that correlated with clinical outcome. The ATRX helicase had an inverse correlation with expression level of RUNX3. Cases with poor outcome had elevated RUNX3 expression and low expression of ATRX.
We next queried the FLT3-WT set (n = 36 samples from diagnosis and n = 9 samples from relapse) to evaluate prognostic value of the RUNX3 cluster and the ATRX gene. The profile did not show any statistically significant difference in EFS or OS between 2 main branches defined by hierarchic clustering with the RUNX3-cluster genes (data not shown).
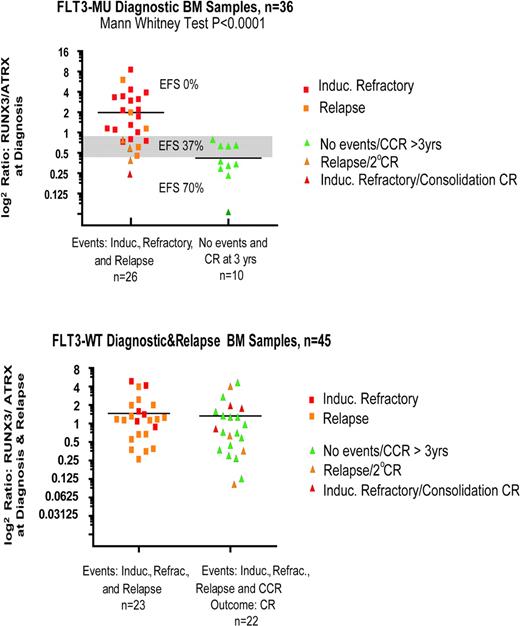
These data suggest that overexpression of the RUNX3 cluster (12 genes), and underexpression of ATRX, with FLT3-MU correlates with high risk of treatment failure (Figure 4). Furthermore, when the ratios of RUNX3 gene and the ATRX were calculated and plotted, we found a significant difference in their mean values. Specimens with log2 [RUNX3/ATRX] ratio more than 0.8 had an EFS of 0%; those with a ratio of 0.4 to 0.8 had an EFS of 37%; and those with a ratio less than 0.4 had an EFS of 70% (Figure 5). The expression of the RUNX3 and ATRX genes was quantified using real-time RT-PCR, and the ratios were recalculated. This analysis revealed similar results to those found by using the microarray data (Figure 6).
Gene expression array: RUNX3/ATRX ratio versus outcome. In children with de novo AML who harbor FLT3-MUs, the ratio of RUNX3 to ATRX may determine outcome. This same ratio in children with de novo AML and FTL3-WT (n = 45) did not correlate with outcome. In the top figure, EFS (%) is noted for samples with a ratio more than 0.8 (0%), from 0.4 to 0.8 (37%), and less than 0.4 (70%). Induc indicates induction.
Gene expression array: RUNX3/ATRX ratio versus outcome. In children with de novo AML who harbor FLT3-MUs, the ratio of RUNX3 to ATRX may determine outcome. This same ratio in children with de novo AML and FTL3-WT (n = 45) did not correlate with outcome. In the top figure, EFS (%) is noted for samples with a ratio more than 0.8 (0%), from 0.4 to 0.8 (37%), and less than 0.4 (70%). Induc indicates induction.
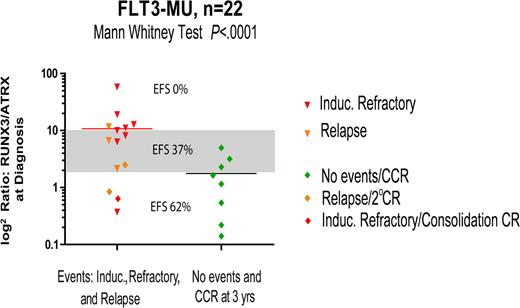
Quantitative PCR: RUNX3/ATRX ratio versus outcome. In children with de novo AML who harbor FLT3-MUs (n = 22), the ratio of RUNX3 to ATRX predicted outcome using quantitative RT-PCR from unamplified total RNA treated with DNAse I. A ratio more than 10 correlated with an EFS of 0%, a threshold ratio of 2 to 10 correlated with an EFS of 37%, and a ratio less than 2 correlated with an EFS of 62%. This same ratio in children with de novo AML and FTL3-WT did not correlate with outcome.
Quantitative PCR: RUNX3/ATRX ratio versus outcome. In children with de novo AML who harbor FLT3-MUs (n = 22), the ratio of RUNX3 to ATRX predicted outcome using quantitative RT-PCR from unamplified total RNA treated with DNAse I. A ratio more than 10 correlated with an EFS of 0%, a threshold ratio of 2 to 10 correlated with an EFS of 37%, and a ratio less than 2 correlated with an EFS of 62%. This same ratio in children with de novo AML and FTL3-WT did not correlate with outcome.
Discussion
We used genomewide analysis to identify gene expression patterns that correlate with poor outcomes in childhood AML. Our statistical office randomly identified, among a set of 272 diagnostic and relapsed AML bone marrow specimens with high-quality total RNA, 3 groups of samples (IR, R, and CCR), with differing clinical outcomes. Patterns of gene expression were analyzed according to FLT3 status (WT vs MU [ITD or ALM]) and clinical outcome.
With pairwise comparisons of our subgroups we found no statistically significant differences in EFS between FLT3-WT and FLT3-MU (P = .12 when defined by mutation status or P = .37 when defined by gene expression profiles) because the effect of FLT3-MUs on outcome is not universal. However, when FLT3-ITD was compared with FLT3-WT by mutation status, we observed a statistically significant difference in EFS (46% WT vs 22% ITD, P = .045), as in other publications.3,36 In our analysis, there was no difference in EFS between FLT3-WT vs FLT3-ALM or FLT3-ITD vs FLT3-ALM gene expression signatures (Figure 1A-C).
We reviewed incidence and outcome of FLT3-MU and found that these mutations were not a universally poor prognostic factor because 22 of 36 FLT3-MUs achieved remission with induction therapy, and 10 of 22 remained in CCR more than 3 years. The distribution of samples with ITD and ALM suggests that at diagnosis there are cassettes of gene expression that, with the FLT3-MUs, effect relapse or failure to achieve remission during induction therapy. Likewise, for patients in CCR, cassettes of gene expression at diagnosis may enhance sensitivity to the therapy used in POG-9421 with or without FLT3-MU.
A few FLT3-WT samples (n = 6) coclustered with FLT3-MU signature samples despite having an FLT3-WT receptor (Figure 2). This strongly suggests that other genetic abnormalities (eg, other RTK mutations), besides FLT3-MUs, can result in profiles similar to those determined by FLT3-MUs; or alternatively these samples harbor an alternate FLT3-ALM other than that seen in codon 835/836. Given the redundancy of signal transduction pathways, mutations or deletions of other targets can result in similar gene expression profiles and phenotypes. Only one case of FLT3-ITD coclustered with the FLT3-WT signature samples. This suggests that cassettes of gene expression or alternate mutations/deletions can result in a FLT3-WT phenotype in spite of the presence of a FLT3-MU. The predictive value of a single mutation can be further refined by looking at the gene expression profile related to its phenotype or clinical outcome.
Analysis by the PAM and SAM methods between FLT3-MU and FLT3-WT samples yielded a misclassification error of 15%; perhaps explained by the lack of correlation between a specific mutation and its predominant downstream gene expression. Moreover, the segregation of samples into MU versus WT group based on this cluster of 40 genes did not correlate significantly with clinical outcome because both groups had patients who relapsed or had CCRs. One gene expression signature differentiated FLT3-ITD from FLT3-WT samples (Figure 1A), and when all diagnostic samples were included we found a strong correlation between samples that were induction refractory and ITD positive (see web supplement). This analysis identified several genes with increased expression known to be part of the signal transduction pathway downstream from the FLT3 receptor. For example, patients with inferior outcome had increased expression of PI-3 kinase, which activates the antiapoptotic AKT pathway.48
Supervised hierarchic clustering of the FLT3-MU specimens by differences in clinical outcome (death vs CCR or remission) also divided these specimens into groups with markedly different survivals. Supervised analysis of FLT3-MU based on clinical outcome identified a similar list of genes with PAM and SAM analysis. Evaluation of each of 2 branches derived form hierarchic clustering revealed inferior EFS and OS in samples with increased expression of RUNX3 (also known as AML2), which is involved in binding several enhancers, and forms a heterodimer that may act as a proto-oncogene or tumor suppressor gene.49-53
Elevated expression of RUNX3 and decreased expression of ATRX correlated with inferior outcome. A subcluster of 13 genes that included RUNX3 and ATRX more specifically identified FLT3-MU AML patients with high risk of failure and death. The relative increased expression (n = 12) and lower expression (n = 1) of these genes when coexpressed with an FLT3-ITD or ALM identified cases with high risk of treatment failure. Further analysis of RUNX3 expression, validated by quantitative PCR, showed a strong correlation with poor clinical outcome in patients who harbored FLT3-MU. However, the correlation of poor outcome with increased expression of RUNX3 was not found in FLT3-WT cases, suggesting a possible relationship between constitutive activation of FLT3 receptor and transcriptional/tumor suppressive effects of RUNX3 product. The predictive value of increased RUNX3 expression was more evident when we analyzed the ratio of its expression to ATRX (Figures 5, 6). High, intermediate, and low ratios had statistically significant differences in EFS and OS (P < .0001). These preliminary results suggest that further investigation is warranted on the relationship of RUNX3 and ATRX to FLT3-MUs, and perhaps other receptor tyrosine kinases.
ATRX modifies gene expression by affecting chromatin. Mutations in ATRX cause changes in the DNA methylation pattern.54-56 Underexpression of ATRX with FLT3-MUs and RUNX3 overexpression may favor proliferation of AML blasts. Our data suggest that increased ATRX expression may interfere with the proliferative advantage of FLT3-MU and higher expression of RUNX3. We are interested in effects of various FLT3 inhibitors on expression and function of these genes in cell lines derived from clinical samples and in established AML cell line models with FLT3-MU.20,33,57
The relationship between FLT3-MU and the RUNX3 and ATRX should be tested retrospectively in banked specimens of adult and childhood AML blasts to verify their predictive value of clinical outcome in patients who received various regimens. Good outcomes in acute promyelocytic leukemia,58 frequently associated with FLT3-ITD,59-62 may be associated with low RUNX3/ATRX ratios, which might nullify the proliferative advantage conferred by the FLT3-ITD and its adverse gene expression profile.
Identifying patients with FLT3-MUs and gene expression profiles that correlate with high risk of failure will be useful in risk stratification and assessment of efficacy of FLT3 inhibitors in clinical trials,4,63-67 or in trials of cytarabine dose-intense regimens reported to overcome the poor prognostic effects of FLT3-ITDs68 in adults with AML. The relationship suggested between FLT3 receptor constitutive activation and changes secondary to increased expression of RUNX3 and decreased expression of ATRX may be important for development of novel treatments. Retrospective and prospective use of gene expression analysis in clinical trials69-71 will refine risk stratification and provide a useful tool for the rational development of targeted therapies in the future.
Prepublished online as Blood First Edition Paper, July 13, 2004; DOI 10.1182/blood-2003-12-4449.
Supported in part by National Institutes of Health (NIH) grants R01 CA90916 (G.V.D. and B.I.S.), R01 CA 92474 (B.I.S.), and M01 RR 00070 (General Clinical Research Center, Stanford University School of Medicine). N.J.L. is supported by a Clinical Associate Physician Award from the General Clinical Research Centers (M01 RR 00070). The Children's Oncology Group and its member institutions are supported by the following NIH grant: CA 98543.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Shaun Mason for editorial support.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal