Abstract
Immunoglobulin light chain amyloidosis (AL) is characterized by a clonal expansion of plasma cells within the bone marrow. Gene expression analysis was used to identify a unique molecular profile for AL using enriched plasma cells (CD138+) from the bone marrow of 24 patients with AL and 28 patients with multiple myeloma (MM) and 6 healthy controls. Class prediction analysis (PAM) revealed a subset of 12 genes, which included TNFRSF7 (CD27), SDF-1, and PSMA2, that distinguished between these 2 groups with an estimated and observed accuracy of classification of 92%. This model was validated with an independent dataset of 11 patients with AL and 12 patients with MM with 87% accuracy. Differential expression for the most discriminant genes in the 12-gene subset was validated using quantitative real-time polymerase chain reaction and protein expression analysis, which upheld the observations from the micro-array expression data. Functional analyses using a novel network mapping software revealed a number of potentially significant pathways that were dysregulated in patients with AL, with those regulating proliferation, apoptosis, cell signaling, chemotaxis, and migration being substantially represented. This study provides new insight into the molecular profile of clonal plasma cells and its functional relevance in the pathogenesis of light chain amyloidosis.
Introduction
Immunoglobulin light chain amyloidosis (AL) is an unusual hematologic dyscrasia with a protein deposition phenotype. The small, monoclonal plasma cell population within the bone marrow produces immunoglobulin, and the light chain component forms insoluble amyloid fibrils that deposit in vital organs, causing severe organ dysfunction and failure.1,2 Gene expression profiling to determine classes and subclasses of tumors as well as pathways that contribute to pathogenesis and prognosis already has been reported for a number of hematologic malignancies, including multiple myeloma (MM),3,4 non-Hodgkin lymphoma,5-8 chronic lymphocytic leukemia,9 and hairy cell leukemia.10 AL has been thought to have mechanisms of pathogenesis similar to myeloma, albeit possessing a light chain with the propensity to form amyloid fibrils. Although 5% to 10% of AL has been described as being associated with MM, true MM-associated AL is quite rare.11 Only about 18% of patients with AL have more than 20% plasma cells in the bone marrow, while most (60%) have less than 10% plasmacytosis.12 AL also shares certain clinical features such as low clonal plasma cell numbers in the bone marrow with monoclonal gammopathy of undetermined significance (MGUS), which is a potential precursor lesion for both MM and AL.13,14
It is therefore important and relevant to determine the molecular signature and functional role of the clonal plasma cells from the bone marrow (BM) in the pathogenesis of AL and MM by analyzing differentially expressed genes. We have identified a set of distinguishing genes that were individually differentially expressed in AL and MM, and we functionally characterized these genes.
Patients, materials, and methods
Sample collection and cell isolation
Bone marrow aspirates were collected from patients with AL and patients with MM. The BM aspirates of 24 patients with AL (19 λ, 5 κ) and 28 MM (10 λ, 18κ) patients were collected along with BM from 6 age-matched (range, 50-70 years) healthy individuals (healthy controls) in accordance with institutional review board (IRB) and Health Insurance Portability Accountability Act (HIPAA) regulations. All clinical samples used for research purposes (bone marrow and blood) were obtained from patients who consented to provide extra material for research at the time of clinical collection. A separate sample was not collected for research purposes. The samples were collected using an IRB-approved consent form and in accordance with HIPAA and IRB regulations. The mononuclear cells (MNCs) from BM were enriched for plasma cells using immunomagnetic bead selection with monoclonal mouse antihuman CD138 (syndecan-1, Stem Cell Technologies, Vancouver, BC, Canada) magnetic beads on an automated cell sorter (AutoMACS; Miltenyi-Biotec, Auburn, CA). Plasma cell (PC) purity was ascertained by immunocytochemistry analysis for cytoplasmic immunoglobulin light chain (LC).
Clinical classification of patients
Patients were classified as having light chain amyloidosis (AL) if they met the previously defined criteria of having amyloid syndrome.2,15,16 Patients were classified in the MM category if they had more than 10% bone marrow plasma cells (BMPCs) along with anemia or hypercalcemia, M-spike more than 3 g/dL, or lytic bone lesions. Of the 24 patients with AL, 10 had greater than 10% BMPCs, but all were negative for bone lesions. Of the 28 patients with MM, 27 had BMPC numbers greater than 10% (24 of 28 had greater than 20%), 21 of the 28 had anemia (< 12 g/dL in women and 13 g/dL in men), and 19 of 28 had lytic bone lesions. All patients with MM were negative for amyloid by Congo red staining of BM biopsies.
RNA isolation, purification, and micro-array hybridization and analysis
The enriched plasma cells (PCs) were stored in RNAlater (Ambion, Austin, TX) for RNA extraction and preparation for the gene chip experiments. Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) and the RNeasy Mini kit (Qiagen, Valencia, CA). The integrity of the RNA was assessed for each sample using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA). Double-stranded complementary DNA (cDNA) and labeled complementary RNA (cRNA) were synthesized from the total RNA and hybridized to the Affymetrix Human U133A gene chips (Affymetrix, Santa Clara, CA). The chips were further processed and scanned according to the manufacturer's protocol. The arrays were scanned with a laser scanner, and the data were visualized using the MAS 5.0 Affymetrix software (Affymetrix) to check for obvious failures in the hybridizations.
Quantitative real-time PCR analysis
Quantitative real-time polymerase chain reaction (qPCR) analysis was performed to validate the micro-array experiments for 4 target genes in CD138+ PCs from an additional 15 patients with AL, 10 patients with MM, and 6 healthy controls. CCND1 (cyclin D1), SDF-1, CD27 (TNFRSF7), PSMA2 (target genes), and β-actin (control housekeeping gene) were simultaneously amplified in the same reaction vials. Reverse transcriptase-polymerase chain reaction (RT-PCR) products were amplified using SuperScript III (Invitrogen) according to manufacturer's instructions for 10 cycles to enhance product concentrations. Two microliters of the sample was removed and amplified using the LightCycler FastStart DNA Master-PLUS SYBR Green I (Roche, Indianapolis, IN). Each of the target genes was amplified separately. All of the samples were removed from the capillaries, and equal volumes were separated on a 1% agarose gel. Densitometry was performed using the AlphaImager software (Alpha Innotech, San Leandro, CA). The ratio of the individual target gene to β-actin was calculated and compared to normal samples to determine expression levels of the appropriate transcripts.
Statistical analysis
Each Affymetrix gene probe set consists of a set of perfect match and mismatch probe pairs, typically 11 pairs in a probe set for the U133A Gene Chip. For our analysis, we used only the perfect match probes. The probe level data were normalized using Fastlo,17 a modified version of cyclic loess normalization. Differentially expressed genes between the AL and MM samples were determined using a linear mixed model similar to that described by Chu et al.18 Gene expression values were determined by a model of the individual perfect match probes within the gene probe set, a group variable to indicate an AL or MM sample, and an interaction term for a gene by group (AL or MM) effect. Genes were ranked according to the magnitude (ie, absolute value) of the grouping variable test statistic, which is a type of 2-sample t-statistic. This is equivalent to ranking genes according to their P value with respect to differential expression between the groups. Genes with a grouping variable test statistic of magnitude 3 or greater (∼ corresponds to P value < .01) were considered statistically significantly differentially expressed between the groups. These test statistic values were used for the pathway analysis performed with Ingenuity software (Ingenuity Systems, Mountain View, CA). A class prediction approach was used to determine a set of genes that best distinguished among the 2 groups (AL and MM). Specifically, the prediction analysis of micro-array (PAM) technique19 was used. This method is based on a shrunken centroid approach and uses cross-validation to choose the set of genes with the smallest estimated misclassification error (ie, the greatest estimated accuracy). The PAM analysis was validated using an independent set of 11 AL and 12 patients with MM not used in the gene chip studies. Also, gene expression analysis was performed on CD138+ plasma cells from 6 healthy individuals as healthy controls, and expression profiles were compared with the 24 patients with AL.
Gene function analysis
The functional analysis to determine the biologic relevance of the data and to identify novel, dysregulated genes was performed using a functional annotation and network-mapping tool, Ingenuity Systems software (Ingenuity Systems), which enables the discovery, visualization, and exploration of biologically and therapeutically relevant networks of gene interactions from the experimental data. A threshold of 3 for the magnitude of the test statistic value was set to identify genes whose expression was significantly differentially regulated (Focus Genes, Ingenuity Pathways Analysis, 2003 release, Ingenuity Systems, Mountain View, CA).
Immunocytochemistry for protein expression
Immunocytochemical analyses to determine protein expression for CD27 and SDF-1 were performed on cytospin slides of bone marrow mononuclear cells (BMNCs) from 4 patients with AL, 4 patients with MM, and 1 healthy control. Antibodies against κ or λ light chains were used to distinguish plasma cells. For CD27 staining, cytospin slides were fixed in 95% ethanol and incubated sequentially with antihuman CD27 fluorescein isothiocyanate (FITC)-conjugated antibody (BD Biosciences, San Diego, CA) and affinity-purified 7-aminomethyl coumarin acetic acid (AMCA)-labeled antihuman κ or λ antibody (Vector Laboratories, Burlingame, CA). For SDF-1, the cytospin slides were fixed in acetone then washed in 50 mM Tris (tris(hydroxymethyl)aminomethane)-buffered saline (TBS) and incubated with nonimmune serum for 30 minutes. Antihuman SDF-1α antibody (R&D Systems, Minneapolis, MN) was then added at a 1:5 dilution and incubated for 1 hour and revealed with the FITC-conjugated secondary antibody (goat anti-mouse IgG; Vector Laboratories). Slides were washed with 1 × TBS and incubated with affinity-purified AMCA-labeled antihuman κ or λ antibody (Vector Laboratories) for 30 minutes.
Scoring of plasma cells and confocal quantitation for protein expression
For each patient, 10 plasma cells (PCs) were counted in each field and 5 representative PCs were photographed under identical conditions using a fluorescent microscope and Leica Q-FISH software (Leica, Wetzlar, West Germany). Confocal analysis was done for each image using the KS400 Image Analysis software (Carl Zeiss, Oberkochen, Germany), and the mean fluorescence intensity (MFI) in the green channel was calculated for each cell and averaged over the 5 PCs for each patient and control.
Fluorescence in situ hybridization (FISH) for chromosomal translocation and deletion
To test for the deletion of chromosome 13 (13q14) in the 24 patients with AL and 27 patients with MM (BM cytospin slides were not available on 1 patient), the commercial probes LSI13 (Rb) (SpectrumGreen) and D13S319 (SpectrumRed, Vysis, Downers Grove, IL) were used. The normal pattern was the presence of 2 red and 2 green signals (2R, 2G). The loss of 1 or more signals was indicative of deletion (2R, 1G; 1R, 2G; or 1R, 1G). For the t(11;14) translocation, 24 patients with AL and 26 patients with MM were tested, and the commercial probe LSI IGH SG/CCND1-XT-So (Vysis) was used. The absence of a translocation (normal pattern) was indicated by 2 pairs of closely associated red and green signals. The translocation was indicated by the presence of a split signal more than 3 signal lengths apart.
Results
In this study, we determined the gene expression profile of plasma cells from the bone marrow of 24 patients with AL and compared them to 28 patients with MM and 6 age-matched healthy controls. To ensure the clonal nature of the samples that were collected for analysis, we enriched BM mononuclear cells for plasma cells using magnetic bead sorting and evaluated the purity using immunoglobulin (Ig) light chain staining (data not shown). With the exception of 2 patients with AL (50%-60% purity), all the patients with AL and patients with MM tested had more than 95% purity for clonal plasma cells within the enriched fraction.
Differential categorization of AL and MM using class prediction
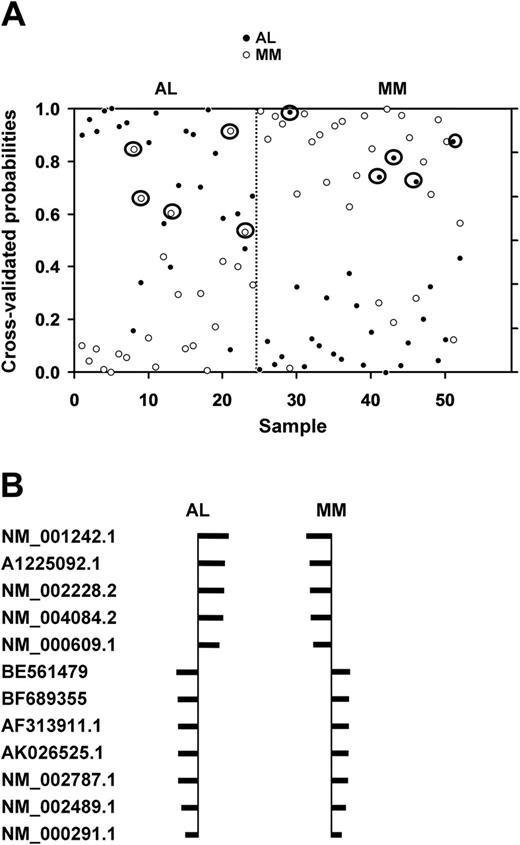
The normalized data from all 22 215 genes on the U133A chip were subjected to a class prediction analysis to identify a set of genes that could differentially classify AL and MM. The initial class prediction analysis for the AL and MM groups revealed a set of 29 genes that best differentiated between the groups with an observed accuracy of 90% and an estimated accuracy of 81% (Figure 1A). Of the 24 patients with AL, 19 were accurately predicted; and 23 of the 28 patients with MM were correctly classified (Figure 1A). The clinical features of the misclassified patients with relation to BMPC, anemia, lytic bone lesions, and M-spike are listed in Table 1.
Shrunken centroid analysis with a 29-gene subset. (A) Class prediction analysis using 29 genes derived from the 22 215 genes. The class prediction analysis was able to accurately classify 19 of the 24 patients with AL and 23 of the 28 patients with MM with an observed accuracy of 90% and a cross-validated accuracy of 81%. The y-axis shows the threshold value with samples closer to 1 having the highest probability of being an AL or MM sample, respectively. The x-axis denotes each of the 24 AL and 28 MM samples. Circled symbols indicate misclassified patients. (B) Identification of the 12 genes used to classify patients in the 2 groups. The shrunken differences for the 12 genes used for class prediction are shown. The size of the bars indicates relative distance from the centroid, with the larger bars having more significance in predicting the class. The set of 12 genes that could classify patients with AL or patients with MM with 92% observed accuracy is listed by their Affymetrix probe identification numbers. The probe sets included (in order) TNFRSF7, SDF-1 (CXCL12), JUN, PSMA2, DEFA1, NDUFA4, PGK1, and TXN.
Shrunken centroid analysis with a 29-gene subset. (A) Class prediction analysis using 29 genes derived from the 22 215 genes. The class prediction analysis was able to accurately classify 19 of the 24 patients with AL and 23 of the 28 patients with MM with an observed accuracy of 90% and a cross-validated accuracy of 81%. The y-axis shows the threshold value with samples closer to 1 having the highest probability of being an AL or MM sample, respectively. The x-axis denotes each of the 24 AL and 28 MM samples. Circled symbols indicate misclassified patients. (B) Identification of the 12 genes used to classify patients in the 2 groups. The shrunken differences for the 12 genes used for class prediction are shown. The size of the bars indicates relative distance from the centroid, with the larger bars having more significance in predicting the class. The set of 12 genes that could classify patients with AL or patients with MM with 92% observed accuracy is listed by their Affymetrix probe identification numbers. The probe sets included (in order) TNFRSF7, SDF-1 (CXCL12), JUN, PSMA2, DEFA1, NDUFA4, PGK1, and TXN.
Clinical characteristics of misclassified patients with AL and patients with MM
Misclassified patients . | . | . | . | . | . | . | |
|---|---|---|---|---|---|---|---|
| 29-gene subset . | 12-gene subset . | Light chain isotype . | BMPC, % . | Hb, g/dL . | Serum M-spike, g/dL . | Lytic bone lesions . | |
| AL-1 | NM | κ | 3 | 10.4 | Negative | Negative | |
| AL-2 | NM | λ | 16 | 13.6 | Negative | Negative | |
| AL-3 | AL-3 | λ | 25 | 11.2 | Negative | Negative | |
| AL-4 | NM | λ | 17 | 11.8 | Negative | Negative | |
| AL-5 | NM | λ | 9 | 7.7 | 1.9 | Negative | |
| ND | AL-6† | λ | 13 | 10.7 | 1.0 | Negative | |
| ND | AL-7† | λ | 11.6 | 13.2 | 2.2 | Negative | |
| MM-1 | MM-1 | λ | 31 | 12.0 | 3.3 | Negative | |
| MM-2 | MM-2 | λ | 30 | 11.3 | Hypogamma* | Negative | |
| MM-3 | NM | λ | 38 | 9.5 | 1.2 | Positive | |
| MM-4 | NM | λ | 42 | 8.5 | 0.4 | Positive | |
| MM-5 | NM | λ | 25 | 9.7 | 0.4 | Positive | |
| NM | MM-6 | κ | 24 | 12 | 2.1 | Negative | |
| ND | MM-7† | κ | 5.4 | 11 | 0.6 | Positive | |
Misclassified patients . | . | . | . | . | . | . | |
|---|---|---|---|---|---|---|---|
| 29-gene subset . | 12-gene subset . | Light chain isotype . | BMPC, % . | Hb, g/dL . | Serum M-spike, g/dL . | Lytic bone lesions . | |
| AL-1 | NM | κ | 3 | 10.4 | Negative | Negative | |
| AL-2 | NM | λ | 16 | 13.6 | Negative | Negative | |
| AL-3 | AL-3 | λ | 25 | 11.2 | Negative | Negative | |
| AL-4 | NM | λ | 17 | 11.8 | Negative | Negative | |
| AL-5 | NM | λ | 9 | 7.7 | 1.9 | Negative | |
| ND | AL-6† | λ | 13 | 10.7 | 1.0 | Negative | |
| ND | AL-7† | λ | 11.6 | 13.2 | 2.2 | Negative | |
| MM-1 | MM-1 | λ | 31 | 12.0 | 3.3 | Negative | |
| MM-2 | MM-2 | λ | 30 | 11.3 | Hypogamma* | Negative | |
| MM-3 | NM | λ | 38 | 9.5 | 1.2 | Positive | |
| MM-4 | NM | λ | 42 | 8.5 | 0.4 | Positive | |
| MM-5 | NM | λ | 25 | 9.7 | 0.4 | Positive | |
| NM | MM-6 | κ | 24 | 12 | 2.1 | Negative | |
| ND | MM-7† | κ | 5.4 | 11 | 0.6 | Positive | |
The major clinical features of the misclassified patients with AL and patients with MM in Figure 1A and Figure 3 with respect to light chain isotype, bone marrow plasma cells (BMPCs), anemia, serum M-protein level, and lytic bone lesions are recorded.
NM indicates not misclassified; ND, not done.
Urine M-spike = 3.8 g/24 hours
Patients misclassified in independent validation analysis
Functional annotation of the 29 genes revealed that 17 of the 29 were associated with the Ig λ light chain locus. Since there is a substantial over-representation of λ light chains in AL,20,21 it was unclear whether the a priori bias of λ light chains artificially skewed the results of the analysis. Therefore, we repeated the centroid analysis with the 12 non-Ig-associated genes (including SDF-1, TNFRSF7 [CD27], PSMA2 [proteasome alpha 2], JUN, DEFA1 [defensin], NDUFA4 [ubiquinone], TXN [thioredoxin], and PGK1 [phosphoglycerate kinase 1]) from the original set of 29 genes to determine if these could still predict the class of the samples. The genes with higher average expression or lower average expression in patients with AL compared to patients with MM are depicted in Figure 1B. We also compared the gene expression profile of AL PCs to normal bone marrow PCs (Tables 2, 3) for the set of genes that were significantly overexpressed or underexpressed in AL PCs compared to MM PCs (Tables 4, 5).
Expression of genes that have significantly higher expression of ALPCs as compared with MMPCs
Gene expression in AL PCs compared to healthy PCs . | Value of test statistic . |
|---|---|
| Rb | – 5.0 |
| RUNX2 | + 3.1 |
| FGFR1 | + 4.5 |
| CCND1 | + 3.2 |
| CD36 | + 4.7 |
| IGFBP5 | + 6.0 |
| APOE | + 3.00 |
| BCL6 | + 3.8 |
| PDGFRB | + 3.5 |
| HMOX1 | – 3.5 |
Gene expression in AL PCs compared to healthy PCs . | Value of test statistic . |
|---|---|
| Rb | – 5.0 |
| RUNX2 | + 3.1 |
| FGFR1 | + 4.5 |
| CCND1 | + 3.2 |
| CD36 | + 4.7 |
| IGFBP5 | + 6.0 |
| APOE | + 3.00 |
| BCL6 | + 3.8 |
| PDGFRB | + 3.5 |
| HMOX1 | – 3.5 |
The cutoff is ± 3 or greater.
+ indicates genes with significantly higher average expression in AL PCs compared to healthy PCs; –, genes with significantly lower expression in AL PCs compared to healthy PCs.
Expression of genes that have significantly lower expression of ALPCs as compared to MMPCs
Gene expression in AL PCs compared to normal PCs . | Value of test statistic . |
|---|---|
| PSEN1 | – 4.9 |
| CD47 | – 4.1 |
| TGFB1 | + 4.1 |
| CASP3 | – 4.2 |
| TCF3 (E2A) | + 5.8 |
| IL-6ST | + 4.7 |
Gene expression in AL PCs compared to normal PCs . | Value of test statistic . |
|---|---|
| PSEN1 | – 4.9 |
| CD47 | – 4.1 |
| TGFB1 | + 4.1 |
| CASP3 | – 4.2 |
| TCF3 (E2A) | + 5.8 |
| IL-6ST | + 4.7 |
Cutoff is ± 3 or greater.
+ indicates genes with significantly higher average expression in AL PCs compared to normal PCs; –, genes with significantly lower expression in AL PCs compared to normal PCs.
Genes with significantly higher average gene expression in AL plasma cells compared to MM
Gene symbol . | Name of gene . | Genes the focus gene regulates . | Genes by which the focus gene is regulated . | Genes the focus gene binds . | Value of the test statistic . |
|---|---|---|---|---|---|
| RARA | Retinoic acid receptor, alpha MMP11, FOS | RXRA, MYOD1, POU1F1, PKC, STAT5B, IL1B | IL3, NCOA3, RXRA, RARB, OXT, MYB | RXRA, NCOA1, NCOR1, 2, | 3.60 |
| RB1/Rb* | Retinoblastoma MAPK8 | HBP1, RNA Pol III, cyclin A, CDKn2A, p53, EGF | E2F1, TGFB1, IL3, BCL2, RBAK, MYC | E2F4; HDAC1, 2, 3; BRCA1 | 3.73 |
| CCND1* | Cyclin D1 CDK4, CCND3, CDKN1A | Cyclin A, RB1 (Rb), CDK2, TNF, CDKN1A, IL3, NFκB, PCNA, TGFBR2 | CDX1, CDKN1B, TGFB1, Rb, ER, CDK6, RELA ubiqultin | CDK4; CDKN1C, B, A; CDK2 | 5.07 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B | CDK4, CCDN1, IL4, MYC, Rb, CDKN1A | TGFB1, CCNE1, DC, DC25A, IL4, LMP1, MYC, IL3, IL7, p53, BCL2 | CCNA2, YWHAE, GRB2, CCND1, CDK4, XPO1, KIS | 3.95 |
| CDC25B | Cell division cycle 25B | CCND1, CDK2, MYC, Cyclin A, CDC2, p53 | CDKN1A, p53, CDC2, Proteasome | CCNB1, YWHAZ, CDC2 | 3.96 |
| JUN | v-jun sarcoma virus homolog | BCL3, CDKN1A, p53, BAX, MDM2, MSH2, SERPINE1, IGFBP1 | IL6; CEPBA; EGF; IL4; MAPK10; TGFB1, 3 | HOXD12; MMP1; TNFRSF6; ATF2; MAPK8, 9, 10; FOS; PGR | 6.58 |
| MAPK12 | Mitogen-activated protein kinase 12 | TNFSF6, p53, IL2, JUN, TNF, FOS, CCL5, THPO | EGF, TGFB1, CXCL12, VEGF, TNF, IL3, CCL5, THPO | MAP2K4, GRB2, MAP3K1, JUN, ATF2, p53, JUNB | 4.77 |
| RUNX2 | Runt-related transcription factor 2 | FN1, COL1A1, MYOD1, TNFRSF11B, MMP13, TPS1 | BMP2, TGFB1, VEGF, FGF2, TNF, MAPK, IGFBP5, IL7 | MADH3, MADh1, Osteocalcin, JUN, FOS, CEBPA, MMP13 | 3.74 |
| NFκB1A* | Inhibitor of nuclear factor of κ light chain gene enhancer in B cells | NFκB, ICAM1, TNF, CASP3, RELA, RELB | IL1B, TNF, Hsp70, IKBKB, IL2, p53 | MYOD1, RELA, XPO1, RELB, p53, JUN, IKK, IKBKB | 3.29 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | AKT1, BCL2, IGF1R, BAX, BAD, BCL2L1 | IL1b, p53, TGFB1, IL6, EGF, PTEN | IGF2, p53, IGF1R, THBS1 | 5.38 |
| IGFBP5 | Insulin-like growth factor binding protein 5 | AKT, BCL2, CASP3, BCL2L1, Erk 1/2, p 38MAPK, RUNX2 | IGF1, TGFB1, IGFBP3, SERPING1, STAT3, PTEN, AKT1 | IGF1, 2; MYB; MYBL2; SERPINE2 | 3.87 |
| FGFR1 | Fibroblast growth factor receptor 1 | FGFR1, JUN, BMP4, AKT, PLCγ | IL3; FGF2, 1; VEGF; MMP2; CCND1; CDKN2A | FGFR2; FGF1, 7, 4; GRB7; CD44; FGF13 | 3.90 |
| TNFRSF1A | Tumor necrosis factor receptor superfamily 1A | CASP3, NFκB, IL1B, CXCL12, TNF, VEGF, TGFB1 | IFNγ, TNF, TRADD, TRAD2, p53, RELB | TRAF1, TRAF2, JAK1, JAK2, TNF, TRADD, FADD, DAXX, STAT1 | 3.78 |
| PDGRFB | Platelet derived growth factor receptor, beta polypeptide | SRC, SYK, FOS, PIP3K, Vav, PKC | FN1, GPCR, MYC, KRAS2 | PIK3R1, PDGFRA, PI3K, STAT5, GPRK2L | 3.53 |
| BCL6 | B-cell CLL/Lymphoma 6 | BAX; p38MAPK; BCL2; CCL-2, 6, and 7 | BCL6, MYOD1, MAPK | IRF4, JUN, BCL11A, BCL2L1 | 3.40 |
| APOE | Apolipoprotein E | APP, Fibrin, Ig, HMOX1, PPARγ, MMP9, CREB1 | APP, APOA1, RXRA, IL1B, LPL | APP, APOA1, LPL, TAU40, SERPINA3C | 4.82 |
| CD36 | Thrombospondin receptor | INS1 PPARγ, PPARα, CYCL12 | TGFB1, IL4, NFκB, IL6, | FYN, LYN, CD9, ITGB1, ITGA3 | 4.54 |
| CD44 | CD44 antigen | MMP9, FGFR1, LYN, MMP7, ETV5 | NFκB, MYC, THPO, JUN, IGF1, TNF, OSM, KRAS2 | GRB2, LCK, FYN, FGFR1, RELA, FN1 | 3.19 |
| CXCL12* | SDF-1 | AKT, ERK 1/2LCK, JAK1, JAK3, STAT3, STA5B, PI3K | TNF, TAT, KRAS2, MMP2, TGFB1, VEGF, MMP9, MMP1 | CXCR4, MMP2, FN1 | 5.95 |
| HMOX1 | Heme Oxygenase 1 | MAPK1, HMOX1, TNF, IL6 | TGFB1, IL1B, IL10, TNF | APP, NFκB, Ap2 | 4.12 |
| DCN | Decorin | ERBB3, ERBB4, CDKN1A, CDKN1B, Cyclin A, VEGF, AKT1, TGFB | LPL; TNF; DES; MMP2, 3, 7, 9; TGFB1; IL10; IL1B | WISP1, EGFR, Collagen, TNX, C1QA, Apoa | 3.94 |
| BTRC | Beta-transducin repeat containing | NFκB1A, NFκBIB, IκBKE, ATF4 | TNF, IκBKB, NFκBIA | NFκBIA, NFκB1, ATF4, UBQLN2, TNFRSF5 | 4.28 |
| APP | Amyloid beta (A4) precursor protein, appican | APOE, CACNL, CASP9, TNF, MMP9, SPNA2 | APOA1, SERPINA1, CASP8, APOE, PSEN1, TNF, IL1B | PSEN1, RXRA, APOA1, APBA2, LRP1 | 3.72 |
| A2M | Alpha 2-macroglobulin | IGFBP1, APP, PDGRFB, CREB1 | LIF, IL6, INS1, APP, IL1B, STAT3 | VEGF, APOE, IL2, IL6, TGFB1 | 3.91 |
| SERPINA1 | Serine proteinase inhibitor (α-anti-trypsin) | CCL2, IL2, APP, TGFB1, PDGF, NFκB | MMP9, ELA2, Proteasome, XBP1, PPARγ, MMP1 | NR5A2, CANX, ELA2, HSPA5 | 5.24 |
| TNFRSF6 | Tumor necrosis factor receptor superfamily, 6 | CASP3, CASP8, CASP9, NFκB1, MAPK8, TNFSF6, BID | IFNg, IL15, PTEN, TNF, MYC, NFκB, p53, IL10, LMP1 | TNFSF6, JUN, STAT3, p53, FADD, CASP10 TRADD | 4.49 |
Gene symbol . | Name of gene . | Genes the focus gene regulates . | Genes by which the focus gene is regulated . | Genes the focus gene binds . | Value of the test statistic . |
|---|---|---|---|---|---|
| RARA | Retinoic acid receptor, alpha MMP11, FOS | RXRA, MYOD1, POU1F1, PKC, STAT5B, IL1B | IL3, NCOA3, RXRA, RARB, OXT, MYB | RXRA, NCOA1, NCOR1, 2, | 3.60 |
| RB1/Rb* | Retinoblastoma MAPK8 | HBP1, RNA Pol III, cyclin A, CDKn2A, p53, EGF | E2F1, TGFB1, IL3, BCL2, RBAK, MYC | E2F4; HDAC1, 2, 3; BRCA1 | 3.73 |
| CCND1* | Cyclin D1 CDK4, CCND3, CDKN1A | Cyclin A, RB1 (Rb), CDK2, TNF, CDKN1A, IL3, NFκB, PCNA, TGFBR2 | CDX1, CDKN1B, TGFB1, Rb, ER, CDK6, RELA ubiqultin | CDK4; CDKN1C, B, A; CDK2 | 5.07 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B | CDK4, CCDN1, IL4, MYC, Rb, CDKN1A | TGFB1, CCNE1, DC, DC25A, IL4, LMP1, MYC, IL3, IL7, p53, BCL2 | CCNA2, YWHAE, GRB2, CCND1, CDK4, XPO1, KIS | 3.95 |
| CDC25B | Cell division cycle 25B | CCND1, CDK2, MYC, Cyclin A, CDC2, p53 | CDKN1A, p53, CDC2, Proteasome | CCNB1, YWHAZ, CDC2 | 3.96 |
| JUN | v-jun sarcoma virus homolog | BCL3, CDKN1A, p53, BAX, MDM2, MSH2, SERPINE1, IGFBP1 | IL6; CEPBA; EGF; IL4; MAPK10; TGFB1, 3 | HOXD12; MMP1; TNFRSF6; ATF2; MAPK8, 9, 10; FOS; PGR | 6.58 |
| MAPK12 | Mitogen-activated protein kinase 12 | TNFSF6, p53, IL2, JUN, TNF, FOS, CCL5, THPO | EGF, TGFB1, CXCL12, VEGF, TNF, IL3, CCL5, THPO | MAP2K4, GRB2, MAP3K1, JUN, ATF2, p53, JUNB | 4.77 |
| RUNX2 | Runt-related transcription factor 2 | FN1, COL1A1, MYOD1, TNFRSF11B, MMP13, TPS1 | BMP2, TGFB1, VEGF, FGF2, TNF, MAPK, IGFBP5, IL7 | MADH3, MADh1, Osteocalcin, JUN, FOS, CEBPA, MMP13 | 3.74 |
| NFκB1A* | Inhibitor of nuclear factor of κ light chain gene enhancer in B cells | NFκB, ICAM1, TNF, CASP3, RELA, RELB | IL1B, TNF, Hsp70, IKBKB, IL2, p53 | MYOD1, RELA, XPO1, RELB, p53, JUN, IKK, IKBKB | 3.29 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | AKT1, BCL2, IGF1R, BAX, BAD, BCL2L1 | IL1b, p53, TGFB1, IL6, EGF, PTEN | IGF2, p53, IGF1R, THBS1 | 5.38 |
| IGFBP5 | Insulin-like growth factor binding protein 5 | AKT, BCL2, CASP3, BCL2L1, Erk 1/2, p 38MAPK, RUNX2 | IGF1, TGFB1, IGFBP3, SERPING1, STAT3, PTEN, AKT1 | IGF1, 2; MYB; MYBL2; SERPINE2 | 3.87 |
| FGFR1 | Fibroblast growth factor receptor 1 | FGFR1, JUN, BMP4, AKT, PLCγ | IL3; FGF2, 1; VEGF; MMP2; CCND1; CDKN2A | FGFR2; FGF1, 7, 4; GRB7; CD44; FGF13 | 3.90 |
| TNFRSF1A | Tumor necrosis factor receptor superfamily 1A | CASP3, NFκB, IL1B, CXCL12, TNF, VEGF, TGFB1 | IFNγ, TNF, TRADD, TRAD2, p53, RELB | TRAF1, TRAF2, JAK1, JAK2, TNF, TRADD, FADD, DAXX, STAT1 | 3.78 |
| PDGRFB | Platelet derived growth factor receptor, beta polypeptide | SRC, SYK, FOS, PIP3K, Vav, PKC | FN1, GPCR, MYC, KRAS2 | PIK3R1, PDGFRA, PI3K, STAT5, GPRK2L | 3.53 |
| BCL6 | B-cell CLL/Lymphoma 6 | BAX; p38MAPK; BCL2; CCL-2, 6, and 7 | BCL6, MYOD1, MAPK | IRF4, JUN, BCL11A, BCL2L1 | 3.40 |
| APOE | Apolipoprotein E | APP, Fibrin, Ig, HMOX1, PPARγ, MMP9, CREB1 | APP, APOA1, RXRA, IL1B, LPL | APP, APOA1, LPL, TAU40, SERPINA3C | 4.82 |
| CD36 | Thrombospondin receptor | INS1 PPARγ, PPARα, CYCL12 | TGFB1, IL4, NFκB, IL6, | FYN, LYN, CD9, ITGB1, ITGA3 | 4.54 |
| CD44 | CD44 antigen | MMP9, FGFR1, LYN, MMP7, ETV5 | NFκB, MYC, THPO, JUN, IGF1, TNF, OSM, KRAS2 | GRB2, LCK, FYN, FGFR1, RELA, FN1 | 3.19 |
| CXCL12* | SDF-1 | AKT, ERK 1/2LCK, JAK1, JAK3, STAT3, STA5B, PI3K | TNF, TAT, KRAS2, MMP2, TGFB1, VEGF, MMP9, MMP1 | CXCR4, MMP2, FN1 | 5.95 |
| HMOX1 | Heme Oxygenase 1 | MAPK1, HMOX1, TNF, IL6 | TGFB1, IL1B, IL10, TNF | APP, NFκB, Ap2 | 4.12 |
| DCN | Decorin | ERBB3, ERBB4, CDKN1A, CDKN1B, Cyclin A, VEGF, AKT1, TGFB | LPL; TNF; DES; MMP2, 3, 7, 9; TGFB1; IL10; IL1B | WISP1, EGFR, Collagen, TNX, C1QA, Apoa | 3.94 |
| BTRC | Beta-transducin repeat containing | NFκB1A, NFκBIB, IκBKE, ATF4 | TNF, IκBKB, NFκBIA | NFκBIA, NFκB1, ATF4, UBQLN2, TNFRSF5 | 4.28 |
| APP | Amyloid beta (A4) precursor protein, appican | APOE, CACNL, CASP9, TNF, MMP9, SPNA2 | APOA1, SERPINA1, CASP8, APOE, PSEN1, TNF, IL1B | PSEN1, RXRA, APOA1, APBA2, LRP1 | 3.72 |
| A2M | Alpha 2-macroglobulin | IGFBP1, APP, PDGRFB, CREB1 | LIF, IL6, INS1, APP, IL1B, STAT3 | VEGF, APOE, IL2, IL6, TGFB1 | 3.91 |
| SERPINA1 | Serine proteinase inhibitor (α-anti-trypsin) | CCL2, IL2, APP, TGFB1, PDGF, NFκB | MMP9, ELA2, Proteasome, XBP1, PPARγ, MMP1 | NR5A2, CANX, ELA2, HSPA5 | 5.24 |
| TNFRSF6 | Tumor necrosis factor receptor superfamily, 6 | CASP3, CASP8, CASP9, NFκB1, MAPK8, TNFSF6, BID | IFNg, IL15, PTEN, TNF, MYC, NFκB, p53, IL10, LMP1 | TNFSF6, JUN, STAT3, p53, FADD, CASP10 TRADD | 4.49 |
Twenty-six genes, identified for further analysis from 5 networks using the Ingenuity Analysis software, had higher average expression in patients with AL. The table shows a partial list of dynamic interactions between these 26 focus genes and other genes that critically influence their function. The value of the test statistic is shown as a comparison of AL and MM plasma cells.
Genes in these rows may be particularly relevant to the pathogenesis of AL, including RB, CCND1, NFκB1A, and CXCL12
Genes with significantly lower average gene expression in AL plasma cells than in MM cells
Gene symbol . | Name of gene . | Genes the focus gene regulates . | Genes by which the focus gene is regulated . | Genes the focus genes binds . | Value of the test statistic . |
|---|---|---|---|---|---|
| MYC* | v-myc myelocytomatosis viral oncogene homolog | CSK2, CDKN1A, NFκB, CDC25A, PDGFRB | RGFB1, IL3, CDKN1B, EPO, KRAS2, RELA | TRADD; RBL1; E2F1, 2, 3, 4, and 5 | –3.78 |
| p53* | Tumor protein p53 | JUN, CDKN1A, IGFBP3, NFκB1, RELA, CREBBP | IL3, ATM, ETS1, NFκB, MAPK8 | MDM2, CREB, CDKN2A, CREBBP | –3.54 |
| E2A* | E2A Ig enhancer binding factor (E12/E47) | CDKN1a, PAX-5, RAG1, VPREB1, CD79A | EBP, PAX3, IL6, PIP, NOTCH1 | HOXB7, HOXB8, HOXA5, MYOG, MYOD1, RB1, EBF | –4.17 |
| XRCC4 | X-ray repair factor 4 | POLM, LIG4 | POLM, KITLG | LIG4, XRCC5, PRKDC, DNA Ligase | –3.43 |
| XRCC5* | Ku 80 | Osteocalcin, DNA-PK, RNA Pol II | LOX, CXCL12, p53, DNA-PK | XRCC4, PCNA, Telomerase | –3.99 |
| TXN | Thioredoxin | VEGF, CD4, PTEN, IL2, NFκB, BCL2, TRAF6 | ILlb, MYC, ATM | CREB3, PTEN, NFκB | –5.88 |
| IL6ST* | IL6 signal transducer | IL6, JUNB, JAK1, 2, STAT1, 3, 5B | TNF, IL6, OSM, IL6R, SOCS | STAT3; SOCS3; IL11; IL6; JAK1, 2; STAT1 | –3.71 |
| TGFB1 | Transforming growth factor B1 | CDK2, CDKN1A, Cyclin D, CDK6, CDKN1B, MYC | IL4, IL13, TNF | TGFBR, EGR1, ITGAV, A2M | –3.21 |
| CASP3* | Caspase 3 | CASP8, 9, BID, CASP6, SSB | AKT1, BID, TNF | CASP1, CDKN1A, BCL2L1, STAT1, CCND3 | –3.00 |
| RELA | NFκB p65 subunit | JUNB, CD86, CD44, TNFRSF5, CCR7, ICAM, BCL2L1, TNFRSF6 | PPARγ, TNF, FADD, TRADD, TNFRSF1A, p53, IKBKB | MMP9, NFKB1A, IFNG, IL6, IL4, IL12B | –3.91 |
| CD47 | CD47 antigen | THBS1, PTPN2 | ERBB2, HOXa11, CSCL12 | UBQLN1, SRC, LICAM | –4.33 |
| CDK4* | Cyclin-dependent kinase 4 | RBL1; CDK2; MYC; RB1; CDKN1B, C; DKN1A; CDK6 | IL4, MYC, CDKN1A, CDKN1B, BCL2, PPAR6, BCL2L1 | PCNA, MDM2, p53, CCND1, CDKN1C, CDKN1A, CDKN1B, CDK6, CCND2 | –4.69 |
| CDK5 | Cyclin-dependent kinase 5 | MYC, JUN, MAPK10, APP | CDK5R1, Laminin 1, PHO80 | CCND1, SET, ACTN1, PHO80 | –3.61 |
| CDK9 | Cyclin-dependent kinase 9 | MYC, RB1, MBP, MYOD1 | Alkaline phosphatase, PTEN, IL6, TAT | TAT, RB1, MYC, RELA, Hsp90, MYOD1, NFκB1 | –4.42 |
| CDC37 | Cell division cycle 37 homolog | CDK4, Hsp90 | CDKN2A, LCK, CCL5 | CCND1, CDK6, Hsp90, TRAF3, SRC, IKBKG | –3.63 |
| CCNB1 | Cyclin B1 | RB1, CDC25A, CDKN1B, p53, GRO | TGFB1, CDC25A, CDKN1A, MYC, PTEN, p53 | CDC2, CDC25A, CDKN1A, RELA, PAP, MYC, ERBB2 | –4.06 |
| PSEN1 | Presenilin 1 | LRP1, APP, Gamma secretase, JUN, CCND1, NOTCH1 | ETS1, p53, PSEN2, CASP3, IL1B, GSK3B | APBA1, APBA2, NOTCH1, APP, Alpha catenin, NOTCH2 and 3 | –3.82 |
| PSEN2 | Presenilin 2 | APP, NOTCH1, ERBB4, BAX, PSEN1, p53, BCL2, CASP3 | PSEN1, p53, CASP3, Ubiquilin, NCSTN | NOTCH1, APP, PEN2, PSEN1, Gamma secretase, PSEN2, BCL2L1 | –3.36 |
| UCHL1 | Ubiquitin carboxyl-terminal esterase L1 | Ubiquitin | HNF4a, MYCN | Ubiquitin, CDKN1B, SNCA | –3.12 |
| PTEN | Phosphatase and tensin homolog | BCL2, CASP3, TNFRSF6, AKT1, CSN2 | ERBB2, BCL2, TGFB1, HBX, EGR1 | MAGI-3, AIP1, TXN, p53 | –3.90 |
Gene symbol . | Name of gene . | Genes the focus gene regulates . | Genes by which the focus gene is regulated . | Genes the focus genes binds . | Value of the test statistic . |
|---|---|---|---|---|---|
| MYC* | v-myc myelocytomatosis viral oncogene homolog | CSK2, CDKN1A, NFκB, CDC25A, PDGFRB | RGFB1, IL3, CDKN1B, EPO, KRAS2, RELA | TRADD; RBL1; E2F1, 2, 3, 4, and 5 | –3.78 |
| p53* | Tumor protein p53 | JUN, CDKN1A, IGFBP3, NFκB1, RELA, CREBBP | IL3, ATM, ETS1, NFκB, MAPK8 | MDM2, CREB, CDKN2A, CREBBP | –3.54 |
| E2A* | E2A Ig enhancer binding factor (E12/E47) | CDKN1a, PAX-5, RAG1, VPREB1, CD79A | EBP, PAX3, IL6, PIP, NOTCH1 | HOXB7, HOXB8, HOXA5, MYOG, MYOD1, RB1, EBF | –4.17 |
| XRCC4 | X-ray repair factor 4 | POLM, LIG4 | POLM, KITLG | LIG4, XRCC5, PRKDC, DNA Ligase | –3.43 |
| XRCC5* | Ku 80 | Osteocalcin, DNA-PK, RNA Pol II | LOX, CXCL12, p53, DNA-PK | XRCC4, PCNA, Telomerase | –3.99 |
| TXN | Thioredoxin | VEGF, CD4, PTEN, IL2, NFκB, BCL2, TRAF6 | ILlb, MYC, ATM | CREB3, PTEN, NFκB | –5.88 |
| IL6ST* | IL6 signal transducer | IL6, JUNB, JAK1, 2, STAT1, 3, 5B | TNF, IL6, OSM, IL6R, SOCS | STAT3; SOCS3; IL11; IL6; JAK1, 2; STAT1 | –3.71 |
| TGFB1 | Transforming growth factor B1 | CDK2, CDKN1A, Cyclin D, CDK6, CDKN1B, MYC | IL4, IL13, TNF | TGFBR, EGR1, ITGAV, A2M | –3.21 |
| CASP3* | Caspase 3 | CASP8, 9, BID, CASP6, SSB | AKT1, BID, TNF | CASP1, CDKN1A, BCL2L1, STAT1, CCND3 | –3.00 |
| RELA | NFκB p65 subunit | JUNB, CD86, CD44, TNFRSF5, CCR7, ICAM, BCL2L1, TNFRSF6 | PPARγ, TNF, FADD, TRADD, TNFRSF1A, p53, IKBKB | MMP9, NFKB1A, IFNG, IL6, IL4, IL12B | –3.91 |
| CD47 | CD47 antigen | THBS1, PTPN2 | ERBB2, HOXa11, CSCL12 | UBQLN1, SRC, LICAM | –4.33 |
| CDK4* | Cyclin-dependent kinase 4 | RBL1; CDK2; MYC; RB1; CDKN1B, C; DKN1A; CDK6 | IL4, MYC, CDKN1A, CDKN1B, BCL2, PPAR6, BCL2L1 | PCNA, MDM2, p53, CCND1, CDKN1C, CDKN1A, CDKN1B, CDK6, CCND2 | –4.69 |
| CDK5 | Cyclin-dependent kinase 5 | MYC, JUN, MAPK10, APP | CDK5R1, Laminin 1, PHO80 | CCND1, SET, ACTN1, PHO80 | –3.61 |
| CDK9 | Cyclin-dependent kinase 9 | MYC, RB1, MBP, MYOD1 | Alkaline phosphatase, PTEN, IL6, TAT | TAT, RB1, MYC, RELA, Hsp90, MYOD1, NFκB1 | –4.42 |
| CDC37 | Cell division cycle 37 homolog | CDK4, Hsp90 | CDKN2A, LCK, CCL5 | CCND1, CDK6, Hsp90, TRAF3, SRC, IKBKG | –3.63 |
| CCNB1 | Cyclin B1 | RB1, CDC25A, CDKN1B, p53, GRO | TGFB1, CDC25A, CDKN1A, MYC, PTEN, p53 | CDC2, CDC25A, CDKN1A, RELA, PAP, MYC, ERBB2 | –4.06 |
| PSEN1 | Presenilin 1 | LRP1, APP, Gamma secretase, JUN, CCND1, NOTCH1 | ETS1, p53, PSEN2, CASP3, IL1B, GSK3B | APBA1, APBA2, NOTCH1, APP, Alpha catenin, NOTCH2 and 3 | –3.82 |
| PSEN2 | Presenilin 2 | APP, NOTCH1, ERBB4, BAX, PSEN1, p53, BCL2, CASP3 | PSEN1, p53, CASP3, Ubiquilin, NCSTN | NOTCH1, APP, PEN2, PSEN1, Gamma secretase, PSEN2, BCL2L1 | –3.36 |
| UCHL1 | Ubiquitin carboxyl-terminal esterase L1 | Ubiquitin | HNF4a, MYCN | Ubiquitin, CDKN1B, SNCA | –3.12 |
| PTEN | Phosphatase and tensin homolog | BCL2, CASP3, TNFRSF6, AKT1, CSN2 | ERBB2, BCL2, TGFB1, HBX, EGR1 | MAGI-3, AIP1, TXN, p53 | –3.90 |
Twenty genes, identified for further analysis from 5 networks using the Ingenuity Analysis software, had lower average expression in patients with AL (or higher average expression in patients with MM). The table is a partial list of dynamic interactions between these 20 focus genes and other genes that critically influence the function of the focus genes. The value of the test statistic is shown as a comparison of AL and MM plasma cells.
Genes in these rows may be particularly relevant to the pathogenesis of AL and MM, including MYC, p53, E2A, XRCC5 (Ku 80), IL6-ST, CASP3, and CDK4
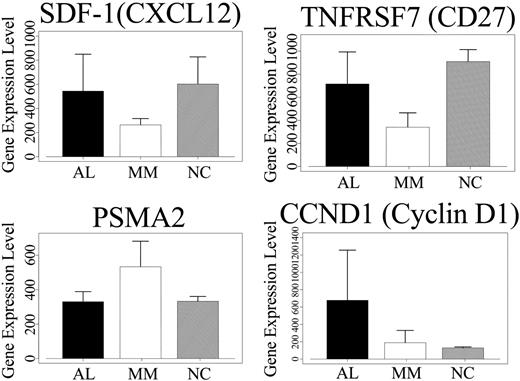
Zhan et al3 in their comparison of gene expression in MM PCs to normal bone marrow PCs demonstrated that SDF-1/CXCL12, DEFA1 (defensin), and TNFRSF7 are significantly down-regulated in MM PCs. We find that these same genes are significantly up-regulated with a test statistic value of 5.95, 5.83, and 7.46, respectively, in AL PCs compared to MM PCs. In AL PCs compared to normal PCs, average gene expression levels for SDF-1 (544 and 602) and TNFRSF7 (CD27) (716 and 909) show a difference of 58 and 193, respectively (Figure 2), while for DEFA1, the expression is substantially lower (data not shown). Munshi et al22 have shown that PSMA2 is significantly up-regulated in MM PCs when compared to normal PCs.23 PSMA2, PGK1, ubiquinone (NDUFA4), and TXN are down-regulated in AL PCs compared to MM PCs, with test statistic values of -7.022, -7.43, -5.622, and -5.88, respectively. In AL PCs compared to normal PCs, there is either comparable expression levels for PSMA2 (Figure 2) or slightly lower expression for TXN and slightly higher expression for NDUFA4 (data not shown). Interestingly, the cell cycle oncogene JUN and inhibitor of NFκB, NFκBIA, which is up-regulated in MM PCs (compared to normal controls),22 have a higher average expression in AL PCs with a test statistic value of 6.58 and 3.29, respectively. The expression of JUN in AL PCs is about 1- to 2-fold higher than normal PCs (data not shown). Therefore, the overall pattern of gene expression for most of the 12 genes shows intermediate levels in AL PCs, compared to MM PCs and normal PCs.
Microarray gene expression analysis in AL, MM, and normal PCs. Average gene expression levels for the 3 most discriminating genes (SDF-1, CD27, PSMA2) are shown for the 24 AL, 28 MM, and 6 healthy controls. In addition, gene expression levels for CCND1 are shown for these 3 groups. For SDF-1 and CD27, the average gene expression is higher in AL than MM PCs, but lower than normal PCs (SDF-1, AL = 544, MM = 264, NC = 602; CD27, AL = 716, MM = 340, NC = 909). For PSMA2, gene expression levels are higher in MM PCs compared to AL and normal PCs. CCND1 expression is the highest in AL PCs compared to MM and normal PCs. Error bars indicate standard deviation.
Microarray gene expression analysis in AL, MM, and normal PCs. Average gene expression levels for the 3 most discriminating genes (SDF-1, CD27, PSMA2) are shown for the 24 AL, 28 MM, and 6 healthy controls. In addition, gene expression levels for CCND1 are shown for these 3 groups. For SDF-1 and CD27, the average gene expression is higher in AL than MM PCs, but lower than normal PCs (SDF-1, AL = 544, MM = 264, NC = 602; CD27, AL = 716, MM = 340, NC = 909). For PSMA2, gene expression levels are higher in MM PCs compared to AL and normal PCs. CCND1 expression is the highest in AL PCs compared to MM and normal PCs. Error bars indicate standard deviation.
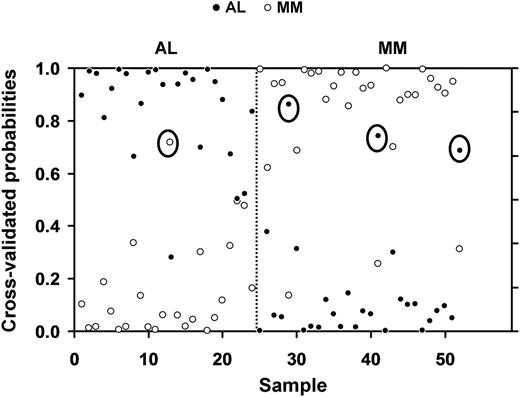
The 12-gene subset was able to substantially improve the observed and cross-validated accuracy of classification to 92% (Figure 3). With the revised analysis, 23 of the 24 patients with AL were correctly predicted using the set of 12 genes, and 25 of the 28 patients with MM also were accurately classified (Figure 3). The single misclassified AL patient (AL-3) and 2 of the MM (MM-1 and MM-2) patients were represented among the 5 misclassified patients in each group in the previous analysis (Table 1). The third misclassified patient (MM-6, Table 1) was diagnosed with a Stage IIA myeloma.
Class prediction analysis using 12 genes derived from the 29 genes used for the initial analysis. The class prediction analysis after the removal of the Ig λ LC genes, which could artificially skew the accuracy of classification, resulted in an improvement in the observed accuracy rate to 92%. The disease category of 23 of the 24 patients with AL and 25 of the 28 patients with MM was accurately predicted. The y-axis shows the threshold value, with samples closer to 1 having the highest probability of being an AL or MM sample, respectively. The x-axis denotes each of the 24 AL and 28 MM samples. Circled symbols indicate patients that were misclassified by the shrunken centroids analysis.
Class prediction analysis using 12 genes derived from the 29 genes used for the initial analysis. The class prediction analysis after the removal of the Ig λ LC genes, which could artificially skew the accuracy of classification, resulted in an improvement in the observed accuracy rate to 92%. The disease category of 23 of the 24 patients with AL and 25 of the 28 patients with MM was accurately predicted. The y-axis shows the threshold value, with samples closer to 1 having the highest probability of being an AL or MM sample, respectively. The x-axis denotes each of the 24 AL and 28 MM samples. Circled symbols indicate patients that were misclassified by the shrunken centroids analysis.
To validate the significance and role of the 12-gene subset in predicting the disease category of the patients, the centroid approach (PAM analysis) was used with the 12 discriminating genes to predict an independent set of 11 patients with AL and 12 patients with MM. With the validation set, we found that the 12 genes were able to accurately classify 87% of the patients with AL and patients with MM (Table 6). Only 2 of the 11 patients with AL (AL-6, AL-7) and 1 of the 12 patients with MM (MM-7) were misclassified and are listed in Table 1.
Independent validation of class prediction analysis
Classification based on gene expression profile . | Diagnosis, no. . | . | |
|---|---|---|---|
| . | AL . | MM . | |
| AL | 9 | 2 | |
| MM | 1 | 11 | |
Classification based on gene expression profile . | Diagnosis, no. . | . | |
|---|---|---|---|
| . | AL . | MM . | |
| AL | 9 | 2 | |
| MM | 1 | 11 | |
The 12-gene subset of discriminating genes was used to classify an independent cohort of AL and patients with MM using the PAM method. This subset of genes was able to accurately classify 87% of the AL and patients with MM. Only 2 of 11 AL and 1 of 12 patients with MM were misclassified.
Validation of differential gene expression for the most discriminating genes by real-time PCR analysis
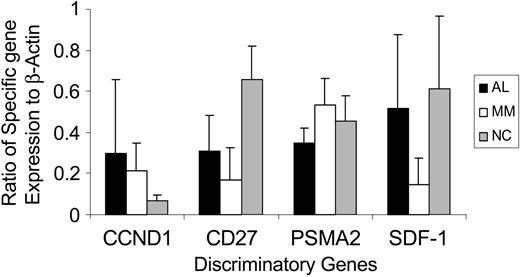
To confirm the differential gene expression pattern for 3 of the most discriminating genes in the 12-gene subset (SDF-1, TNFRSF7 [CD27], PSMA2, and, additionally, cyclin D1 [CCND1]), we used PCR analysis to evaluate transcript levels for these genes and compared them to the expression of a steady state housekeeping gene, β-actin (Figure 4), in 15 patients with AL, 10 patients with MM, and 6 normal controls. The qPCR data (Figure 4) confirmed the differential expression pattern seen with the micro-array analysis for each of these 4 genes (Figure 2).
QPCR validation of discriminating genes. The bar graph depicts the real-time PCR analysis of transcript levels for CCND1, CD27, PSMA2, and SDF-1 in 15 AL, 10 MM, and 6 healthy controls. The y-axis shows the ratio of specific transcript expression to a housekeeping gene, β-actin. As seen with the micro-array analysis, gene expression levels for CCND1 are highest in AL PCs compared to MM and normal PCs. For CD27 and SDF-1, gene expression is higher in AL PCs compared to MM, but lower than in normal PCs. PSMA2 gene expression is higher in MM PCs compared to AL and normal PCs. Error bars indicate standard deviation.
QPCR validation of discriminating genes. The bar graph depicts the real-time PCR analysis of transcript levels for CCND1, CD27, PSMA2, and SDF-1 in 15 AL, 10 MM, and 6 healthy controls. The y-axis shows the ratio of specific transcript expression to a housekeeping gene, β-actin. As seen with the micro-array analysis, gene expression levels for CCND1 are highest in AL PCs compared to MM and normal PCs. For CD27 and SDF-1, gene expression is higher in AL PCs compared to MM, but lower than in normal PCs. PSMA2 gene expression is higher in MM PCs compared to AL and normal PCs. Error bars indicate standard deviation.
Evaluation of CCND1 and Rb1 expression in the context of t(11;14) and del 13q
To determine whether the increased expression of CCDN1 in patients with AL was related to the presence of the t(11;14), we performed fluorescence in situ hybridization (FISH) analysis on the patients with AL and patients with MM. Interestingly, we found that 12 (50%) of the 24 patients with AL had the translocation,24 while only 1 (4%) of the 26 patients with MM tested had the t(11;14), which is in contrast to what has been reported.25 While this could suggest a selection bias, there was no a priori preselection of these patients for any criteria other than sample availability and clinical classification as AL or MM. Since there were insufficient numbers for adequate comparison of the t(11;14)-positive AL and MM, we compared the t(11;14)-negative patients for CCND1 and Rb1 expression (Table 7). We found that the average gene expression level of CCND1 was still substantially higher in the translocation-negative AL group than in MM, indicating an intrinsic difference in gene expression.
Average gene expression in t(11:14)-negative AL and MM
. | AL-negative t(11;14) . | MM-negative t(11;14) . |
|---|---|---|
| No. patients | 12 | 25 |
| CCND1 | 401 | 183 |
| Rb1 | 298 | 283 |
. | AL-negative t(11;14) . | MM-negative t(11;14) . |
|---|---|---|
| No. patients | 12 | 25 |
| CCND1 | 401 | 183 |
| Rb1 | 298 | 283 |
The CCND1 expression was substantially higher in the AL PCs compared to the MM PCs, even in the translocation-negative group, while the Rb1 expression was only slightly higher.
The Rb1 expression was slightly higher in the AL group than in the MM group (Table 7). The deletion of chromosome 13 q was assessed in these patients and correlated with the Rb1 expression. This abnormality has been reported in 33% of patients with AL in one study.26 The incidence of del 13q in this study was 21% (5 of 24) for AL and 44% (12 of 27) for MM. The Rb1 levels were higher in the patients with AL and patients with MM without del 13q compared to those with del 13q (Table 8), suggesting that in this case, the Rb levels are related to the presence or absence of chromosome 13.
Correlation of Rb1 gene expression and loss of chromosome 13 (del 13q)
. | AL0 . | AL1 . | MM0 . | MM1 . |
|---|---|---|---|---|
| No. patients | 19 | 5 | 12 | 15 |
| Average Rb1 gene expression | 248 | 187 | 235 | 194 |
. | AL0 . | AL1 . | MM0 . | MM1 . |
|---|---|---|---|---|
| No. patients | 19 | 5 | 12 | 15 |
| Average Rb1 gene expression | 248 | 187 | 235 | 194 |
Average Rb1 gene expression in AL and patients with MM was correlated with their del 13q status. AL0 and MM0 are the groups without the deletion 13q, while AL1 and MM1 are the groups that are positive for del 13q. The average gene expression was higher in patients without the del 13q, both in AL and MM, compared to patients with the del 13q, indicating a positive correlation between Rb1 expression and chromosome 13 status.
Gene network mapping and pathway analyses
We next sought to ascribe biologic function and interactions to the differentially expressed genes between AL PCs and MM PCs with the Ingenuity Systems software, which is capable of mapping gene networks and identifying potentially dysregulated pathways in a specific disease.
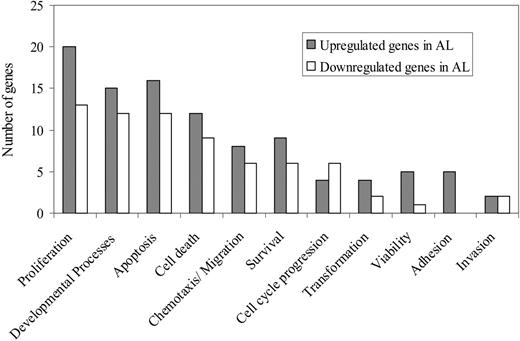
The use of a stringent test statistic threshold of magnitude 3, comparing AL PCs to MM PCs, resulted in the identification of 1051 focus genes, from the original 22 215 genes. From the first 5 highest scoring networks with 163 genes, 47 genes were selected for further analysis; 26 genes with significantly higher expression (test statistic value > 3), and 20 genes with significantly lower expression (test statistic value > -3) in patients with AL compared to patients with MM. These genes were divided into 11 functional categories (Figure 5).
Functional categories of genes differentially expressed in patients with AL and patients with MM. The bar graph depicts the major functional classes of the genes in the 3 highest scoring networks obtained by the Ingenuity analysis. There were 26 genes that were selected for further analysis that had a significantly higher average expression level ▦ in AL compared to patients with MM and 21 genes selected for further analysis that had a significantly lower average expression level (□) in patients with AL compared to patients with MM.
Functional categories of genes differentially expressed in patients with AL and patients with MM. The bar graph depicts the major functional classes of the genes in the 3 highest scoring networks obtained by the Ingenuity analysis. There were 26 genes that were selected for further analysis that had a significantly higher average expression level ▦ in AL compared to patients with MM and 21 genes selected for further analysis that had a significantly lower average expression level (□) in patients with AL compared to patients with MM.
Genes with significantly higher average expression in AL plasma cells compared with MM PCs
Further analysis of these 26 genes with higher average expression in AL plasma cells derived from the Ingenuity networks revealed that there were a number of genes critical to cell regulatory function, cell cycle, protein processing and folding, and certain proto-oncogenes. The complete list is provided in Table 4. The gene expression levels for these genes between AL and normal PCs (cutoff test statistic value = 3) is shown in Table 2. Among the genes important for cell regulatory function, of particular interest is the chemokine-SDF-1/CXCL12 (which has previously been shown to be down-regulated in myeloma PCs relative to normal PCs).3 Plasma cells have increased expression of SDF-1 in patients with AL (but lower levels than normal PCs), while bone marrow stromal cells are the main source of this chemokine in patients with MM (Abraham et al, unpublished data, May 2003). The only known biologic receptor for SDF-1 is CXCR4,27 whose average expression is higher in AL plasma cells compared to patients with MM (test statistic value of 2, data not shown). CXCR4 also has been shown to be significantly up-regulated in MM PCs compared to normal PCs.22,23
Two important cell cycle checkpoint genes, cyclin D1 (CCND1) and Rb1 (tumor suppressor retinoblastoma 1), had higher average expression levels in patients with AL compared to patients with MM as described in the section on CCND1 and Rb1 expression, and in Table 4. Also, AL PCs had a significantly higher average expression of CCND1 (Figure 2) compared to normal PCs, with a test statistic value of 3.2 (Table 2). On the other hand, Rb1 expression is significantly lower in AL compared to normal PCs, with a test statistic value of -5.10 (Table 2). CCND1 has been shown to be up-regulated substantially in 15% of patients with MM and can be a consequence of the t(11;14) (q13;q32) translocations.3 However, the ectopic expression of CCND1 in the absence of the translocation has been reported,28 and therefore this is not the sole factor in determining CCND1 gene expression level. Also, overexpression of CCND1 does not in itself correlate with cell cycle progression, and it has been reported for poorly proliferating tumors.29-31
Rb1 has been shown to be down-regulated in MM PCs compared to normal controls32 and in AL PCs compared to normal controls, and this is related to the fact that this gene is present on chromosome 13q, which has been shown to be deleted in a large number of patients with MM, MGUS,33,34 and AL (Harrison et al26 ; Table 8).
Of special interest to the pathogenesis of AL are genes that are related to amyloid formation and deposition, particularly those involved in protein processing and clearance and protein folding. Our analysis showed that genes in this category, with higher average expression in AL, include APP, the amyloid beta precursor protein, A2M, and SERPINA1 (alpha-1-antitrypsin), all protease inhibitors. APP and SERPINA1 belong to the family of serine protease inhibitors, while A2M is a wide-spectrum protease inhibitor. APP and A2M are implicated in amyloid pathogenesis in neuronal tissue.35,36 These genes that code for protease inhibitors may play a common role in the pathogenesis of other abnormal protein folding diseases including AL.
BTRC, which encodes an F-box protein family member as a subunit of the ubiquitin protein ligase complex, has higher expression in AL PCs compared to MM (Table 2). This protein associates specifically with phosphorylated IκBA and beta-catenin destruction motifs, probably functioning in inhibiting the beta-catenin pathway.37,38 The regulation of beta-catenin stability is essential for Wnt signal transduction during development and tumorigenesis. Morphologic changes associated with Wnt signaling have been implicated in the migration and metastatic potential of myeloma cells.39 Antagonism of Wnt by DKK1 (Dickkopf 1) is associated with the development of bone lesions in myeloma.40 Additionally, negative regulation of the WNT pathway by FRZB has been implicated in the transition from MGUS to MM.32
Genes with significantly lower average expression in AL plasma cells compared to MM PCs
These 20 genes selected for further analysis had statistically significant lower average expression values in AL plasma cells (compared to MM) (Table 5). The gene expression level of these genes in AL PCs compared to normal controls is listed in Table 3. Among these genes, of relevance to understanding the differential pathogenesis of AL and MM, were genes encoding for DNA repair factors (XRCC4 and XRCC5), B-cell transcription factors in immunoglobulin rearrangement (E2A/TCF3), cell cycle proteins (CDK4, CDK5, and CDK9), and proteins involved in protein processing and folding (PSEN 1 and 2, UCHL1, CASP3) (Table 3).
XRCC5 or the 80-kDa subunit of the Ku protein (DNA helicase II) and XRCC4 function together with DNA ligase IV in the repair of DNA double-stranded breaks by nonhomologous end-joining (NHEJ) and the completion of V (D) J recombination events. XRCC5 has been shown to be up-regulated in MM PCs compared to normal PCs,22 and it has been postulated that the activation of this complex may be involved in the facilitation of chromosomal translocations.3
There is significantly lower expression of several genes related to the cyclin-dependent kinase family in AL, including CDK4, CDK5, and CDK9. CDK4, in particular, is a catalytic subunit of the protein kinase complex that is important for cell cycle G1 phase progression and is responsible for the phosphorylation of Rb along with cyclin D1.41,42
In the category of genes that regulate protein processing and folding and have average lower expression in AL PCs compared to MM are UCHL1, PSEN1 and 2 (Presenilin 1 and 2), and CASP3 (caspase 3) (Table 3). UCHL1 is a ubiquitin thiolesterase, which, interestingly, has been reported to be significantly up-regulated in MM PCs compared to normal PCs in one study,3 while it has been shown to be down-regulated in MM PCs in another study.22 UCHL1 has been shown to be important in clearing and regulating the amount of misfolded proteins, in particular, alpha synuclein, which can otherwise accumulate intracellularly and cause neurological dysfunction associated with familial Parkinson disease.43-45
CASP 3 is involved in proteolysis and peptidolysis and has been shown to be the predominant caspase involved in the cleavage of the amyloid-beta precursor protein, which is involved in the pathogenesis of Alzheimer disease.46,47 PSEN genes (PSEN 1 and 2) also are associated with Alzheimer disease, where mutations in these proteins result in increased production of the longer form of amyloid-beta, which forms the main component of amyloid deposits in brain tissue.48-50 The deregulation of these genes may be important in aberrant protein processing and folding in other amyloid diseases, including AL.
Quantitative analysis of protein expression
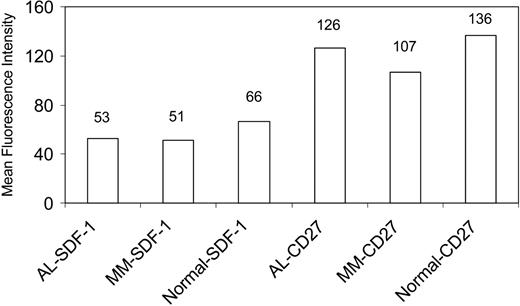
In addition to determining gene expression levels by micro-array analysis and validating the results by qPCR studies, it also was important to determine whether the protein levels correlated with transcript message, at least for specific genes that may be particularly relevant to the pathobiology of disease. We used immunocytochemical and quantitative confocal analysis to determine protein expression for CD27 and SDF-1 in plasma cells of AL and patients with MM and healthy controls (Figure 6). The levels of SDF-1 protein were only slightly different between AL and MM PCs (in contrast to the larger difference seen in the gene expression data), due to the considerable size variation and large morphology of the MM PCs used for the SDF-1 protein analysis. Due to limitation of cytospin slides of BM mononuclear cells from a given patient, we used different AL and patients with MM for the SDF-1 and CD27 staining. Despite this, the protein expression patterns follow the same trend as seen with the gene expression patterns, further confirming the results of the micro-array analysis.
Quantitative confocal analysis of protein expression. Protein levels of SDF-1 and CD27 were determined for plasma cells from AL, MM, and healthy controls. The y-axis shows the MFI for each group. The numbers above the bars depict the actual MFI values.
Quantitative confocal analysis of protein expression. Protein levels of SDF-1 and CD27 were determined for plasma cells from AL, MM, and healthy controls. The y-axis shows the MFI for each group. The numbers above the bars depict the actual MFI values.
Discussion
The objective of this study was 2-fold: the first, to demonstrate that AL is a molecularly distinct entity from MM with a unique molecular signature; and the second, to broadly identify networks of genes or pathways that may be of significance in the pathogenesis of AL. In support of the first objective, a subset of 12 genes was shown to distinguish between these 2 groups of patients with 92% accuracy (Figure 1B and 3; Table 6). This subset included several genes (TNFRSF7, SDF-1, and DEFA1) that have been previously reported to be down-regulated in MM PCs compared to normal PCs.3 The comparison of gene expression profiles between AL plasma cells and normal BM plasma cells (Tables 2, 3) revealed that AL had an intermediate expression pattern, with transcript levels in between that of MM and normal, for most of the significantly differentially expressed genes (between AL and MM, Tables 4, 5). To achieve the second objective of identifying deregulated gene networks, we used a novel software tool, Ingenuity Systems Pathway Analysis, for functional annotation, classification, and identification of clusters or networks of interrelated genes that could potentially be of significance in the pathogenesis of disease.
Of these genes, a few may be of particular relevance in understanding the differences in the pathobiology of these 2 disease entities. One of these, TNFRSF7, a member of the tumor necrosis factor (TNR) receptor superfamily, which codes for CD27, a marker expressed on memory B cells51 and important in controlling maturation and apoptosis of plasma cells,52-54 has a higher average expression in AL PCs. CD27 has been postulated to be important in the oncogenesis of myeloma,55 since MM PCs do not express this marker, whereas normal PCs do, and the expression of CD27 declines with the more advanced stages of MM.55 CD27 interacts with its ligand CD70,56 and this interaction is thought to be important in the differentiation of plasma cells.53 Interestingly, the tail of CD27 binds a proapoptotic protein, Siva,54 and CD27-70 interaction may activate a death signal that determines the life span of plasma cells.55 The higher expression of CD27 on AL PCs may explain one of the major differences between the 2 diseases, which is the extent of bone marrow plasmacytosis.
Another gene that was significantly different between AL and MM was the chemokine SDF-1, which is comparatively highly expressed in AL PCs. However, SDF-1 levels in normal PCs are higher than those expressed in AL (Figures 2, 4, and 6). Whereas overexpression of SDF-1 has been implicated in preventing apoptosis, promoting proliferation and metastatic spread in a number of neoplastic diseases through interactions with CXCR4,57-62 it is apparent that the relatively high levels of SDF-1 in normal and AL PCs have a paradoxical effect. This paradox can be explained by invoking the regulatory mechanisms that curtail SDF-1/CXCR4 interactions. The binding of SDF-1 to CXCR4 results in activation of the suppressors of cytokine signaling (SOCS) proteins, in particular, SOCS-3,63 which can negatively regulate CXCR4 function without interfering with surface receptor expression.63 It is plausible to speculate that the interaction of SDF-1 and CXCR4 in MM aberrantly does not induce the regulatory SOCS proteins, which then allows the downstream signaling events of this interaction to proceed unchecked, resulting in reduced apoptosis and enhanced proliferation of MM cells. On the other hand, in AL and normal PCs, the activation of SOCS proteins blocks continued signaling as a result of binding of CXCR4 by SDF-1.
Whereas the higher expression of TNFRSF7 and SDF-1 and their interactions with other regulatory genes may help us understand some of the differences between AL and MM, it was equally important to determine if the gene expression and network analysis could explain certain unique features of AL, such as the preponderance of λ light chain isotype in the clonal PCs1,12 and the restricted use of light chain germ-line genes.15,64,65 While the class prediction analysis clearly demonstrates that the presence of λ light chains (Figures 2A and 3) is not the main distinguishing feature between AL and MM, it is nonetheless relevant to determine what contributes to the isotype bias and what the implications of this might be for the pathogenesis of amyloid disease and early events in the biology of AL. This is particularly relevant in light of recent data in patients with MM from Magrangeas et al,4 showing significant differences in gene expression based on light chain isotype usage.
An interesting hypothesis that emerges from the network analysis (Figure 4; Tables 2, 3) is the interaction of cyclin D1, CDK4, and Rb and its potential role in the rearrangement of the Ig light chain locus, which is regulated by several B-cell transcription factors. It has been shown that Pax-5 or BSAP, a B-cell transcription factor that is crucial to B-cell differentiation,66 binds underphosphorylated Rb in order to be released from a binding site proximal to the RAG-binding site on the κ light chain locus, and this then allows for κ light chain rearrangement.67 The phosphorylation status of Rb is regulated by the CDK4-cyclin D1 complex.68 Cyclin D1 is overexpressed in AL PCs (Tables 2 and 7), therefore it seems plausible to speculate that constitutive activation resulting in increased expression of cyclin D1 would prevent the dephosphorylation of Rb, thus preventing Pax-5 from binding to it, which in turn would preclude κ locus transcription. This would result in increased rearrangement of the λ locus. However, while this hypothesis needs to be experimentally validated (work in progress), it also remains to be determined as to how the increase in cyclin D1 expression affects late stages of B-cell development. These data offer new insights and hypotheses into genes and pathways that may be critical to the evolution of B-cell differentiation events that characterize the pathogenesis of AL.
Lastly, there are a number of deregulated genes and pathways in AL PCs that appear to feed into a common loop related to protein processing and folding including APP, A2M, PSEN1 and 2, UCHL1, and CASP3. This finding raises the intriguing possibility that there may be deregulation of common pathways related to protein clearance, degradation, and intracellular folding that are shared by all amyloid diseases, irrespective of the nature of the protein that forms amyloid. Therefore, these genes or related others in these specific pathways may offer appropriate targets for therapeutic intervention, either circumventing amyloid formation or hastening the removal of deposited amyloid protein, preventing irreversible organ dysfunction.
In summary, we have been able to demonstrate that AL can be differentially classified from MM using a small set of discriminating genes. Also, the use of a novel network and pathway mapping software has permitted us to identify genes that are of potential significance in AL biology, though this needs to be validated in a larger cohort of patients. In addition, comparison of normal and AL plasma cells reveal that for most of the genes that are dramatically dysregulated between AL and MM, AL PCs have an expression that is intermediate to the overt malignant state and the normal state, which is in keeping with the clinical phenotype observed in these patients.
Prepublished online as Blood First Edition Paper, September 23, 2004; DOI 10.1182/blood-2004-04-1424.
Supported in part by the Hematologic Malignancies Fund, Mayo Clinic (R.S.A.), Public Health Service (PHS) grant R21-CA91561 (A.D.), National Cancer Institute grant RO1-CA83724 (R.F.), and the Fund to Cure Myeloma (R.F.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Drs Robert A. Kyle and Philip R. Greipp for providing samples through the Dysproteinemia Sample Bank. R.F. is a clinical investigator of the Damon Runyon Cancer Research Fund. The authors thank and acknowledge Jim Tarara for excellent assistance with confocal microscopy analysis and quantitation, and Asghar Ali for help with statistical data analysis. The authors also thank Ms Karen Best for assistance with manuscript preparation.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal