Abstract
Interaction between the platelet glycoprotein Ibα (GPIbα) receptor and its adhesive ligand von Willebrand factor (VWF) has a critical role in the process of hemostasis. Platelet-type von Willebrand disease (PT-VWD) is a rare bleeding disorder that results from gain-of-function mutations in the GPIBA gene. We studied this gene from 5 members of a previously unreported family with a PT-VWD phenotype. We identified a novel in-frame deletion of 27 base pair (bp) in the macroglycopeptide region. This deletion was not found in the unaffected family members or in 50 healthy controls. The patients' platelets expressed normal quantities of GPIb/IX/V complex on their surface and the mutant (Mut) GPIbα was expressed at levels indistinguishable from the wild-type (WT) receptor on the surface of transfected Chinese hamster ovary (CHO) β/IX cells. Analysis of ristocetin-mediated 125I-VWF binding showed that the Mut receptor binds VWF in the absence of ristocetin and displays an increased sensitivity to lower concentrations of the modulator. This is the first report of a gain-of-function mutation in the GPIbα receptor outside the VWF-binding domain in patients with PT-VWD. The mutation provides a molecular basis for the PT-VWD phenotype and supports a role for the macroglycopeptide region in receptor function.
Introduction
Platelet-type von Willebrand disease (PT-VWD) is a rare autosomal dominant bleeding disorder first described in 1982.1 It results from an abnormally high affinity interaction between the platelet membrane glycoprotein (GP) Ib/IX/V complex and von Willebrand factor (VWF), leading to characteristic platelet hyperaggregability.1,2 Patients with PT-VWD have frequent and severe nosebleeds and excessive bleeding following tooth extraction, tonsillectomy, and other surgical operations.3 The condition shares most of the clinical and laboratory features of type IIB VWD and the final discrimination between them requires either platelet-mixing studies or a molecular genetic approach. Rapid clearance of high-molecular-weight (HMW) VWF multimers from plasma together with thrombocytopenia caused by an increased removal of the aggregated platelets contribute to the bleeding diathesis in PT-VWD.3,4
The GPIb/IX/V platelet receptor comprises 4 transmembrane polypeptides, each of which belongs to the leucine-rich repeat (LRR) family of proteins. They are GPIbα disulfide linked to GPIbβ, GPIX, and GPV, each of which is noncovalently associated with the complex with a stoichiometry of 2:2:2:1.5,6 The 2 proteins, GPIbβ and GPIX, are required for efficient cell-surface expression of GPIbα.7
GPIbα is the largest component of the receptor (610 amino acids) and carries the VWF-binding site. The structure of GPIbα reveals 4 functional domains: the 45-kDa amino-terminal domain; a heavily glycosylated mucinlike stalk known as the macroglycopeptide region, a single transmembrane domain, and an intracytoplasmic domain.8-10 Four regions within the amino-terminal domain contribute to VWF binding: N-terminal flank (His1-lle35), LRRs (Leu36-Ala200 residues), C-terminal disulfide loop (Phe201-Gly268), and an anionic tyrosine sulfated region (Asp269-Glu282).11-13
The interaction between VWF and its platelet receptor GPIbα is essential for platelet adhesion at sites of high-shear stress. Structural changes affecting GPIbα are responsible for the PT-VWD phenotype.14,15 To date, 5 families and one isolated case of PT-VWD have been reported and in these cases the responsible mutations reside within the VWF-binding region of the gene encoding GPIbα. Three missense mutations have been described: a Gly233Val reported in a white family,2 a Met239Val reported in 2 Japanese16 and one Puerto Rican family17 and as a de novo mutation in a white individual,18 and recently, Gly233Ser19 reported in another Japanese family. Recombinant protein expression and functional studies have shown that these mutations result in a platelet receptor with an increased affinity for VWF and are responsible for the disease phenotype.14,15,19
In the current study we sequenced the GPIBA gene from 5 members of a white family with PT-VWD to determine the molecular basis for the disease phenotype. We identified a novel 27–base pair (bp) in-frame deletion in the GPIBA gene region coding for the mucinlike macroglycopeptide region (nucleotides 4374-4400) encoding for 9 amino acids (PTILVSATS). All 3 affected members are heterozygous for the deletion and homozygous for the GPIbα VNTR (variable number of tandem repeats) C polymorphism.31 We showed that the mutation did not affect surface expression of GPIbα or cause a major alteration in the conformation of the ligand-binding domain of the GPIbα. However, we present evidence that the mutant receptor shows enhanced binding of VWF under low concentrations of ristocetin and that binding is significant even in the absence of ristocetin. This is the first report of a family with PT-VWD where a mutation in GPIbα, outside of the VWF-binding domain, can result in a hyperresponsive receptor and a gain-of-function phenotype.
Patients, materials, and methods
Patient material and clinical and laboratory data
Five members of a British family with PT-VWD were studied; 3 affected (the propositus, mother, and daughter) and 2 unaffected (the husband and son). The propositus, who was 37 years old at the time of the study, had a long history of bleeding since the age of 4. She experienced recurrent attacks of epistaxis for which repeated cauterization was performed without effective response and also had recurrent postoperative and dental bleeding. With her long-standing and troublesome history of menorrhagia, a series of hematologic investigations led to a diagnosis of type IIB VWD. In her first pregnancy, a moderate thrombocytopenia developed and she was admitted to the hospital late in pregnancy with an antepartum hemorrhage that failed to respond to the infusion of VWF/FVIII concentrate. Her platelet count dropped dramatically in response to VWF/FVIII infusion and only increased when platelet concentrates were given, suggestive of PT-VWD rather than type IIB VWD. Accordingly, hematologic re-evaluation was carried out with FVIII assay, VWF antigen, VWF:Ag ristocetin cofactor (RCoF), VWF multimer analysis, and ristocetin-induced platelet aggregation (RIPA) studies. Approval was obtained from the University of Southampton Institutional Review Board for these studies. Informed consent was provided according to the Declaration of Helsinki.
PCR amplification and sequencing of the GPIBA gene
gDNA was isolated from blood from the patient, her family members, and controls. The entire coding region of the patient's GPIBA gene was amplified in 2 overlapping fragments: fragment I (N-terminal region), amplified using primers modified from Takahashi et al16 (numbering according to GenBank accession no. M22403) and fragment II (COOH-terminal region). In addition, primers were designed to amplify an area within the macroglycopeptide-coding region including the VNTR polymorphism. All amplifications were carried out using Gold buffer (Applied Biosystems, Warrington, United Kingdom), 30 pmol each primer, 0.2 mmol/L deoxynucleotide triphosphate (dNTP), 1.5 mM MgCl2 (1.3 mM for fragment I) 500 ng gDNA, and 2.5 U Taq Gold (Applied Biosystems). Primers and polymerase chain reaction (PCR) conditions are shown in Table 1. PCR products were purified, cloned into pGEM-T Easy Vector (Promega, Southampton, United Kingdom) and sequenced on an ABI Prism 377 DNA sequencer using the Big Dye Termination Cycle Sequencing Ready Reaction kit (Applied Biosystems).
Primer sequences and PCR conditions for GPIbα amplifications
Region amplified . | Size, bp . | Primer sequence . | PCR conditions . |
|---|---|---|---|
| Fragment I | 976 | 5′–CCAGGGGATGCAGGGGGATCC–3′ (3019–3039) | Denaturation 94°C, 5 min |
| 5′–CTTGTGGCACGCACCTTATC–3′ (3975–3994) | Annealing 52°C, 1 min | ||
| Extension 72°C, 5 min | |||
| Fragment II | 1160 | 5′–GTGATGAAGGTGACACAGACC–3′ (3919–3939) | Denaturation 94°C, 5 min |
| 5′–GACAGGGCTTCTGATACAGAA–3′ (5066–5086) | Annealing 60°C, 1 min | ||
| Extension 72°C, 5 min | |||
| Macroglycopeptide region | 320 or 359 | 5′–CCATCACATTCTCCAAAACTCC–3′ (4174–4195) | Denaturation 94°C, 5 min |
| 5′–TGGCTGATCAAGTTCAGGGATG–3′ (4472–4493) | Annealing 57°C, 1 min | ||
| Extension 72°C, 5 min | |||
| Entire GPIbα-coding region | 1882 | 5′–ACTAGTATGCCTCTCCTCCTCTTGCTG–3′ | Denaturation 92°C, 30 s |
| 5′–TCAGAGGCTGTGGCCAGAGTA–3′ | Annealing 61°C, 30 s | ||
| Extension 68°C, 20 min |
Region amplified . | Size, bp . | Primer sequence . | PCR conditions . |
|---|---|---|---|
| Fragment I | 976 | 5′–CCAGGGGATGCAGGGGGATCC–3′ (3019–3039) | Denaturation 94°C, 5 min |
| 5′–CTTGTGGCACGCACCTTATC–3′ (3975–3994) | Annealing 52°C, 1 min | ||
| Extension 72°C, 5 min | |||
| Fragment II | 1160 | 5′–GTGATGAAGGTGACACAGACC–3′ (3919–3939) | Denaturation 94°C, 5 min |
| 5′–GACAGGGCTTCTGATACAGAA–3′ (5066–5086) | Annealing 60°C, 1 min | ||
| Extension 72°C, 5 min | |||
| Macroglycopeptide region | 320 or 359 | 5′–CCATCACATTCTCCAAAACTCC–3′ (4174–4195) | Denaturation 94°C, 5 min |
| 5′–TGGCTGATCAAGTTCAGGGATG–3′ (4472–4493) | Annealing 57°C, 1 min | ||
| Extension 72°C, 5 min | |||
| Entire GPIbα-coding region | 1882 | 5′–ACTAGTATGCCTCTCCTCCTCTTGCTG–3′ | Denaturation 92°C, 30 s |
| 5′–TCAGAGGCTGTGGCCAGAGTA–3′ | Annealing 61°C, 30 s | ||
| Extension 68°C, 20 min |
The underlined sequence represents a SpeI restriction site generated to facilitate cloning.
Reverse transcriptase–PCR
Blood (15 mL) was taken from affected and unaffected family members. Platelet-rich plasma was obtained by centrifugation at 400g for 8 minutes and platelets were pelleted by centrifugation at 1200g for 5 minutes at 20°C. The pellet was then washed twice with phosphate-buffered saline (PBS) and RNA was isolated from platelets using Qiagen RNA miniprep (Qiagen, Crawley, United Kingdom). Reverse transcription was carried out using M-MLV reverse transcriptase kit (Promega) according to the manufacturer's instructions. Amplification of the macroglycopeptide-coding region was carried out as described (see “PCR amplification and sequencing of the GPIBA gene”).
Quantitative analysis of the platelet GPIb/IX complex using flow cytometry
Quantitation of the platelet GPIb/IX/V receptor was performed using the ADIAflo kit (Axis-Shield Diagnostics, Dundee, United Kingdom). Blood samples were first incubated with the specific monoclonal antibodies (mAbs) SZ2, SZ1, and SW16 against GPIb, GPIX, and GPV, respectively, and then mixed with a polyclonal anti–mouse fluorescein isothiocyanate (FITC)–labeled IgG and incubated for 10 minutes. The mean fluorescence intensity (MFI) was measured for samples as well as for calibration standard beads and a standard curve was plotted against their corresponding number of mAb molecules (provided). The number of molecules of GPIb, GPIX, and GPV on the surface of the platelets in the tested samples was determined by converting the MFI (after subtracting the negative control value) into the corresponding number of antigenic sites per platelet based on the calibration bead standard curve.
Methods used for functional characterization
Generation of wild-type and mutant GPIbα constructs. The entire coding region of the GPIBA gene was amplified using Expand 20-Kbplus PCR system (Roche Diagnostics, Mississauga, CA). Amplification involved 2 sets of PCR cycles: 10 cycles followed by a further 25 cycles; each had similar conditions except for an additional 2 minutes' extension in the second cycle set. PCR primers and conditions are shown in Table 1. PCR products were cloned into PCR 2.1 vector using the TA cloning kit (Invitrogen, Carlsbad, CA) and the integrity of the DNA sequence was confirmed by direct sequencing. The SpeI and XhoI fragments were subcloned into the zeocin-selectable pZeoSV2 (+) mammalian expression vector (Invitrogen) using a T4 DNA ligation kit (Gibco BRL, Carlsbad, CA). Plasmid DNA was purified using the Maxi Plasmid purification kit (Qiagen).
Transfection of CHO β/IX cells. The Chinese hamster ovary (CHO) β/IX cells (a generous gift from Dr Jose Lopez, Baylor College of Medicine, Houston, TX) stably express the GPIbβ and GPIX units of the receptor complex.7 Cells were grown in minimal essential media (MEM) supplemented with 10% fetal bovine serum (FBS) 1% penicillin/streptomycin and 1% l-glutamine. To maintain high expression of the GPIbβ and GPIX polypeptides the media was also supplemented with methotrexate (MTX) 80 mmol/L and geneticin (Invitrogen) 400 μg/mL (Gibco BRL). The cells were incubated at 37°C in 5% CO2. To develop a stable cell line that expresses the GPIbα polypeptide, 2 μg of the purified, zeocin-selectable plasmid pZeoSV2 (+) carrying either the wild-type (WT) or the mutant (Mut) GPIbα was introduced into CHO β/IX cells using a Lipofectamine 2000 kit (Invitrogen). Control CHO β/IX cells were transfected with the empty vector and the process of cell selection was carried out in media supplemented with 400 μg/mL zeocin (Invitrogen).
Cell sorting using magnetic beads. Transfected and zeocin-selected CHO cells that express GPIbα with the GPIb/IX complex were sorted using CELLection Dynabeads (Dynal Biotech, Lake Success, NY). Polystyrene magnetic beads coated with human anti–mouse immunoglobulin were first washed and incubated with the GPIbα mAb VM16d. The cells were then positively selected using a magnetic concentrator (Dynal Biotech) according to the manufacturer.
mAb binding to the CHO cells expressing the WT and Mut GPIbα
To determine the effect of the deletion on surface expression of the receptor, we transiently expressed WT and Mut GPIbα in CHO cells without drug selection. We performed flow cytometry, 48 hours after transfection, using specific mAbs that bind different epitopes on the GPIbα molecule.
Cells were detached from culture plates using 0.53 mM EDTA (ethylenediaminetetraacetic acid), washed with PBS, and resuspended in PBS-1% bovine serum albumin (BSA). Cells (5 × 105) were incubated with 2 μg of each of the mAbs AK2 and WM23 (kind gift from Dr Michael Berndt, Baker Medical Research Institute, Melbourne, Victoria, Australia) as well as VM16d (distributed by Cedarlane Labs, Hornby, CA) for 60 minutes at room temperature (rt). Cells were then washed with PBS and incubated with FITC-labeled rabbit anti–mouse IgG (Chemicon, Temecula, CA) at a dilution of 1:175, for 30 minutes at rt. After a second wash with PBS the cells were fixed with 4% paraformaldehyde and analyzed by a FACScan flow cytometer (Beckman Coulter Epics Altra; Beckman Coulter, Mississauga, CA) with 488-nm laser light and collecting light emitted at more than 530 excitation signal.
Ristocetin-mediated binding of 125I-VWF to CHO cells expressing WT and Mut GPIbα
Purified human VWF (a gift from Dr Michael Berndt) was iodinated with sodium 125I by the chloramine-T method.11 In phosphate buffer, 150 μg protein was mixed with chloramine-T and 0.2 mCi (7.4 MBq) sodium 125I for 10 minutes on ice after which the reaction was stopped by sodium metabisulfite. The iodinated protein was purified using a Sephadex G-25 column (GE Healthcare, QC, Canada) and the protein concentration was assessed by Bio-Rad protein assay (Bio-Rad Laboratories, Mississauga, CA). Ristocetin-induced 125I-VWF binding was performed as previously described.32 Briefly, detached CHO cells expressing WT or Mut GPIbα were washed in Ca2+- and Mg2+-free Tyrode buffer (138 mM NaCl, 5.5 mM glucose, 12 mM Na HCO3, 0.36 mM NaH2PO4, 2.9 mM KCl, pH, 7.4) and resuspended in Tyrode buffer containing 1% BSA to a final concentration of 4 × 106 cells/mL. Each time the cells were used for a binding experiment, the level of expression of the GPIbα receptor was assessed by flow cytometry to normalize for differences in surface receptor levels. To assess VWF binding, 25 μL cell suspension was mixed with different concentrations of ristocetin (0-1 mg/mL; Sigma-Aldrich, Oakville, CA) and increasing quantities of 125I-VWF (0.2-1.6 μg/mL). The final volume of the reaction mixture was adjusted to 100 μL with Tyrode buffer. The mixture was incubated for 30 minutes at rt and then loaded onto a mini-column containing 20% sucrose in Tyrode buffer. CHO cells bound to VWF were separated from the nonbound VWF by centrifugation through the mini-column for 5 minutes at rt at 10 000g. The bottom of the tube containing the cell pellet was cut off and the radioactivity was measured in a γ counter. Specific binding was determined by subtracting the amount of nonspecific binding (binding of 125I-VWF to CHO β/IX cells) and then normalizing for differences in surface receptor levels as determined by flow cytometry.
Statistics
Data were analyzed using the Student t test. P values less than .05 were considered statistically significant. Data were represented as the mean of triplicate determinations.
Results
A British family with PT-VWD phenotype
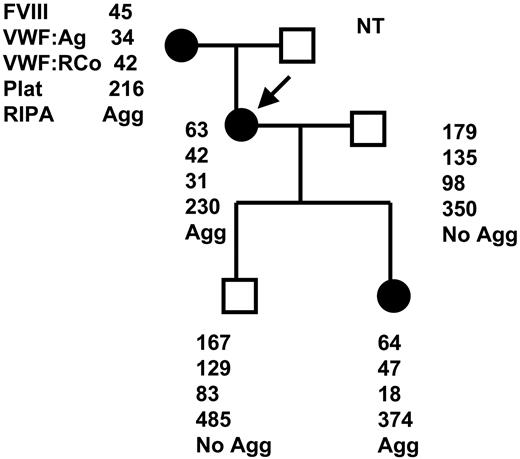
The family pedigree with 3 affected (the propositus, mother, and daughter) and 2 unaffected (the husband and son) members is shown in Figure 1. Affected individuals had laboratory features including VWF antigen in the range of 34 to 47 U/dL, FVIII 45 to 64 U/dL, and ristocetin cofactor activity (VWF:Ag RCo) ranging from 18 to 42 U/dL together with loss of the HMW VWF multimers from plasma. Platelet aggregation studies revealed an enhanced RIPA to 0.5 mg/mL ristocetin (Figure 2) that was independent of increasing the concentration of VWF. Also, washed normal platelets did not show an enhanced RIPA when mixed with the patient's plasma, indicating that the defect is localized to the patient's platelets rather than the VWF itself.
Novel 27-bp deletion in the GPIBA gene coding for the macroglycopeptide region
The entire coding region of the GPIBA gene in the propositus was amplified, cloned, and sequenced. No mutations were found in the N-terminal region including the 2 previously identified point mutations associated with PT-VWD (Figure 3A). The 3 affected family members—propositus, mother, and daughter—were heterozygous for an in-frame deletion of 27 bp (nucleotides 4374-4400; amino acid residues 421-429; Figure 3B) coding for 9 amino acids (PTILVSATS) in the macroglycopeptide region of GPIbα. In total, of the 4 to 6 sequenced clones for each affected family member, at least 2 showed the deletion and at least 2 were normal. This deletion was not found in the unaffected son or husband. No other mutations were detected elsewhere in the GPIBA gene. All family members were homozygous for the VNTR C allele (2 copies of 39-bp repeats) in the macroglycopeptide region. Sequencing revealed a threonine at residue 145 and the common - 5C allele at the Kozak sequence (C at position - 5 from the ATG translation start codon). The 27-bp deletion was not identified in any of the 50 healthy controls. PCR amplification of the macroglycopeptide region showed that all controls gave bands of 320 bp (D allele) or 359 bp (C allele), whereas the affected family members gave bands of 359 bp (wild-type C allele) and 332 bp (deleted C allele) and the unaffected family members gave only the 359-bp nondeleted C allele (Figure 4A-B). Analysis of platelet RNA from the affected family members showed that both the WT and the 27-bp deleted Mut alleles are transcribed (data not shown).
Pedigree of PT-VWD family. Affected members (•) and the unaffected members (□). The arrow indicates the propositus. Hemostatic data including FVIII (U/dL), VWF:Ag (U/dL), VWF:Ag RCoF (U/dL), platelet count (× 1000/mm2), and RIPA to 0.5 mg/mL are shown in order. Plat indicates platelet; Agg, aggregation.
Pedigree of PT-VWD family. Affected members (•) and the unaffected members (□). The arrow indicates the propositus. Hemostatic data including FVIII (U/dL), VWF:Ag (U/dL), VWF:Ag RCoF (U/dL), platelet count (× 1000/mm2), and RIPA to 0.5 mg/mL are shown in order. Plat indicates platelet; Agg, aggregation.
RIPA in patients with PT-VWD. Platelet aggregation in the presence of low concentration ristocetin (0.5 mg/mL) shows that the propositus (P), mother (M), and daughter (D) have a heightened response in comparison to lack of response of the control (C).
RIPA in patients with PT-VWD. Platelet aggregation in the presence of low concentration ristocetin (0.5 mg/mL) shows that the propositus (P), mother (M), and daughter (D) have a heightened response in comparison to lack of response of the control (C).
Normal quantities of GPIb/IX/V are expressed on the platelet surface
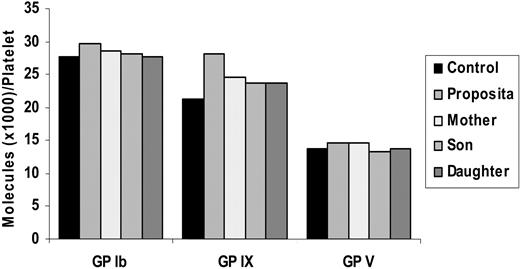
To examine if the GPIbα deletion has an effect on the synthesis or surface expression of the GPIbα protein, we performed flow cytometric quantitative analysis of the platelet GPIb/IX/V complex. The number of molecules for each of GPIb, GP IX, and GPV, in each of the affected and unaffected family members fell within the normal range (Figure 5) for the 3 glycoproteins (34.000 ± 9000, 26.000 ± 6000, and 12.000 ± 4000, respectively).
DNA sequences of the GPIBA gene showing the 27-bp deletion in the macroglycopeptide region. (A) Cloned fragment I from the GPIBA gene in the propositus showing a glycine codon at position 233 and methionine at 239 in both normal and mutant alleles. (B) Cloned fragment II from the GPIBA gene in the propositus showing the normal allele (no deletion [the 4373-4400 27 bp are boxed]) and the mutant allele (site of 27-bp deletion junction shown by an arrow).
DNA sequences of the GPIBA gene showing the 27-bp deletion in the macroglycopeptide region. (A) Cloned fragment I from the GPIBA gene in the propositus showing a glycine codon at position 233 and methionine at 239 in both normal and mutant alleles. (B) Cloned fragment II from the GPIBA gene in the propositus showing the normal allele (no deletion [the 4373-4400 27 bp are boxed]) and the mutant allele (site of 27-bp deletion junction shown by an arrow).
PCR analysis of the macroglycopeptide region of the GPIBA gene in 5 members of the PT-VWD family. (A) Schematic of macroglycopeptide region amplified in the PCR showing the position of the 27-bp deletion relative to the VNTR region (1 copy D allele). (B) Gel electrophoresis of PCR products from amplification of the macroglycopeptide region. A 100-bp ladder (200, 300, 400, 500, and 600 bp) is shown in lane 1. C1 is a normal control, homozygous for VNTR D allele (single 320-bp band). C2 is a normal control, homozygous for VNTR C allele (single 359-bp band). P, M, and D are the propositus, mother, and daughter, respectively, each with 2 bands representing the normal C allele (359 bp) and the C allele with the accompanying 27-bp deletion (332 bp). S and H refer to the healthy son and healthy husband, respectively, both of whom are homozygous for VNTR C allele and show a single 359-bp band.
PCR analysis of the macroglycopeptide region of the GPIBA gene in 5 members of the PT-VWD family. (A) Schematic of macroglycopeptide region amplified in the PCR showing the position of the 27-bp deletion relative to the VNTR region (1 copy D allele). (B) Gel electrophoresis of PCR products from amplification of the macroglycopeptide region. A 100-bp ladder (200, 300, 400, 500, and 600 bp) is shown in lane 1. C1 is a normal control, homozygous for VNTR D allele (single 320-bp band). C2 is a normal control, homozygous for VNTR C allele (single 359-bp band). P, M, and D are the propositus, mother, and daughter, respectively, each with 2 bands representing the normal C allele (359 bp) and the C allele with the accompanying 27-bp deletion (332 bp). S and H refer to the healthy son and healthy husband, respectively, both of whom are homozygous for VNTR C allele and show a single 359-bp band.
Expression of the WT and Mut GPIbα receptor in CHO β/IX cells
Plasmids that contained each of the WT GPIbα, the Mut 27-bp deletion, and a control plasmid that contains no GPIbα cDNA were generated and transfected into CHO β/IX cells that stably express GPIbβ and GPIbIX to produce CHO α/β/IX WT cells, CHO α/β/IX Mut cells, and CHO β/IX, respectively. We examined the expression of WT and Mut GPIbα receptors on the surface of transfected CHO β/IX cells by means of flow cytometry using 3 mAbs that bind to different sites on the GPIbα molecule. The mAb AK2 binds at the first LRR, residues 36-59.20,21 The mAb VM16d binds at residues 201 to 268 of the C-terminal region13,21 and WM23 binds at the macroglycopeptide region downstream to residue 28220 (Figure 6A). The binding of AK2 and VM16d were compared to that of WM23, which binds outside the VWF-binding region, to adjust for differences in surface expression of GPIbα between transfected cell lines. Data presented in Figure 6B-C demonstrate that each of the 3 mAbs bound WT and Mut GPIbα to a similar extent (n = 3, P > .05). Nevertheless, due to the limited extent of these data, we cannot exclude the possibility of conformational changes occurring to the mutant receptor that might enhance the binding of VWF. The binding of the mAbs to GpIbα was shown to be specific because binding to negative control cells (CHO β/IX) was negligible.
Ristocetin-induced 125I-VWF binding
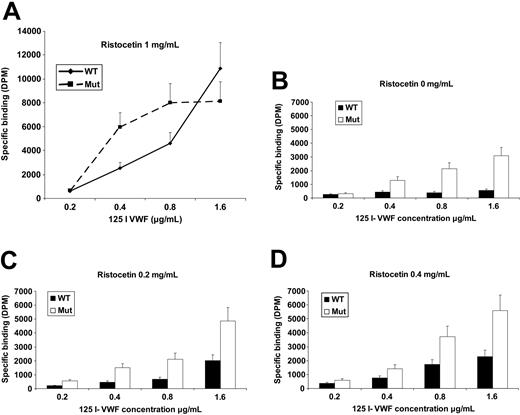
A defining feature of PT-VWD platelets is their ability to bind VWF at much lower ristocetin concentrations than those supporting binding of normal platelets.2,16,17 The same effect has been observed for recombinant GPIbα containing the PT-VWD mutations.14,15 To determine the effect of the 27-bp deletion on VWF binding, we first analyzed the binding between CHO cells expressing the Mut and WT GPIbα receptor and 125I-VWF in the presence of 1 mg/mL ristocetin. The Mut receptor bound significantly more VWF than the WT (n = 3, P = .01 and .008 at VWF levels of 0.4 and 0.8 μg/mL, respectively). With a very low (0.2 μg/mL) and a saturating concentration of VWF (1.6 μg/mL), no significant difference was observed between the Mut and WT receptor (Figure 7A). We then examined the binding under lower ristocetin concentrations (0, 0.2, and 0.4 mg/mL) and we varied the VWF concentration (0.2-1.6 μg/mL) with each level of ristocetin (Figure 7B-D). We found that the Mut receptor binds significantly more VWF than the WT in the absence of ristocetin (n = 3, P = .02-.002 at various VWF levels) and also it displayed heightened sensitivity to low ristocetin concentrations compared to the WT cells. At ristocetin concentrations of 0.2 and 0.4 mg/mL (n = 3), P was less than .001 at various VWF levels. At a VWF concentration of 0.2 μg/mL, no significant difference in binding was observed between WT and Mut GPIbα under all ristocetin concentrations (n = 3, P > .05). These results indicate that the Mut GPIbα shows enhanced binding to VWF compared to the WT receptor and that this enhanced binding is most marked with low concentrations of ristocetin.
Quantitative flow cytometric analysis of the GPIb/IX/V complex on the platelet surface. The mean fluorescence intensity (MFI) values were converted into number of molecules per platelet based on the calibration bead standard curve. A normal control as well as a standard bead is run alongside with each patient. The number of molecules of each of the 3 glycoproteins per platelet in both affected and unaffected members fall within the ranges obtained for normal control subjects (GPIb, 34.000 ± 9000; GPIX, 26.000 ± 6000; GPV, 12.000 ± 4000; n = 1).
Quantitative flow cytometric analysis of the GPIb/IX/V complex on the platelet surface. The mean fluorescence intensity (MFI) values were converted into number of molecules per platelet based on the calibration bead standard curve. A normal control as well as a standard bead is run alongside with each patient. The number of molecules of each of the 3 glycoproteins per platelet in both affected and unaffected members fall within the ranges obtained for normal control subjects (GPIb, 34.000 ± 9000; GPIX, 26.000 ± 6000; GPV, 12.000 ± 4000; n = 1).
Binding of anti-GPIbα mAbs to transfected CHO cells. (A) Schematic representation of GPIbα showing the deletion mutation and epitopes of anti-GPIbα mAbs. (B-C) The binding of AK2, VM16d, and WM23 to CHO cells expressing WT (▪) is indistinguishable from that of the Mut GPIbα (□; n = 3, P > .05). Data are represented by the mean channel fluorescence. The mAb binding is specific because the binding to CHO β/IX cells (▦; cells lacking GPIbα) is negligible. The binding of AK2 and VM16d was compared with that of WM23, whose epitope is outside the VWF-binding domain.
Binding of anti-GPIbα mAbs to transfected CHO cells. (A) Schematic representation of GPIbα showing the deletion mutation and epitopes of anti-GPIbα mAbs. (B-C) The binding of AK2, VM16d, and WM23 to CHO cells expressing WT (▪) is indistinguishable from that of the Mut GPIbα (□; n = 3, P > .05). Data are represented by the mean channel fluorescence. The mAb binding is specific because the binding to CHO β/IX cells (▦; cells lacking GPIbα) is negligible. The binding of AK2 and VM16d was compared with that of WM23, whose epitope is outside the VWF-binding domain.
Ristocetin-mediated 125I-VWF binding. (A) Transfected CHO cells are incubated for 30 minutes with increasing concentrations of 125I-VWF in the presence of ristocetin 1 mg/mL. Cell-surface GPIbα levels were measured by flow cytometry on a separate aliquot simultaneously with each binding experiment. The Mut cells bound significantly higher amounts of VWF than WT (n = 3, P = .01 and 0.008 at VWF levels of 0.4 and 0.8, respectively). (B-D) CHO cells incubated with increasing concentrations of 125I-VWF (0.2-1.6 μg/mL) under ristocetin levels of 0, 0.2, and 0.4 mg/mL. The Mut cells bound significantly higher amounts of VWF in the absence of ristocetin (n = 3, P = .02-.002 at various levels of VWF) and bound significantly more VWF than WT under different low levels of ristocetin. At ristocetin 0.2 and 0.4 mg/mL, P was .04 to .000 004 at various VWF levels. DPM indicates disintegrations per minute.
Ristocetin-mediated 125I-VWF binding. (A) Transfected CHO cells are incubated for 30 minutes with increasing concentrations of 125I-VWF in the presence of ristocetin 1 mg/mL. Cell-surface GPIbα levels were measured by flow cytometry on a separate aliquot simultaneously with each binding experiment. The Mut cells bound significantly higher amounts of VWF than WT (n = 3, P = .01 and 0.008 at VWF levels of 0.4 and 0.8, respectively). (B-D) CHO cells incubated with increasing concentrations of 125I-VWF (0.2-1.6 μg/mL) under ristocetin levels of 0, 0.2, and 0.4 mg/mL. The Mut cells bound significantly higher amounts of VWF in the absence of ristocetin (n = 3, P = .02-.002 at various levels of VWF) and bound significantly more VWF than WT under different low levels of ristocetin. At ristocetin 0.2 and 0.4 mg/mL, P was .04 to .000 004 at various VWF levels. DPM indicates disintegrations per minute.
Discussion
PT-VWD results from an increased affinity of the GPIbα platelet receptor to HMW VWF. To date, the identified mutations that cause the phenotype have been localized in the VWF-binding region of GPIbα.14,15 These mutations do not affect surface expression of the receptor but rather induce a conformational change that results in increased affinity of the GPIbα receptor for VWF at low ristocetin concentrations. Here, we present the first report of a patient with PT-VWD where the mutation does not occur within the VWF-binding region of the GPIBA gene, but rather results from an in-frame deletion within the macroglycopeptide region.
Clinical and hematologic observations as well as a laboratory re-evaluation of the propositus in this family suggested a PT-VWD phenotype rather than the previously diagnosed type IIB VWD. This was the trigger for studying the GPIBA gene. We could not find any of the previously reported mutations responsible for this disorder. However, the analysis of the GPIbα coding sequence revealed a 27-bp deletion in the macroglycopeptide region distant to the VWF-binding region. The Mut GPIbα was expressed on the surface of the platelets at normal levels and when CHO β/IX cells were transfected with a plasmid that encodes for GPIbα, the Mut receptor was expressed at levels similar to that of the WT receptor. Similar to the previously reported PT-VWD mutations Gly233Val and Met239Val, the mutant GPIbα in this study had an increased affinity for binding to VWF, an interaction that does not require the presence of ristocetin and is sensitive to low concentrations of the modulator. Although our data provide evidence for the molecular pathology of the phenotype, a question still remains as to how a deletion of 9 amino acids in the macroglycopeptide region, located at some distance from the VWF-binding domain, leads to a hyperresponsive receptor.
The crystal structure of the N-terminal domain of GPIbα22 and that of the VWF A1 domain/GPIbα complex23,24 have revealed a possible mechanism of receptor binding to VWF and shed light on the mechanism of the gain-of-function PT-VWD mutations. The positively charged VWF A1 domain contacts GPIbα at the negatively charged LRRs. The anionic sulfated region of GPIbα may also be involved in this interaction based on surface charge representation,22 but this role has not been confirmed.23 An extended discontinuous interaction at the LRR as well as the NH2-terminal B finger without involving the anionic region was also revealed.23 Evidence also suggests that 4 residues at the C-terminal flank, amino acids 225, 226, 228, and 241, bind the VWF A1 domain directly.22 Conformational changes in both the receptor and its ligand seem to be a prerequisite for binding to remove the steric hindrance made by the highly flexible loop projecting from the concave face (R-loop) and allow the fitting of the positively charged VWF A1 domain into the negatively charged concavity made by the LRR. Both Gly233Val and Met239Val mutations fall within the R-loop, a part of the C-terminal flank region, and it has been suggested that they act by favoring a more open conformation of the receptor22 or by stabilizing the interaction between the 2 molecules.23,24
The precise role of the macroglycopeptide region in receptor function is still not known. Proteolytic studies of glycocalicin, the native fragment of the α chain of GPIbα have suggested that the 84-kDa fragment, corresponding to the macroglycopeptide, may play a role in ristocetin-dependent VWF binding.25 An indication of the importance of the macroglycopeptide region can be derived from the species-specific interaction between murine platelets and human VWF. The sequence of the murine GPIbα polypeptide is 108 residues longer than its human counterpart due to extra tandem copies of a 16-residue motif in the macroglycopeptide. It has been suggested that this sequence divergence, together with the sequence variability in the VWF-binding domain, may contribute to the inability of mouse platelets to interact with human VWF.26
The GPIbα 2-bp deletion described in this report will result in decreased glycosylation in the macroglycopeptide region. This region, which is rich in proline, serine, and threonine, is heavily glycosylated on threonine and serine residues and it is known that this posttranslational modification is a potential regulator for the GPIb/IX/V-dependent function.27 It is estimated that in the normal protein there are 5 O-linked sites for every 13 amino acids.5 We speculate that, as a result of the 9-amino acid deletion in this case, 4 glycosylation sites will be lost due to the deletion of 2 serine and 2 threonine residues. This loss may have an influence on the conformation of the receptor because glycosylation has a functional role related to protein folding, stability, and receptor binding.28-30
The macroglycopeptide region forms a rigid stalk that extends the ligand-binding domain a distance of 45 nm from the platelet surface.5 Based on the GPIbα VNTR polymorphism,31 each 13-amino-acid repeat will add an extra length of 32 Å (3.2 nm) to the protein.5 Accordingly, deletion of 9 amino acids may cause a shortening of about 22 Å (2.2 nm) to the extracellular region of the receptor. It is possible that this may elicit an effect on VWF binding by restricting the mobility of the extracellular domain, which, in turn, increases sensitivity to ristocetin-induced VWF binding. A previous study has correlated an increased mobility of the extracellular domain with decreased ristocetin-induced VWF binding.32 The results of our anti-GPIbα antibody-binding studies show no differences between the WT and Mut GPIbα receptor for epitopes recognized in the LRR, C-terminal flank, and a region within the macroglycopeptide. Apparently, the epitope for WM23 is mapped outside the deleted area (residues 421-429) because it binds normally to both Mut and WT receptor. These results suggest that any conformational change of GPIbα induced by the 9-amino acid deletion is likely to be relatively subtle.
In contrast to our findings is the observation that deletion of the entire macroglycopeptide region does not affect ristocetin-mediated binding of VWF to the mutant receptor.33 This latter study, however, documented the importance of the region for shear stress-induced binding and it is possible that the specific deletion of these 9 amino acids within the macroglycopeptide region might result in a different effect on the receptor binding under static conditions. We have not studied this deletion mutant under flow conditions.
PT-VWD is a rare disease but it may be underdiagnosed because it shares most of the clinical and laboratory features with type IIB VWD and requires a molecular genetic approach to reliably discriminate between the 2 entities. In clinical practice, patients with type IIB disease are managed by VWF/FVIII infusions, but if the bleeding is severe, the patient may be given transfusions with platelets as well. This may mask the platelet abnormality and the patient may not be diagnosed as having PT-VWD. The optimum treatment for bleeding in PT-VWD is the infusion of platelet concentrates.3 Clearly, the possibility of PT-VWD should be considered in each case of type IIB VWD because the optimum treatment differs.
We have demonstrated a novel gain-of-function GPIbα mutation outside the VWF-binding domain in all affected members of a family with PT-VWD phenotype. The mutation provides a molecular basis for this phenotype and supports the role of the macroglycopeptide region in the regulation of VWF binding.
Prepublished online as Blood First Edition Paper, February 10, 2005; DOI 10.1182/blood-2002-09-2942.
Supported by funding from Southampton University Trust Hematology and Molecular Pathology Departments, Southampton, United Kingdom, and from the Canadian Institutes for Health Research (grant: MOP 42467). M.O. is a recipient of a PhD scholarship from the Egyptian Ministry of Higher Education. D.L. is the recipient of a Career Investigator Award from the Heart and Stroke Foundation of Ontario and a Canada Research Chair in Molecular Hemostasis.
An Inside Blood analysis of this article appears at the front of the issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Dr John L. Smith, Dr Elizabeth Hodges, and Dr Bernie Morley for their contribution to the early stages of the project.


![Figure 3. DNA sequences of the GPIBA gene showing the 27-bp deletion in the macroglycopeptide region. (A) Cloned fragment I from the GPIBA gene in the propositus showing a glycine codon at position 233 and methionine at 239 in both normal and mutant alleles. (B) Cloned fragment II from the GPIBA gene in the propositus showing the normal allele (no deletion [the 4373-4400 27 bp are boxed]) and the mutant allele (site of 27-bp deletion junction shown by an arrow).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/11/10.1182_blood-2002-09-2942/6/m_zh80110579110003.jpeg?Expires=1767703922&Signature=Vh9UQYKwZECC4Enl3FuF37ZpJlPfhcSsS58uOcSES4o1z21IlCTr37Mu33RNVYB-MKBJh41qKkTUchacCU9sKuDDovANE-mKLV4SBl~7Y~T3Ukgs-G3BCoyaLa~KDDjgD4Kfxf1b0JegNerR20zO3vPkMQk4kZo8YzY8KlBg3QL9MIp3Bw4elyNtXngKobPGvVCGL1N2dYPUgAtAlnWc8cFhF40hxgSCDcn0urW6BMv64MMV9SdT4EQf8~MiIc44ubMYhx85jRir~EeRnAiXBaVyJRas2DPS2z5UJ1a3rVsXwUrHrIhYgVo55b2nOf0Y9H0DyWf0Io1Bn4Vp9DZ1IA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal