Abstract
Paroxysmal nocturnal hemoglobinuria (PNH) is caused by phosphatidylinositol glycan–class A (PIG-A) mutations in hematopoietic stem cells (HSCs). PIG-A mutations have been found in granulocytes from most healthy individuals, suggesting that these spontaneous PIG-A mutations are important in the pathogenesis of PNH. It remains unclear if these PIG-A mutations have relevance to those found in PNH. We isolated CD34+ progenitors from 4 patients with PNH and 27 controls. The frequency of PIG-A mutant progenitors was determined by assaying for colony-forming cells (CFCs) in methylcellulose containing toxic doses of aerolysin (1 × 10-9 M). Glycosylphosphatidylinositol (GPI)–anchored proteins serve as receptors for aerolysin; thus, PNH cells are resistant to aerolysin. The frequency of aerolysin resistant CFC was 14.7 ± 4.0 × 10-6 in the bone marrow of healthy donors and was 57.0 ± 6.7 × 10-6 from mobilized peripheral blood. DNA was extracted from individual day-14 aerolysin-resistant CFCs and the PIG-A gene was sequenced to determine clonality. Aerolysin-resistant CFCs from patients with PNH exhibited clonal PIG-A mutations. In contrast, PIG-A mutations in the CFCs from controls were polyclonal, and did not involve T cells. Our data confirm the finding that PIG-A mutations are relatively common in normal hematopoiesis; however, the finding suggests that these mutations occur in differentiated progenitors rather than HSCs.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is a rare, clonal disease of hematopoietic stem cells (HSCs) that causes intravascular hemolysis, venous thrombosis, and bone marrow failure.1-3 The hallmark of PNH blood cells is a deficiency or absence of proteins that utilize glycosylphosphatidylinositol (GPI) anchors for their attachment to the plasma membrane. The absence of certain GPI-anchored complement regulatory proteins, such as CD55 and CD59, accounts for the complement-mediated hemolysis that characterizes PNH. In all patients with PNH to date, the GPI-anchor protein deficiency has been shown to be a direct consequence of PIG-A gene mutations; the PIG-A gene product is necessary for the first step in GPI anchor biosynthesis.4-8 The human PIG-A gene contains 6 exons, 5 introns, and extends over 17 kb. Most PIG-A mutations are small insertions or deletions that result in a frameshift and early termination of transcription.9 More than 100 mutations spanning the PIG-A coding region have been described with very few repeated mutations.9
Subclinical PNH clones are found in a high percentage of patients with acquired aplastic anemia, suggesting a pathophysiologic link between these 2 diseases.10-13 Moreover, many patients with aplastic anemia will have expansion of the PNH clone and develop clinical manifestations of PNH.14 A 2-step model has been proposed to explain the close relationship between PNH and aplastic anemia. This model proposes that HSCs randomly and spontaneously acquire PIG-A mutations at a very low frequency. Step 2 in this model proposes that an immunologic attack, such as that which occurs in aplastic anemia, targets “normal” HSCs, but spares PNH cells leading to clonal expansion.15,16 In support of step 1, PIG-A mutant blood cells (granulocytes and T lymphocytes) have been found in most healthy adults at a frequency of 2 to 4 per 100 000 cells.17-19 However, other observations raise questions whether these PIG-A mutations in healthy individuals arise from mutated HSCs. A mutational frequency of 2 to 4 per 100 000 cells exceeds the best estimates of spontaneous somatic mutation rates in humans by several logs.20 Moreover, gene marking studies in nonhuman primates and other large animals estimate that only 1000 HSCs contribute to hematopoiesis at a given time.21-23 If these estimates are valid, a mutational frequency even as high as 1 in 10 000 cells would be expected to produce a HSC with a PIG-A mutation in only about 10% of healthy controls.
In this study we used aerolysin, a channel-forming toxin secreted by the human pathogen Aeromonas hydrophila,24 to identify hematopoietic colony-forming cells (CFCs) with PIG-A mutations in patients with PNH and healthy controls. PNH cells are unique in their resistance to aerolysin because the glycan portion of GPI-anchored proteins serves as the receptor for aerolysin.25-27 In contrast, cells expressing GPI-anchored surface proteins are killed within minutes by nanomolar concentrations of aerolysin. We found PIG-A mutations in CFCs from hematopoietic progenitors in healthy individuals at a similar frequency as that found in previous studies in granulocytes and T lymphocytes.17,18 However, unlike patients with PNH, in whom the PIG-A mutations are clonal, the PIG-A mutations in CFCs from non–PNH patients were polyclonal and did not involve T cells. On the basis of these data, we suggest that PIG-A mutations in healthy individuals do not occur at the level of HSCs, but rather occur at the level of differentiated progenitor cells.
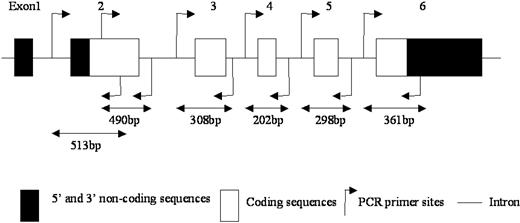
Schematic representation of the PIG-A gene and PCR primers. □ and ▪ represent coding and noncoding regions, respectively; arrows, PCR primer sites; and —, introns.
Schematic representation of the PIG-A gene and PCR primers. □ and ▪ represent coding and noncoding regions, respectively; arrows, PCR primer sites; and —, introns.
Methods
Isolation of hematopoietic cells
Bone marrow cells were obtained from posterior iliac crest aspiration from patients with PNH and healthy donors. Excess peripheral blood CD34+ cells from patients who underwent peripheral blood stem cell mobilization as part of their autologous transplantation were also used in these studies. All patients and volunteers gave informed consent approved by the Johns Hopkins Medical Institution's Internal Review Board. Bone marrow mononuclear cells were recovered by Ficoll/Hypaque (density < 1.077) centrifugation. To recover purified CD34+ cells, mononuclear cells were washed twice in phosphate-buffered saline (PBS; BioFluids; Biosource International, Camarillo, CA) incubated with the anti-CD34 microbeads and processed through a magnetic-activated cell sorting (MACS) magnetic separation column as per the manufacturer's instructions (Miltenyi Biotec, Auburn, CA).
Selection of aerolysin-resistant CFCs
CD34+ cells (106 to 107) were plated in MethoCult complete medium with cytokines (StemCell Technologies, Vancouver, BC, Canada) supplemented with 1 nM proaerolysin at a concentration of 1 × 105 per dish. In order to determine the cloning efficiency from each healthy donor, a control plate containing 1 × 103 CD34+ cells was plated in semisolid growth medium without proaerolysin. After 14 to 16 days of incubation, plates were scored for CFCs, granulocyte-macrophage–colony-forming units (CFU-GMs), and erythroid burst-forming units (BFU-Es), using an inverted phase contrast microscope (Leitz, Wetzlar, Germany). The frequency of aerolysin-resistant CFCs equaled the number of aerolysin resistant CFCs divided by the total number of CFCs. The total number of cultured CD34+ cells times the percent cloning efficiency determined the total number of CFCs.28 Cell-surface GPI-anchored proteins from day-14 pooled BFU-Es and CFU-GMs were assessed by flow cytometry after staining with phycoerythrin (PE)–conjugated glycophorin A (Immunotech, Marseille, France) and fluorescein isothiocyanate (FITC)–conjugated anti-CD59 (Immunotech) for BFU-Es, and PE-conjugated anti-CD15 (Immunotech) and fluorescent proaerolysin variant (FLAER) (Protox Biotech, Victoria, BC, Canada) for CFU-GMs.
Selection of aerolysin-resistant T cells
Aerolysin-resistant T-cell clones were isolated as previously described with minor modifications.18 Briefly, bone marrow or peripheral blood mononuclear cells (2 × 107) were incubated at 2 × 106 cells/mL in RPMI 1640 supplemented with 20% HL-1 medium, 1 μg/mL phytohemagglutinin (PHA; Murex, Danford, England), and 5% fetal bovine serum (FBS) in T-25 culture flasks at 37°C. After overnight mitogenic stimulation, the cells were placed in T-25 flasks containing selection medium consisting of RPMI 1640 supplemented with 20% HL-1 medium, 0.125 μg/mL PHA, 5% FBS, 1 nM aerolysin, T-stim culture supplement (Collaborative Biomedical Products, Bedford, MA), and LD- feeder cells,29 pretreated with 90 Gy 137Cs γ-irradiation. After 14 to 16 days of incubation, T-cell colonies were harvested and analyzed for PIG-A gene mutations (Figure 1) and for cell surface GPI anchor protein expression by flow cytometry.
Genomic DNA preparation
Sixteen days after plating, CFU-GMs, BFU-Es, and T-cell clones from the patients with PNH and healthy donors were individually plucked and genomic DNA was extracted using InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's recommendation.
PIG-Agene amplication
Each exon of the PIG-A gene was amplified with the proofreading polymerase Pfu Ultra (Stratagene, La Jolla, CA) using the primers shown in Figure 1 and Table 1. Two overlapping fragments generated exon 2. Matrix (20 μL) was used in a reaction containing 1× manufacturer's buffer (Invitrogen, Carlsbad, CA), 0.2 mM each deoxyribonucleotide triphosphates (dNTPs), 0.2 μM each of sense and antisense primer and 2.5 U Pfu Ultra DNA polymerase (Stratagene, La Jolla, CA) in a total volume of 50 μL, using a Biometra T3 thermocycler with the following conditions: initial denaturing at 95°C for 5 minutes, and 35 cycles at 95°C for 30 seconds, 55°C for 30 seconds, 72°C for 1 minute, and with a final extension at 72°C for 10 minutes. The polymerase chain reaction (PCR) products were analyzed by electrophoresis on 1.2% agarose and stained with ethidium bromide.
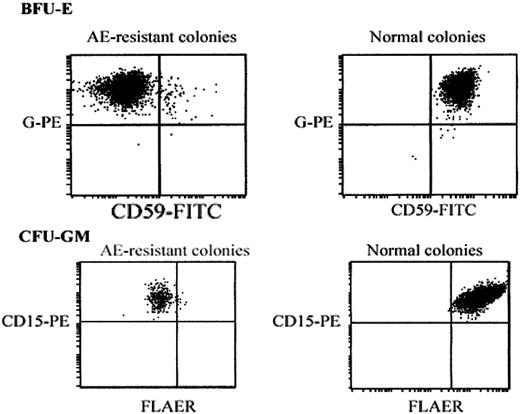
Flow cytometric analysis of pooled aerolysin-resistant colonies. Day-14 BFU-Es (top row) or CFU-GMs (bottom row) were washed from methycellulose-containing plates. BFU-Es were stained with FITC-conjugated anti–glycophorin-A and PE-conjugated anti-CD59. CFU-GMs were stained with PE-conjugated anti-CD15 and FLAER. Vertical and horizontal axes represent fluorescence intensity compared with isotypic controls.
Flow cytometric analysis of pooled aerolysin-resistant colonies. Day-14 BFU-Es (top row) or CFU-GMs (bottom row) were washed from methycellulose-containing plates. BFU-Es were stained with FITC-conjugated anti–glycophorin-A and PE-conjugated anti-CD59. CFU-GMs were stained with PE-conjugated anti-CD15 and FLAER. Vertical and horizontal axes represent fluorescence intensity compared with isotypic controls.
PCR primers
Primer ID . | Sequence 5′-3′ . | Location . | Annealing temp, °C . | Product size, bp . |
|---|---|---|---|---|
| S-2-1 | GTTTCTGAGCTGAGATCCTG | Intron 1 | 58.8 | 513 |
| AS-2-1 | GAGCATCATGGGCCATAGCA | Exon 2 | 68.7 | - |
| S-2-2 | GTCATGTACAACCAGTCTAC | Exon 2 | 50.6 | 490 |
| AS-2-2 | GCCAAACAATCATTATATACAAG | Intron 2 | 59.7 | - |
| S-3 | TGGATTCTCAGTCGTTCTGGTGA | Intron 2 | 69.2 | 308 |
| AS-3 | ATGCAGGAGAAGCAACACAC | Intron 3 | 62.7 | - |
| S-4 | TCACTCCTTTCTTCCCCTCTC | Intron 3 | 64.7 | 202 |
| AS-4 | AATCCCAACCATGAATGCCCTC | Intron 4 | 72 | - |
| S-5 | TCTTCCTGAGGTATGATTATGGTG | Intron 4 | 57 | 298 |
| AS-5 | AAGAGTTCAGACACAATCTTTTCTC | Intron 5 | 63.6 | - |
| S-6 | GGTCATTGTTATCATGGGACAG | Intron 5 | 64.1 | 361 |
| AS-6 | TCTTACAATCTAGGCTTCCTTC | Exon 6 | 60.5 | - |
Primer ID . | Sequence 5′-3′ . | Location . | Annealing temp, °C . | Product size, bp . |
|---|---|---|---|---|
| S-2-1 | GTTTCTGAGCTGAGATCCTG | Intron 1 | 58.8 | 513 |
| AS-2-1 | GAGCATCATGGGCCATAGCA | Exon 2 | 68.7 | - |
| S-2-2 | GTCATGTACAACCAGTCTAC | Exon 2 | 50.6 | 490 |
| AS-2-2 | GCCAAACAATCATTATATACAAG | Intron 2 | 59.7 | - |
| S-3 | TGGATTCTCAGTCGTTCTGGTGA | Intron 2 | 69.2 | 308 |
| AS-3 | ATGCAGGAGAAGCAACACAC | Intron 3 | 62.7 | - |
| S-4 | TCACTCCTTTCTTCCCCTCTC | Intron 3 | 64.7 | 202 |
| AS-4 | AATCCCAACCATGAATGCCCTC | Intron 4 | 72 | - |
| S-5 | TCTTCCTGAGGTATGATTATGGTG | Intron 4 | 57 | 298 |
| AS-5 | AAGAGTTCAGACACAATCTTTTCTC | Intron 5 | 63.6 | - |
| S-6 | GGTCATTGTTATCATGGGACAG | Intron 5 | 64.1 | 361 |
| AS-6 | TCTTACAATCTAGGCTTCCTTC | Exon 6 | 60.5 | - |
- indicates not applicable; bp, base pair.
Cloning and sequencing
The products were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA). Direct sequencing was performed using fragment-specific primers and fragments were analyzed using an automated sequencer ABI PRISM Model 3700 (Applied Biosystems, Foster City, CA). CFCs from unselected (no proaerolysin) plates served as negative controls. CFCs from patients with PNH served as positive controls. To rule out PCR artifact, only mutations found in 2 independent reactions were listed. Sequencing products of the PIG-A gene from females were mixed in a 1:1 ratio with the normal allele, because the PIG-A gene is on the X chromosome and the X chromosome is randomly inactivated in females. In these cases the PCR products were cloned using Zero Blunt TOPO PCR Cloning Kit (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendation. Positive colonies were first identified using blue-white β-galactosidase selection and then confirmed by PCR using flanking M13 forward and reverse primers. Individual clones that contained inserts of the correct size were then grown overnight in an 8-mL culture and the amplified plasmids were purified using QIA pre-Spin Plasmid Kit (Qiagen). In the aim to rule out Escherichia coli DNA polymerase errors, we have listed only mutations identified in at least 3 independent colonies.
Restriction analysis of PCR products
MslI was used to screen aerolysin-enriched T-cell clones. Exon 4 of PIG-A gene was amplified, purified, and digested with MslI. The PCR product from CFU-GM/AE 9/7, which serves as a positive control, was mixed in a 1:1 ratio with normal, because the PIG-A gene is on the X chromosome and the X chromosome is randomly inactivated in females.
Results
Isolation and characterization of GPI-deficient CFCs
The frequency of aerolysin-resistant CFCs from 4 patients with PNH, 18 healthy donors, and 9 patients undergoing autologous stem cell transplantation was assessed by plating isolated CD34+ progenitors in methylcellulose containing 1 nM aerolysin. In patients with PNH, 67% ± 7% of the bone marrow CFCs were aerolysin resistant. The frequency of aerolysin resistant CFCs was 14.7 ± 4.0 × 10-6 in the bone marrow of healthy donors and was 57.0 ± 6.7 × 10-6 from mobilized peripheral blood. To confirm that the aerolysin-resistant colonies were GPI-anchor deficient, individual BFU-Es and CFU-GMs were pooled, washed in PBS, and assessed by flow cytometry after staining with anti-CD59 and FLAER. As shown in Figure 2, the aerolysin-resistant BFU-Es and CFU-GMs from the aerolysin-treated plates were GPI-anchor deficient; however, BFU-Es and CFU-GMs on the control plates were GPI-anchor replete.
PIG-A gene sequencing
To prove that the colonies possessed PIG-A mutations and to assess clonality, individual CFC colonies were isolated for DNA sequencing of the PIG-A gene. No PIG-A mutations were detected from unselected CFCs grown from non–PNH donors on the control plates (n = 10, data not shown). PIG-A gene sequencing was performed on the 4 patients with PNH (Table 2). Five colonies were sequenced from PNH patient 1 and all 5 contained a 1-nucleotide insertion in exon 2 at position 452A. PNH patient 2 had an 8-nucleotide deletion and a 1-nucleotide mutation in exon 4 in 5 of 5 CFCs, which also resulted in a frameshift mutation. PNH patient 3 harbored a 1-nucleotide mutation in exon 2 in 4 of 4 CFCs, which resulted in a stop codon. Patient 4 had a 1-nucleotide mutation in exon 2 in 5 of 5 CFCs, which resulted in an amino acid replacement from His to Tyr.
Isolation and characterization of aerolysin-resistant clones from patients with PNH
Patient no./colony no. . | Colony type . | Exon . | Codon . | Position and mutations . | Consequence . |
|---|---|---|---|---|---|
| 1/1 | CFU-GM/AE | 2 | 123 | 452insA | frame shift |
| 1/2 | CFU-GM/AE | 2 | 123 | 452insA | frame shift |
| 1/3 | CFU-GM/AE | 2 | 123 | 452insA | frame shift |
| 1/4 | BFU/AE | 2 | 123 | 452insA | frame shift |
| 1/5 | BFU/AE | 2 | 123 | 452insA | frame shift |
| 2/1 | CFU-GM/AE | 4 | 296-299 | 972G>A, del 973-980 | frame shift |
| 2/2 | CFU-GM/AE | 4 | 296-299 | 972G>A, del 973-980 | frame shift |
| 2/3 | CFU-GM/AE | 4 | 296-299 | 972G>A, del 973-980 | frame shift |
| 2/4 | BFU/AE | 4 | 296-299 | 972G>A, del 973-980 | frame shift |
| 2/5 | BFU/AE | 4 | 296-299 | 972G>A, del 973-980 | frame shift |
| 3/1 | CFU-GM/AE | 2 | 99 | 379C>A | stop codon |
| 3/2 | CFU-GM/AE | 2 | 99 | 379C>A | stop codon |
| 3/3 | BFU/AE | 2 | 99 | 379C>A | stop codon |
| 3/4 | BFU/AE | 2 | 99 | 379C>A | stop codon |
| 4/1 | CFU-GM/AE | 2 | 126 | 461C>T | His→Tyr |
| 4/2 | CFU-GM/AE | 2 | 126 | 461C>T | His→Tyr |
| 4/3 | CFU-GM/AE | 2 | 126 | 461C>T | His→Tyr |
| 4/4 | BFU/AE | 2 | 126 | 461C>T | His→Tyr |
| 4/5 | BFU/AE | 2 | 126 | 461C>T | His→Tyr |
Patient no./colony no. . | Colony type . | Exon . | Codon . | Position and mutations . | Consequence . |
|---|---|---|---|---|---|
| 1/1 | CFU-GM/AE | 2 | 123 | 452insA | frame shift |
| 1/2 | CFU-GM/AE | 2 | 123 | 452insA | frame shift |
| 1/3 | CFU-GM/AE | 2 | 123 | 452insA | frame shift |
| 1/4 | BFU/AE | 2 | 123 | 452insA | frame shift |
| 1/5 | BFU/AE | 2 | 123 | 452insA | frame shift |
| 2/1 | CFU-GM/AE | 4 | 296-299 | 972G>A, del 973-980 | frame shift |
| 2/2 | CFU-GM/AE | 4 | 296-299 | 972G>A, del 973-980 | frame shift |
| 2/3 | CFU-GM/AE | 4 | 296-299 | 972G>A, del 973-980 | frame shift |
| 2/4 | BFU/AE | 4 | 296-299 | 972G>A, del 973-980 | frame shift |
| 2/5 | BFU/AE | 4 | 296-299 | 972G>A, del 973-980 | frame shift |
| 3/1 | CFU-GM/AE | 2 | 99 | 379C>A | stop codon |
| 3/2 | CFU-GM/AE | 2 | 99 | 379C>A | stop codon |
| 3/3 | BFU/AE | 2 | 99 | 379C>A | stop codon |
| 3/4 | BFU/AE | 2 | 99 | 379C>A | stop codon |
| 4/1 | CFU-GM/AE | 2 | 126 | 461C>T | His→Tyr |
| 4/2 | CFU-GM/AE | 2 | 126 | 461C>T | His→Tyr |
| 4/3 | CFU-GM/AE | 2 | 126 | 461C>T | His→Tyr |
| 4/4 | BFU/AE | 2 | 126 | 461C>T | His→Tyr |
| 4/5 | BFU/AE | 2 | 126 | 461C>T | His→Tyr |
Sequencing of the aerolysin-resistant colonies from the bone marrow of 9 of the healthy donors also revealed PIG-A mutations (Table 3). Only 1 aerolysin-resistant colony was found in donors 2, 3, 6, and 7, making it impossible to assess clonality. However, donors 1, 4, 5, 8, and 9 had 2 or more clones sequenced for PIG-A mutations. In contrast to the patients with PNH, the PIG-A mutations in the aerolysin-resistant CFCs derived from healthy controls were polyclonal. In donor 4, 4 separate PIG-A mutations were found: 3 in exon 2 and one in exon 5. In donor 5, 3 separate PIG-A mutations were found: 2 in exon 2 and 1 in exon 6. Donor 8 had 5 matchless mutations, 3 in exon 5, 1 in exon 2, and 1 in exon 6. Donor 9 had 7 aerolysin-resistant CFCs with 4 different PIG-A mutations; 3 had matching mutations in exon 2, codon 155, and 2 had a 4 base pair (bp) deletion in exon 2.
Isolation and characterization of aerolysin-resistant clones from normal bone marrow
Donor no./colony no. . | Colony type . | Exon . | Codon . | Positions and mutations . | Consequence . |
|---|---|---|---|---|---|
| 1/1 | CFU-GM/AE | 2 | 56 | 252T>C | Leu→Pro |
| 1/2 | CFU-GM/AE | 2 | 25-28 | 158-176 del 10bp | frame shift |
| 2/1 | CFU-GM/AE | 2 | 140 | 503insT | frame shift |
| 3/1 | CFU-GM/AE | 2 | 226 | 761insA | frame shift |
| 4/1 | CFU-GM/AE | 2 | 60 | 264T>C | Leu→Pro |
| 4/2 | CFU-GM/AE | 2 | 57, 58 | 256, 257delTC | frame shift |
| 4/3 | CFU-GM/AE | 2 | 238 | 799delA | frame shift |
| 4/4 | CFU-GM/AE | 5 | 337 | 1095delC | frame shift |
| 5/1 | CFU-GM/AE | 2 | 149 | 531delC | frame shift |
| 5/2 | CFU-GM/AE | 6 | 442 | 1410T>A | stop codon |
| 5/3 | CFU-GM/AE | 2 | 183 | 633G>A | Cys→Tyr |
| 6/1 | CFU-GM/AE | 2 | 145 | 518delA | frame shift |
| 7/1 | CFU-GM/AE | 2 | 57, 58 | 254-257delTCTC | frame shift |
| 8/1 | CFU-GM/AE | 5 | 380 | 1224delT | frame shift |
| 8/2 | CFU-GM/AE | 5 | 332 | 1079A>T | stop codon |
| 8/3 | CFU-GM/AE | 2 | 183 | 632T>C | Cys→Arg |
| 8/4 | CFU-GM/AE | 6 | 448-450 | 1427-1443delGATTCTA | frame shift |
| 8/5 | CFU-GM/AE | 5 | 380 | 1224insTA | frame shift |
| 9/1 | CFU-GM/AE | 2 | 155 | 549C>T | Ser→Phe |
| 9/2 | CFU-GM/AE | 2 | 155 | 549C>T | Ser→Phe |
| 9/3 | CFU-GM/AE | 2 | 155 | 549C>T | Ser→Phe |
| 9/4 | CFU-GM/AE | 2 | 168, 169 | 588-591delACAA | frame shift |
| 9/5 | CFU-GM/AE | 2 | 168, 169 | 588-591delACAA | frame shift |
| 9/6 | CFU-GM/AE | 6 | 421 | 1347delG | frame shift |
| 9/7 | CFU-GM/AE | 4 | 294 | 965delG | frame shift |
Donor no./colony no. . | Colony type . | Exon . | Codon . | Positions and mutations . | Consequence . |
|---|---|---|---|---|---|
| 1/1 | CFU-GM/AE | 2 | 56 | 252T>C | Leu→Pro |
| 1/2 | CFU-GM/AE | 2 | 25-28 | 158-176 del 10bp | frame shift |
| 2/1 | CFU-GM/AE | 2 | 140 | 503insT | frame shift |
| 3/1 | CFU-GM/AE | 2 | 226 | 761insA | frame shift |
| 4/1 | CFU-GM/AE | 2 | 60 | 264T>C | Leu→Pro |
| 4/2 | CFU-GM/AE | 2 | 57, 58 | 256, 257delTC | frame shift |
| 4/3 | CFU-GM/AE | 2 | 238 | 799delA | frame shift |
| 4/4 | CFU-GM/AE | 5 | 337 | 1095delC | frame shift |
| 5/1 | CFU-GM/AE | 2 | 149 | 531delC | frame shift |
| 5/2 | CFU-GM/AE | 6 | 442 | 1410T>A | stop codon |
| 5/3 | CFU-GM/AE | 2 | 183 | 633G>A | Cys→Tyr |
| 6/1 | CFU-GM/AE | 2 | 145 | 518delA | frame shift |
| 7/1 | CFU-GM/AE | 2 | 57, 58 | 254-257delTCTC | frame shift |
| 8/1 | CFU-GM/AE | 5 | 380 | 1224delT | frame shift |
| 8/2 | CFU-GM/AE | 5 | 332 | 1079A>T | stop codon |
| 8/3 | CFU-GM/AE | 2 | 183 | 632T>C | Cys→Arg |
| 8/4 | CFU-GM/AE | 6 | 448-450 | 1427-1443delGATTCTA | frame shift |
| 8/5 | CFU-GM/AE | 5 | 380 | 1224insTA | frame shift |
| 9/1 | CFU-GM/AE | 2 | 155 | 549C>T | Ser→Phe |
| 9/2 | CFU-GM/AE | 2 | 155 | 549C>T | Ser→Phe |
| 9/3 | CFU-GM/AE | 2 | 155 | 549C>T | Ser→Phe |
| 9/4 | CFU-GM/AE | 2 | 168, 169 | 588-591delACAA | frame shift |
| 9/5 | CFU-GM/AE | 2 | 168, 169 | 588-591delACAA | frame shift |
| 9/6 | CFU-GM/AE | 6 | 421 | 1347delG | frame shift |
| 9/7 | CFU-GM/AE | 4 | 294 | 965delG | frame shift |
PIG-A gene sequencing was performed on the aerolysin-resistant CFC colonies on 5 non–PNH patients who underwent peripheral blood stem cell mobilization in preparation for an autologous stem cell transplantation. We sequenced a total of 8, 12, 34, 7, and 12 individual CFCs from these patients, respectively (Table 4). In patient 10, 3 different mutations were found from the 8 CFCs, with 5 CFCs harboring a 10–base pair (bp) deletion in exon 3. Patient 11 had 10 different mutations from 12 CFCs, with 3 CFCs sharing a 4 bp insertion in exon 6. Patient 12 had 15 different mutations from 34 CFCs, and 8 of them shared a 1-nucleotide mutation in exon 2. Patient 13 had 5 different PIG-A mutations in 7 different colonies, and patient 14 harbored 9 different mutations in 12 different colonies.
Isolation and characterization of aerolysin-resistant clones from mobilized peripheral blood
Donor no./colony no. . | Colony type . | Exon . | Codon . | Position and mutations . | Consequence . |
|---|---|---|---|---|---|
| 10/1-10/3 | CFU-GM/AE | 3 | 274-277 | 906-915delTTCGGGAAAG | frame shift |
| 10/4-10/5 | BFU-E/AE | 3 | 274-277 | 906-915delTTCGGGAAAG | frame shift |
| 10/6 | CFU-GM/AE | 2 | 155 | 549C>A | Ser→Tyr |
| 10/7 | BFU-E/AE | 2 | 155 | 549C>A | Ser→Tyr |
| 10/8 | CFU-GM/AE | 5 | 350 | 1134delC | frame shift |
| 11/1-11/3 | BFU-E/AE | 6 | 426 | 1363insGCTA | frame shift |
| 11/4 | CFU-GM/AE | 2 | 155 | 549C>T | Ser→Phe |
| 11/5 | CFU-GM/AE | 2 | 237 | 794A>T | stop codon |
| 11/6 | CFU-GM/AE | 2 | 176 | 613T>A | stop codon |
| 11/7 | CFU-GM/AE | 5 | 350 | 1134delC | frame shift |
| 11/8 | BFU-E/AE | 3 | 265 | 880delA | frame shift |
| 11/9 | BFU-E/AE | 2 | 238 | 799delA | frame shift |
| 11/10 | BFU-E/AE | 2 | 40 | 203G>C | Asp→His |
| 11/11 | BFU-E/AE | 2 | 77 | 314C>T | stop codon |
| 11/12 | BFU-E/AE | 6 | 449 | 1430delT | frame shift |
| 12/1-12/4 | CFU-GM/AE | 2 | 42 | 210delT | frame shift |
| 12/5 | CFU-GM/AE | 2 | 42,164 | 210delT, 576C>A | frame shift |
| 12/6 | CFU-GM/AE | 2 | 42,226 | 210delT, 762insT | frame shift |
| 12/7 | CFU-GM/AE | 2,3 | 42,259 | 210delT, 861delT | frame shift |
| 12/8-12/9 | BFU-E/AE | 2 | 42 | 210delT | frame shift |
| 12/10-12/15 | CFU-GM/AE | 2 | 128 | 468A>G | His→Arg |
| 12/16-12/17 | BFU-E/AE | 2 | 128 | 468A>G | His→Arg |
| 12/18 | CFU-GM/AE | 2 | 226 | 762insT | frame shift |
| 12/19-12/21 | BFU-E/AE | 2 | 226 | 762insT | frame shift |
| 12/22-12/24 | CFU-GM/AE | 5 | 397 | 1274G>C | Val→Leu |
| 12/25-12/26 | CFU-GM/AE | 3 | 259 | 861delT | frame shift |
| 12/27 | CFU-GM/AE | 2,3 | 175,259 | 609T>C, 861delT | frame shift |
| 12/28-12/29 | CFU-GM/AE | 2 | 82 | 331C>A | Tyr→stop codon |
| 12/30 | CFU-GM/AE | 2 | 56 | 252T>C | Leu→Pro |
| 12/31 | CFU-GM/AE | 2 | 90 | 355T>A | Tyr→stop codon |
| 12/32 | CFU-GM/AE | 2 | 113 | 423T>C | Leu→Pro |
| 12/33 | CFU-GM/AE | 5 | 397 | 1274G>A | Val→Ile |
| 12/34 | BFU-E/AE | 2 | 74 | 306,307delAT | frame shift |
| 13/1-13/2 | CFU-GM/AE | 2 | 221 | 746insA | frame shift |
| 13/3-13/4 | CFU-GM/AE | 2 | 40 | 204A>T | Asp→Val |
| 13/5 | CFU-GM/AE | 2 | 37 | 194delA | frame shift |
| 13/6 | CFU-GM/AE | 2 | 155 | 549C>T | Ser→Phe |
| 13/7 | BFU-E/AE | 4 | 298 | 978delT | frame shift |
| 14/1 | CFU-GM/AE | 2 | 113 | 431A>G | Ile→Val |
| 14/2 | CFU-GM/AE | 2 | 36 | 193C>G | Cys→Trp |
| 14/3 | CFU-GM/AE | 2 | 50 | 233G>T | stop codon |
| 14/4 | CFU-GM/AE | 3 | 278 | 919C>G | stop codon |
| 14/5 | CFU-GM/AE | 2 | 188 | 649delT | frame shift |
| 14/6 | CFU-GM/AE | 2 | 199 | 649delT | frame shift |
| 14/7-14/8 | CFU-GM/AE | 2 | 110 | 413delC | frame shift |
| 14/9-14/10 | CFU-GM/AE | 2 | 123,124 | 454-456delGAT | frame shift |
| 14/11-14/12 | CFU-GM/AE | 2 | 183 | 634T>G | Cys→Trp |
Donor no./colony no. . | Colony type . | Exon . | Codon . | Position and mutations . | Consequence . |
|---|---|---|---|---|---|
| 10/1-10/3 | CFU-GM/AE | 3 | 274-277 | 906-915delTTCGGGAAAG | frame shift |
| 10/4-10/5 | BFU-E/AE | 3 | 274-277 | 906-915delTTCGGGAAAG | frame shift |
| 10/6 | CFU-GM/AE | 2 | 155 | 549C>A | Ser→Tyr |
| 10/7 | BFU-E/AE | 2 | 155 | 549C>A | Ser→Tyr |
| 10/8 | CFU-GM/AE | 5 | 350 | 1134delC | frame shift |
| 11/1-11/3 | BFU-E/AE | 6 | 426 | 1363insGCTA | frame shift |
| 11/4 | CFU-GM/AE | 2 | 155 | 549C>T | Ser→Phe |
| 11/5 | CFU-GM/AE | 2 | 237 | 794A>T | stop codon |
| 11/6 | CFU-GM/AE | 2 | 176 | 613T>A | stop codon |
| 11/7 | CFU-GM/AE | 5 | 350 | 1134delC | frame shift |
| 11/8 | BFU-E/AE | 3 | 265 | 880delA | frame shift |
| 11/9 | BFU-E/AE | 2 | 238 | 799delA | frame shift |
| 11/10 | BFU-E/AE | 2 | 40 | 203G>C | Asp→His |
| 11/11 | BFU-E/AE | 2 | 77 | 314C>T | stop codon |
| 11/12 | BFU-E/AE | 6 | 449 | 1430delT | frame shift |
| 12/1-12/4 | CFU-GM/AE | 2 | 42 | 210delT | frame shift |
| 12/5 | CFU-GM/AE | 2 | 42,164 | 210delT, 576C>A | frame shift |
| 12/6 | CFU-GM/AE | 2 | 42,226 | 210delT, 762insT | frame shift |
| 12/7 | CFU-GM/AE | 2,3 | 42,259 | 210delT, 861delT | frame shift |
| 12/8-12/9 | BFU-E/AE | 2 | 42 | 210delT | frame shift |
| 12/10-12/15 | CFU-GM/AE | 2 | 128 | 468A>G | His→Arg |
| 12/16-12/17 | BFU-E/AE | 2 | 128 | 468A>G | His→Arg |
| 12/18 | CFU-GM/AE | 2 | 226 | 762insT | frame shift |
| 12/19-12/21 | BFU-E/AE | 2 | 226 | 762insT | frame shift |
| 12/22-12/24 | CFU-GM/AE | 5 | 397 | 1274G>C | Val→Leu |
| 12/25-12/26 | CFU-GM/AE | 3 | 259 | 861delT | frame shift |
| 12/27 | CFU-GM/AE | 2,3 | 175,259 | 609T>C, 861delT | frame shift |
| 12/28-12/29 | CFU-GM/AE | 2 | 82 | 331C>A | Tyr→stop codon |
| 12/30 | CFU-GM/AE | 2 | 56 | 252T>C | Leu→Pro |
| 12/31 | CFU-GM/AE | 2 | 90 | 355T>A | Tyr→stop codon |
| 12/32 | CFU-GM/AE | 2 | 113 | 423T>C | Leu→Pro |
| 12/33 | CFU-GM/AE | 5 | 397 | 1274G>A | Val→Ile |
| 12/34 | BFU-E/AE | 2 | 74 | 306,307delAT | frame shift |
| 13/1-13/2 | CFU-GM/AE | 2 | 221 | 746insA | frame shift |
| 13/3-13/4 | CFU-GM/AE | 2 | 40 | 204A>T | Asp→Val |
| 13/5 | CFU-GM/AE | 2 | 37 | 194delA | frame shift |
| 13/6 | CFU-GM/AE | 2 | 155 | 549C>T | Ser→Phe |
| 13/7 | BFU-E/AE | 4 | 298 | 978delT | frame shift |
| 14/1 | CFU-GM/AE | 2 | 113 | 431A>G | Ile→Val |
| 14/2 | CFU-GM/AE | 2 | 36 | 193C>G | Cys→Trp |
| 14/3 | CFU-GM/AE | 2 | 50 | 233G>T | stop codon |
| 14/4 | CFU-GM/AE | 3 | 278 | 919C>G | stop codon |
| 14/5 | CFU-GM/AE | 2 | 188 | 649delT | frame shift |
| 14/6 | CFU-GM/AE | 2 | 199 | 649delT | frame shift |
| 14/7-14/8 | CFU-GM/AE | 2 | 110 | 413delC | frame shift |
| 14/9-14/10 | CFU-GM/AE | 2 | 123,124 | 454-456delGAT | frame shift |
| 14/11-14/12 | CFU-GM/AE | 2 | 183 | 634T>G | Cys→Trp |
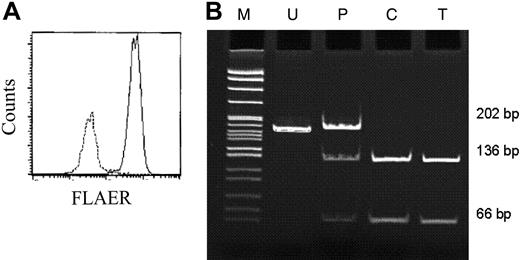
In PNH, the PIG-A defect is known to be present in all cell lineages.30-32 Thus, the demonstration of overlapping mutations in T cells from the same subject would establish that these mutations arose from a PIG-A mutated HSC. Accordingly, we isolated aerolysin-resistant T-cell clones from 2 patients with PNH and extracted DNA for PIG-A gene sequencing (Table 5). In both patients, the aerolysin-resistant colonies had clonal PIG-A mutations (Table 5) that matched the PIG-A mutation found in myeloid cells (data not shown). To determine if myeloid and lymphoid progenitors from controls also shared overlapping mutations, cells from 3 of the controls were simultaneously cultured for T-cell clones in the presence of aerolysin. In 2 of the patients (no. 8, Table 3; and no. 14, Table 4), no aerolysin-resistant T-cell clones were detected. However, there were 8 aerolysin-resistant T-cell clones in one of the controls (no. 9, Table 3). In this subject, 4 distinctive PIG-A mutations were found from 7 aerolysin-resistant CFCs (Table 3). DNA sequencing of exons 2, 4, and 6 from 8 individual aerolysin-resistant T-cell clones from patient 9 found no matching mutations. In addition, CFC 9/7 resulted in the loss of an Ms1I restriction site. Therefore, DNA from the 8 aerolysin-resistant T-cell clones was digested with Ms1I in order to probe for the 965 G deletion (Figure 3B). While Ms1I digestion of DNA from CFC 9/7 confirmed the 965 G deletion, none of the 8 T-cell clones harbored this mutation.
Isolation and characterization of aerolysin-resistant T-cell clones from patients with PNH
Patient no./colony no. . | Colony type . | Exon . | Codon . | Position and mutations . | Consequence . |
|---|---|---|---|---|---|
| 5/1 | T-cell/AE | 3 | 247 | 826insTA | frame shift |
| 5/2 | T-cell/AE | 3 | 247 | 826insTA | frame shift |
| 5/3 | T-cell/AE | 3 | 247 | 826insTA | frame shift |
| 5/4 | T-cell/AE | 3 | 247 | 826insTA | frame shift |
| 6/1 | T-cell/AE | 5 | 335 | 1089G>A | Gly→Glu |
| 6/2 | T-cell/AE | 5 | 335 | 1089G>A | Gly→Glu |
| 6/3 | T-cell/AE | 5 | 335 | 1089G>A | Gly→Glu |
| 6/4 | T-cell/AE | 5 | 335 | 1089G>A | Gly→Glu |
Patient no./colony no. . | Colony type . | Exon . | Codon . | Position and mutations . | Consequence . |
|---|---|---|---|---|---|
| 5/1 | T-cell/AE | 3 | 247 | 826insTA | frame shift |
| 5/2 | T-cell/AE | 3 | 247 | 826insTA | frame shift |
| 5/3 | T-cell/AE | 3 | 247 | 826insTA | frame shift |
| 5/4 | T-cell/AE | 3 | 247 | 826insTA | frame shift |
| 6/1 | T-cell/AE | 5 | 335 | 1089G>A | Gly→Glu |
| 6/2 | T-cell/AE | 5 | 335 | 1089G>A | Gly→Glu |
| 6/3 | T-cell/AE | 5 | 335 | 1089G>A | Gly→Glu |
| 6/4 | T-cell/AE | 5 | 335 | 1089G>A | Gly→Glu |
Discussion
The demonstration of PIG-A mutant granulocytes17 and T lymphocytes18,19 in healthy adults has been offered as support for the hypothesis that many healthy individuals harbor HSCs with PIG-A mutations.15,16 We found that hematopoietic progenitors with PIG-A mutations are also present in CFCs from most healthy individuals, at a cell frequency remarkably similar to that reported in granulocytes and T lymphocytes. The PIG-A mutational frequency in CFCs derived from mobilized peripheral blood CD34+ cells was 3-fold higher than bone marrow–derived CD34+ cells. The reason for this is unclear. It may be that by increasing the production of hematopoietic progenitors by the process of peripheral stem cell mobilization could amplify the frequency of PIG-A mutations that occur during hematopoietic differentiation.33 Alternatively, the patients undergoing peripheral blood stem cell mobilization had a history of prior chemotherapy for their underlying lymphoma or multiple myeloma that may have increased the mutation frequency.
Characterization of aerolysin resistant T-cell clones. (A) Flow cytometric analysis of pooled aerolysin-resistant T-cell clones. Day-16 colonies were washed and stained with FLAER. — represents expanded T-cell clones grown without aerolysin; –, expanded T-cell clones grown in the presence of aerolysin. (B) Mutation from CFU-GM/AE 9/7 lost MslI restriction site, which is CACAA′GGATG. Thus, MslI was used to screen aerolysin-enriched T-cell clones grown from the same donor. A total of 8 aerolysin-resistant T-cell clones were individually plucked and genomic DNA was extracted. A representative example is shown. Exon 4 of the PIG-A gene was amplified with the proofreading polymerase Pfu Ultra using the primers shown in Figure 1 and Table 1. The products were purified and digested with MslI. The PCR product from CFU-GM/AE 9/7 (lane P) which serves as a positive control were mixed in a 1:1 ratio with the normal allele, because the PIG-A gene is on the X chromosome and the X chromosome is randomly inactived in females. The TF1 cell line (lane C) serves as a normal control. Lane U represents uncut DNA from the T-cell clone and lane T shows the consequence of the Ms1I digest of DNA from a representative T-cell clone. M indicates molecular size markers.
Characterization of aerolysin resistant T-cell clones. (A) Flow cytometric analysis of pooled aerolysin-resistant T-cell clones. Day-16 colonies were washed and stained with FLAER. — represents expanded T-cell clones grown without aerolysin; –, expanded T-cell clones grown in the presence of aerolysin. (B) Mutation from CFU-GM/AE 9/7 lost MslI restriction site, which is CACAA′GGATG. Thus, MslI was used to screen aerolysin-enriched T-cell clones grown from the same donor. A total of 8 aerolysin-resistant T-cell clones were individually plucked and genomic DNA was extracted. A representative example is shown. Exon 4 of the PIG-A gene was amplified with the proofreading polymerase Pfu Ultra using the primers shown in Figure 1 and Table 1. The products were purified and digested with MslI. The PCR product from CFU-GM/AE 9/7 (lane P) which serves as a positive control were mixed in a 1:1 ratio with the normal allele, because the PIG-A gene is on the X chromosome and the X chromosome is randomly inactived in females. The TF1 cell line (lane C) serves as a normal control. Lane U represents uncut DNA from the T-cell clone and lane T shows the consequence of the Ms1I digest of DNA from a representative T-cell clone. M indicates molecular size markers.
The CFCs from patients with PNH, where the PIG-A mutation occurs in primitive HSCs, are clonal; matching mutations were found in myeloid, erythroid, and lymphoid progenitors. In contrast, PIG-A mutations in the CFCs from controls are polyclonal (up to 15 mutations from 1 individual). Recent evidence suggests that humans possess only 10 000 primitive HSCs,34 and only 1000 of these cells are thought to contribute to hematopoiesis at any one time.21-23 If this assumption is correct, only about 5% of the population would be expected to harbor any mutant CFCs at a PIG-A mutational frequency of 60 × 10-6, and less than 1 × 10-9 of individuals would harbor 10 or more different PIG-A mutations if 10 000 hematopoietic stem cells were contributing to hematopoiesis (Table 6). Even if 100 000 hematopoietic stem cells were contributing to hematopoiesis, only 8% of individuals would be expected to harbor 10 or more PIG-A mutations. Thus, the high frequency of PIG-A mutations in CFCs from healthy individuals, coupled with their polyclonality, suggests that most PIG-A mutations in healthy controls do not arise at the level of HSCs; this is further supported by the observation that aerolysin-resistant T cells from healthy controls do not harbor the same PIG-A mutations found in CFCs. The finding of 2 or more matching PIG-A mutations in control subjects' CFU-GMs and BFU-Es could be explained by the mutations occurring at the level of differentiated progenitors between HSCs and CFU-GMs/BFU-Es, such as granulocyte-erythrocyte-macrophage-megakaryocyte–colony-forming units (CFU-GEMMs). Unlike HSCs, CFCs have no self-renewal capacity; hence, mutations occurring at this level will not be propogated.
Poisson distribution of mutational frequency
. | . | Chance of n or more PIG-A mutations . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Population of HSCs . | Mutational frequency . | n = 1 . | n = 2 . | n = 3 . | n = 5 . | n = 10 . | ||||
| 1 000 | 15 × 10-6 | 0.02 | 1.11 × 10-4 | 5.56 × 10-7 | 6.25 × 10-12 | 0 | ||||
| 1 000 | 60 × 10-6 | 0.06 | 1.73 × 10-3 | 3.44 × 10-5 | 6.16 × 10-9 | 0 | ||||
| 10 000 | 15 × 10-6 | 0.14 | 0.01 | 5.03 × 10-4 | 5.59 × 10-7 | 1.33 × 10-15 | ||||
| 10 000 | 60 × 10-6 | 0.45 | 0.08 | 0.02 | 3.94 × 10-4 | 9.67 × 10-10 | ||||
. | . | Chance of n or more PIG-A mutations . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Population of HSCs . | Mutational frequency . | n = 1 . | n = 2 . | n = 3 . | n = 5 . | n = 10 . | ||||
| 1 000 | 15 × 10-6 | 0.02 | 1.11 × 10-4 | 5.56 × 10-7 | 6.25 × 10-12 | 0 | ||||
| 1 000 | 60 × 10-6 | 0.06 | 1.73 × 10-3 | 3.44 × 10-5 | 6.16 × 10-9 | 0 | ||||
| 10 000 | 15 × 10-6 | 0.14 | 0.01 | 5.03 × 10-4 | 5.59 × 10-7 | 1.33 × 10-15 | ||||
| 10 000 | 60 × 10-6 | 0.45 | 0.08 | 0.02 | 3.94 × 10-4 | 9.67 × 10-10 | ||||
Table shows chance of n or more clones (n equals 1, 2, 3, 5, or 10, as indicated) in the population size (no. of active HSCs) with the given frequency (Mf), according to a Poisson distribution.
Our data suggest that PIG-A mutations in healthy individuals may be a function of normal hematopoietic differentiation. DNA repair is progressively attenuated during the process of cellular differentiation,35 possibly leading to differentiation-dependent spontaneous mutations.36 It probably should not be surprising that cells undergoing terminal differentiation would exhibit decreased maintenance of genetic integrity, as mutations occurring in cells unable to self-renew will not be propagated. The finding that PIG-A mutant granulocytes in healthy controls are often short-lived17 is certainly consistent with these mutations occurring with differentiation rather than in HSCs. The spontaneous mutation rate in human cells is estimated to be between 1 × 10-8 and 1 × 10-7 per gene per cell division.20,37 The much higher, and surprisingly consistent, frequency of PIG-A mutations found in healthy individuals in our study and others17,18 might also be expected if they were a function of differentiation. Differentiation-associated genetic mutations may also explain the genotypic mosaicism that has been reported in some patients with PNH.38,39 While most studies show that PNH is a monoclonal disease, several studies have found more than one PIG-A mutant clone in patients with PNH; 1 study found 4 distinct somatic mutations from 1 patient, suggesting that PIG-A may be hypermutable.39 However, analysis of CFCs from patients with PNH with more than 1 PIG-A mutation usually reveals 1 dominant clone.40,41 The dominant clone may represent the mutation in an HSC, with the nondominant clones representing mutations in differentiated progenitor cells.
Our findings confirm that PIG-A mutations are relatively common in normal hematopoiesis. Our studies also demonstrate the first objective data that PIG-A mutations in healthy controls may occur in cells as early as CFU-GEMMs. An alternative hypothesis to explain the findings of PIG-A mutations in healthy individuals is that they arise as a function of normal hematopoietic differentiation; this explanation for the frequent occurrence of polyclonal PIG-A mutations in healthy controls calls into question the relevance of PIG-A mutations in healthy individuals to the pathogenesis of PNH.
Prepublished online as Blood First Edition Paper, February 1, 2005; DOI 10.1182/blood-2004-04-1472.
Supported in part by National Institutes of Health grant no. CA70970. R.A.B. is a Clinical Scholar of the Leukemia and Lymphoma Society of America. R.H. is the Thomas E. Sandefur Jr Fellow in Stem Cell Research.
An Inside Blood analysis of this article appears at the front of the issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal