Abstract
In Philadelphia-positive (Ph+) leukemia, point mutations within the Bcr-Abl kinase domain emerged as a major mechanism of resistance to imatinib mesylate. We established a cell-based screening strategy for detection of clinically relevant point mutations using Bcr-Abl-transformed Ba/F3 cells. We identified 32 different single-point mutations within the kinase domain of Bcr-Abl. The pattern and frequency of mutations in this cell culture-based screen resembled the pattern and frequency observed in resistant patients. We then applied this screen to an alternative Abl kinase inhibitor. Using PD166326, the frequency of resistant colonies emerging at 5 to 10 times the median growth inhibition (IC50) of PD166326 was significantly lower than with imatinib. In addition, PD166326 produced a distinct pattern of Bcr-Abl mutations. The majority of mutations that came up with both imatinib and PD166326 could effectively be suppressed by increasing the dose of PD166326 to 50 to 500 nM. In contrast, only a few mutations could be suppressed by increasing the imatinib dose to 5 to 10 μM. However, 3 mutations affecting F317 displayed complete resistance to PD166326, but could be effectively inhibited by standard concentrations of imatinib. Thus, this robust and simple screening system provides a rational basis for combinatorial and sequential treatment strategies in targeted cancer therapy. (Blood. 2005;105: 1652-1659)
Introduction
Imatinib mesylate (STI571, Gleevec) serves as a paradigm for therapeutic application of small molecule kinase inhibitors in cancer treatment. Impressive response rates are achieved treating chronic myeloid leukemia (CML) in chronic phase with imatinib.1,2 However, resistance to imatinib frequently evolves in advanced-phase CML and Philadelphia-positive acute lymphoblastic leukemia (Ph+ ALL).3-5 The characteristic molecular abnormality present in CML and Ph+ ALL is the Philadelphia chromosome, which leads to the expression of a fusion gene encoding the constitutively activated protein tyrosine kinase Bcr-Abl. Imatinib acts by binding to the Bcr-Abl kinase domain, where it blocks the access of adenosine triphosphate (ATP), thereby suppressing Bcr-Abl activity. Point mutations within the Bcr-Abl kinase domain interfere with drug binding while retaining binding of ATP and catalytic activity, and constitute the major mechanism of resistance detected in patients.6-12 In addition, amplification of the Bcr-Abl gene and/or overexpression of the Bcr-Abl protein and the formation of secondary genetic alterations were found in resistant patients.6,12
Pyrido-pyrimidines, initially identified as inhibitors of src family kinases, are potent inhibitors of Bcr-Abl.13,14 Crystal structure analysis of human c-Abl in complex with imatinib and the pyrido-pyrimidine compound PD173955 has demonstrated differences in the binding mode of both compounds, and in positions that are critical for binding.15 Moreover, pyridopyrimidines were demonstrated to inhibit Bcr-Abl with higher potency when compared with imatinib,15 and reportedly suppress some of the most frequently observed mutations of Bcr-Abl that cause resistance to imatinib.16-18 Therefore, pyridopyrimidines show promise as candidate substances for application in future clinical trials.
As both the number of diseases that can be treated using small molecule kinase inhibitors and the number of compounds available for a specific target increase, it will be of utmost importance to determine the pattern of critical resistance mutations for different compounds prior to therapeutic application. This will allow one to establish combinational and sequential strategies using different kinase inhibitors. We intended to establish a cell culture system that allows one to determine the frequency of resistance and to compare the pattern of resistance mutations emerging under treatment with different inhibitors in cells that were transformed by a specific oncogene. We validated this cell-based system for Bcr-Abl-positive leukemia using imatinib, and then applied the system on an alternative Bcr-Abl kinase inhibitor, the pyridopyrimidine PD166326.
Materials and methods
Inhibitors
The pyrido-[2,3-d]pyrimidine analog PD166326 was dissolved at 10 mM in dimethyl sulfoxide (DMSO) and stored at -20°C. Imatinib was dissolved at 10 mM in water and stored at -20°C.
Cell culture and transfection
Parental Ba/F3 cells were obtained from DSMZ (Braunschweig, Germany). Ba/F3 cells were maintained in RPMI 1640 growth media (Gibco, Karlsruhe, Germany) containing 10% fetal calf serum (Gibco) and P/S (200 U penicillin per mL and 200 μg streptomycin per mL; Gibco). Parental cells were cultured in the presence of 2 ng/mL interleukin-3 (IL-3; R&D, Wiesbaden, Germany). Ba/F3 cells were transfected by electroporation and transformed upon withdrawal of IL-3.
Mutagenesis screen
Ba/F3 Mig EGFPp185 wild-type cells were cultured in 96-well plates at a density of 0.5, 1, and 4 × 105 cells per well in the presence of imatinib at 1, 2, and 4 μM or PD166326 at 25, 50, and 100 nM. In addition, Ba/F3 Mig EGFPp185 wild-type cells were plated in soft agar in 6-well plates at a density of 1 × 106 cells per well and maintained in the presence of 1 μM imatinib, and at a density of 2 × 106 cells per well with 2 μM imatinib. Single colonies growing out on the bottom of the well in liquid culture or within the agar layer in solid culture were picked and expanded for analysis. Resulting resistant sublines were cultured in the presence of inhibitor at a concentration corresponding to that used in the screen.
DNA constructs and PCR work
Bcr-Ablp185 wild-type, a 4812-bp human/mouse chimera containing human BCR and murine ABL,19 was cloned into Mig EGFP20 and pcDNA3.1/Zeo(-) (Invitrogen, Leek, the Netherlands). Point mutant forms of Bcr-Abl were engineered in Mig EGFPp185 using the QuickChange mutagenesis kit (Stratagene, Amsterdam, the Netherlands), subcloned into pBluescript SK+ (Stratagene) p185 and subsequently into pcDNA3.1/Zeo(-) (Invitrogen, Leek, the Netherlands). All constructs were verified using automated sequencing. Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA). For reverse-transcription-polymerase chain reaction (RT-PCR) of a 2242-bp fragment of Bcr-Abl encompassing the fusion, the following primers were used: BCR 5′ CCACTAAAGCGAGTGAGCTGGACTTGG 3′ and ABL 5′ GGTGGATGAGTCAAACTGCTTGCCAGC 3′. Using this fragment as template, the following primers were used for nested PCR and sequence analysis: for ABL Src homology 3 (SH3) domain, SH3-SH2 connector, SH2 domain, and CD linker (encompassing the fusion to G227, human cAbl Ia numbering): 5′ GGAGATGAGAAAATGGGTCCTGTCG 3′ and 5′ CCGTAGATACTGGGCTTGTTGCGC 3′; for ABL kinase domain (encompassing R220 through L411): 5′ GCGCAACAAGCCCACTGTCTATGG 3′ and 5′ GCCAGGCTCTCGGGTGCAGTCC 3′.
Proliferation
Proliferation was measured using an MTS (3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium)-based method by absorption of formazan at 490 nm (CellTiter 96; Promega, Madison, WI). Measures were taken as triplicates after 24 and 48 hours of culture without and in the presence of inhibitor at the indicated concentrations.
Western blot
Ba/F3 cells were cultured for 2.5 hours without and in presence of inhibitor at the indicated concentrations. Cell lysis, sodium dodecyl sulfatepolyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting were done as described previously.21 Abl antibodies were obtained from Pharmingen (8E9) (BD Biosciences, Heidelberg, Germany) and Calbiochem-Novabiochem (Ab3) (Schwalbach, Germany). Antibodies to phosphotyrosine were purchased from Upstate Biotechnology (4G10) (Biozol, Eching, Germany) and Transduction (PY20) (BD Biosciences). Anti-phospho-signal transducer and activator of transcription 5 (Stat5; Tyr694) was obtained from Cell Signaling (New England Biolabs, Frankfurt/Main, Germany), and anti-Stat5 (G-2) was obtained from Santa Cruz (Santa Cruz Biotechnology, Heidelberg, Germany). Bands were visualized using the enhanced chemiluminescence (ECL) system (Amersham, Braunschweig, Germany).
Results
A cell culture-based screen for Bcr-Abl mutations
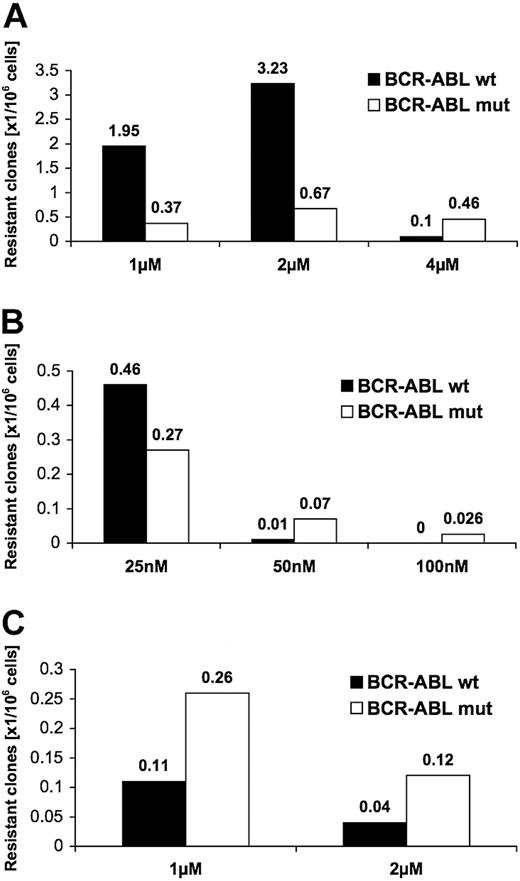
Bcr-Abl wild-type-transfected Ba/F3 cells (Ba/F3-BA-wt) were exposed to imatinib at a density of 0.5, 1, and 4 × 105 cells per well in 96-well plates. We chose 1-, 2-, and 4-μM concentrations that correspond to or exceed plasma concentrations measured in imatinib-treated patients.22 At 1 μM imatinib, resistant colonies grew up after a median of 16 days (minimum, 10 days; maximum, 19 days); at 2 μM imatinib, after a median of 22 days (minimum, 8 days; maximum, 38 days); and at 4 μM imatinib, after a median of 29 days (minimum, 12 days; maximum, 42 days). The highest frequency of resistant colonies occurred at imatinib 2 μM, with 3.9 resistant colonies per 106 cells (Figure 1A). At 4 μM, corresponding to 10 times the concentration, at which 50% of growth inhibition of Ba/F3-BA-wt cells occurred (IC50 value; 0.4 μM for imatinib), there were 0.565 resistant colonies emerging per 106 cells.
Frequency of resistant colonies emerging in the presence of imatinib is higher than with PD166326. Single colonies of Ba/F3 cells growing in the presence of inhibitor were picked and analyzed. Shown is the incidence of resistant colonies that grew up in relation to the number of Ba/F3-Bcr-Abl wild-type (Ba/F3-BA-wt) cells in liquid culture in the presence of imatinib (A) or PD166326 (B), or in solid culture conditions in the presence of imatinib (C). Resistant colonies are itemized according to wild-type (wt; ▪) or mutant (mut; □) sequence of Bcr-Abl.
Frequency of resistant colonies emerging in the presence of imatinib is higher than with PD166326. Single colonies of Ba/F3 cells growing in the presence of inhibitor were picked and analyzed. Shown is the incidence of resistant colonies that grew up in relation to the number of Ba/F3-Bcr-Abl wild-type (Ba/F3-BA-wt) cells in liquid culture in the presence of imatinib (A) or PD166326 (B), or in solid culture conditions in the presence of imatinib (C). Resistant colonies are itemized according to wild-type (wt; ▪) or mutant (mut; □) sequence of Bcr-Abl.
We next determined the frequency of resistant colonies in the presence of the alternative Abl kinase inhibitor PD166326. We used concentrations that correspond to 5 times (25 nM), 10 times (50 nM), and 20 times (100 nM) the cellular IC50 (Figure 1B). Median time to growth of resistant colonies with 25 nM PD166326 was 30 days (minimum, 13 days; maximum, 39 days); 18 days for 50 nM (minimum, 11 days; maximum, 22 days); and 19 days for 2 colonies that grew at 100 nM PD166326 (18 and 20 days, respectively). In contrast to imatinib, resistant colonies appeared with a much lower frequency when Ba/F3-BA-wt cells were cultured in the presence of PD166326 (Figure 1B). At 25 nM, there were 0.73 resistant colonies per 106 cells; at 50 nM, 0.08 resistant colonies per 106 cells; and at 100 nM, only 2 resistant colonies, corresponding to a frequency of 0.026 per 106 cells. When the frequency of resistant colonies was put into relation to the IC50 value for both inhibitors, there were 3.9 resistant colonies per 106 cells (at 2 μM imatinib) opposed to 0.73 per 106 (at 25 nM PD166326; 5 times IC50, Figure 1A-B); there were 0.56 resistant colonies (at 4 μM imatinib) opposed to 0.08 per 106 cells (at PD166326 50 nM; 10 times IC50). Thus, resistant colonies in this cell culture-based screen occur with a higher frequency with imatinib when compared with PD166326.
Resistant colonies exhibit kinase domain mutations
To investigate the molecular mechanism underlying the observed resistance, we analyzed the Bcr-Abl kinase domain for mutations that could interfere with inhibitor binding. Point mutations within the Bcr-Abl kinase domain were detected in 25% of resistant colonies for imatinib and 41% for PD166326 (Figure 1A-B). At higher inhibitor concentrations, the proportion of colonies with mutations in the kinase domain increased for both compounds with 82% at 4 μM imatinib and 86% at 50 nM PD166326. In comparison with liquid culture, soft agar culture with 1 μM and 2 μM imatinib yielded a lower total number of resistant colonies but a higher proportion of colonies with a mutant Bcr-Abl kinase domain (Figure 1C). The occurrence of resistance and the emergence of different single kinase domain mutations within Bcr-Abl were observed in several sublines that were either freshly transfected or that had been maintained for up to 2 years. In all cases, initial sequencing of Bcr-Abl from the fusion to L411 (human cAbl Ia numbering) revealed wild-type sequence.
In order to determine whether selection of single cells with preexisting mutations in the presence of inhibitor has occurred, single Ba/F3-BA-wt cells derived from limited dilution were expanded and exposed to imatinib or PD166326. Again, resistant colonies displayed a variety of different single kinase domain point mutations (data not shown). Thus, in Ba/F3 cells different kinase domain mutations within Bcr-Abl were acquired during a relatively short period of culture and were selected in the presence of inhibitor.
Kinase domain mutations occur at positions that are known from imatinib-resistant patients and at novel positions
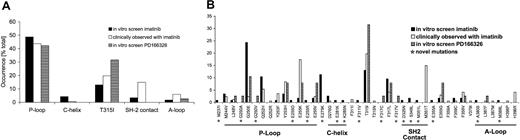
Overall, 32 different exchanges of 22 amino acid residues within Bcr-Abl were detected, each occurring as the sole mutation in a single resistant colony (Figure 2). In only 2 cases, there were single silent mutations (mutations that do not result in an amino acid exchange), and in 1 case a double mutant (Q252H and Y253F) was present. With imatinib, we observed 10 different exchanges at 6 different positions within the nucleotide-binding loop (P-loop). The observed frequency of P-loop mutations of 49% was almost identical to the 44% reported in imatinib-resistant patients (Figure 2A). Positions that were affected included all mutations in the P-loop that were described in patients (Figure 2B). T315I made up 13% in our screen emerging with imatinib (T315I is the most frequently detected mutation in patients [20%]), constituting the second most-affected amino acid. Other positions that were also reported in patients included D276 within C-helix, the imatinib contact sites F311 and F317, the SH2 contact position M351, and positions E355 and F359. While our screen missed only 2 clinically infrequently observed positions (V379, H396), it identified 17 additional exchanges that may be observed in patients in the future (Figure 2B). Among those were changes at M237 and E275 (C-helix), E281 and K285 (amino-terminal lobe), D325 and S348 (SH2 domain contact site), and A380 and M388 (activation loop). We did not find any point mutations within the SH3 domain, SH3-SH2 connector, SH2 domain, and CD linker.
Distribution of point mutations in colonies resistant to imatinib and PD166326 compared with imatinib-resistant patients. Ba/F3 cell colonies that displayed a resistant phenotype to imatinib or PD166326 were analyzed for point mutations within the Bcr-Abl kinase domain, SH3 domain, SH3-SH2 connector, SH2 domain, and CD linker. (A) Distribution of affected positions according to functional domains of Abl. C-Helix and SH2 contact region mutations occurred in imatinib-resistant colonies exclusively. (B) The mutation pattern observed with imatinib, PD166326, and imatinib-resistant patients. With imatinib, 116 resistant colonies with mutant Bcr-Abl contained 117 point mutations, including one colony with 2 point mutations (Q252H and Y253F). In 39 mutant colonies resistant to PD166326, 39 single point mutations were identified. Relative frequencies are itemized according to positions within the Bcr-Abl kinase domain. An asterisk identifies mutations that have not been described so far. For comparison, Bcr-Abl kinase domain mutations reported in 167 patients with CML (chronic phase, accelerated phase, or blast crisis) or Ph+ ALL and primary (4 patients) or acquired imatinib resistance (163 patients) are shown. 6,8-11,23-26
Distribution of point mutations in colonies resistant to imatinib and PD166326 compared with imatinib-resistant patients. Ba/F3 cell colonies that displayed a resistant phenotype to imatinib or PD166326 were analyzed for point mutations within the Bcr-Abl kinase domain, SH3 domain, SH3-SH2 connector, SH2 domain, and CD linker. (A) Distribution of affected positions according to functional domains of Abl. C-Helix and SH2 contact region mutations occurred in imatinib-resistant colonies exclusively. (B) The mutation pattern observed with imatinib, PD166326, and imatinib-resistant patients. With imatinib, 116 resistant colonies with mutant Bcr-Abl contained 117 point mutations, including one colony with 2 point mutations (Q252H and Y253F). In 39 mutant colonies resistant to PD166326, 39 single point mutations were identified. Relative frequencies are itemized according to positions within the Bcr-Abl kinase domain. An asterisk identifies mutations that have not been described so far. For comparison, Bcr-Abl kinase domain mutations reported in 167 patients with CML (chronic phase, accelerated phase, or blast crisis) or Ph+ ALL and primary (4 patients) or acquired imatinib resistance (163 patients) are shown. 6,8-11,23-26
Colonies resistant to PD166326 exhibit an overlapping but distinct pattern of kinase domain mutations
As with imatinib, the P-loop constituted the predominant mutated region within Bcr-Abl in colonies resistant to PD166326 (Figure 2A-B). The exchange observed most frequently within the P-loop was G250E. Other positions known from imatinib-resistant patients included Y253 and E255. The leading exchange with PD166326 was T315I, which was detected in 32% of all mutant colonies, including the only 2 colonies that came up at 100 nM PD166326. F317L was described in patients and appeared in our screen with both inhibitors. In contrast, F317C and F317V occurred with PD166326, but not with imatinib. In line with this, the different F317 mutants displayed a differential behavior toward imatinib and PD166326 (Table 1). In marked contrast to the imatinib screen and clinically observed positions, mutations of C-helix and SH2 domain contact sites did not appear with PD166326. The A-loop, rarely mutated in imatinib-treated patients, was affected in few colonies with both inhibitors, and did not reveal a consistent pattern toward each inhibitor. Similar to the imatinib screen, we did not detect point mutations within the SH3 domain, SH3-SH2 connector, SH2 domain, and CD linker. Thus, while the mutation pattern with imatinib and PD166326 showed some overlap, mutations of helix C and SH2 contact sites emerged with imatinib exclusively.
Cellular IC50 values of Bcr-Ab1 point mutants that were identified
Mutation . | Imatinib, μM . | Fold, IC50 wt . | PD166326, nM . | Fold IC50 wt . |
|---|---|---|---|---|
| Wild-type | 0.4 | — | 5 | — |
| M2371 | 1 | 2.5 | 13 | 2.6 |
| M244V | 2.3 | 5.8 | 33 | 6.6 |
| L248V | 1.5 | 3.8 | 23 | 4.6 |
| G250A | 0.3 | 1 | 6 | 1.2 |
| G250E | 3.9 | 7.5 | 100 | 20 |
| G250V | 0.8 | 2 | 31 | 6.2 |
| Q252H | 1.2 | 3 | 25 | 5 |
| E255D | 0.6 | 1.5 | 14 | 2.8 |
| E255R | 3.5 | 8.8 | 24 | 4.8 |
| E275K | 2.9 | 7.3 | 23 | 4.6 |
| D276G | 1.5 | 3.8 | 19 | 3.8 |
| E281K | 1.2 | 3 | 14 | 2.8 |
| K285N | 0.9 | 2.3 | 5 | 1 |
| F311V | 5.4 | 13.5 | 47 | 9.4 |
| F317C | 1.2 | 3 | 110 | 22 |
| F317L | 1.5 | 3.8 | 70 | 14 |
| F317V | 0.5 | 1.3 | 135 | 27 |
| D325N | 1.5 | 3.8 | 12 | 2.4 |
| S348L | 0.7 | 1.4 | 5 | 1 |
| M351L | 0.9 | 2.3 | 7 | 1.4 |
| E355A | 1.2 | 3 | 16 | 3.2 |
| E355G | 0.4 | 1 | 7 | 1.4 |
| F359C | 1.2 | 3 | 50 | 10 |
| F359V | 1.2 | 3 | 45 | 9 |
| A380S | 6.2 | 15.5 | 22 | 4.4 |
| L387F | 1.1 | 2.8 | 20 | 4 |
| M388L | 1.6 | 4 | 16 | 3.2 |
Mutation . | Imatinib, μM . | Fold, IC50 wt . | PD166326, nM . | Fold IC50 wt . |
|---|---|---|---|---|
| Wild-type | 0.4 | — | 5 | — |
| M2371 | 1 | 2.5 | 13 | 2.6 |
| M244V | 2.3 | 5.8 | 33 | 6.6 |
| L248V | 1.5 | 3.8 | 23 | 4.6 |
| G250A | 0.3 | 1 | 6 | 1.2 |
| G250E | 3.9 | 7.5 | 100 | 20 |
| G250V | 0.8 | 2 | 31 | 6.2 |
| Q252H | 1.2 | 3 | 25 | 5 |
| E255D | 0.6 | 1.5 | 14 | 2.8 |
| E255R | 3.5 | 8.8 | 24 | 4.8 |
| E275K | 2.9 | 7.3 | 23 | 4.6 |
| D276G | 1.5 | 3.8 | 19 | 3.8 |
| E281K | 1.2 | 3 | 14 | 2.8 |
| K285N | 0.9 | 2.3 | 5 | 1 |
| F311V | 5.4 | 13.5 | 47 | 9.4 |
| F317C | 1.2 | 3 | 110 | 22 |
| F317L | 1.5 | 3.8 | 70 | 14 |
| F317V | 0.5 | 1.3 | 135 | 27 |
| D325N | 1.5 | 3.8 | 12 | 2.4 |
| S348L | 0.7 | 1.4 | 5 | 1 |
| M351L | 0.9 | 2.3 | 7 | 1.4 |
| E355A | 1.2 | 3 | 16 | 3.2 |
| E355G | 0.4 | 1 | 7 | 1.4 |
| F359C | 1.2 | 3 | 50 | 10 |
| F359V | 1.2 | 3 | 45 | 9 |
| A380S | 6.2 | 15.5 | 22 | 4.4 |
| L387F | 1.1 | 2.8 | 20 | 4 |
| M388L | 1.6 | 4 | 16 | 3.2 |
Ba/F3 cells transfected with engineered mutant forms of Bcr-Abl were cultured for 24 and 48 hours without and in the presence of imatinib and PD166326 at concentrations as stated in Figure 3. Proliferation was measured in an MTS-based assay. There were 2 independent experiments performed. IC50 values were calculated using representative results of one experiment after 48 hours of incubation. Y253H, E255K, E255V, and T315I have been shown previously.8,18
Resistance can be overcome by switching the inhibitor or increasing the dosage
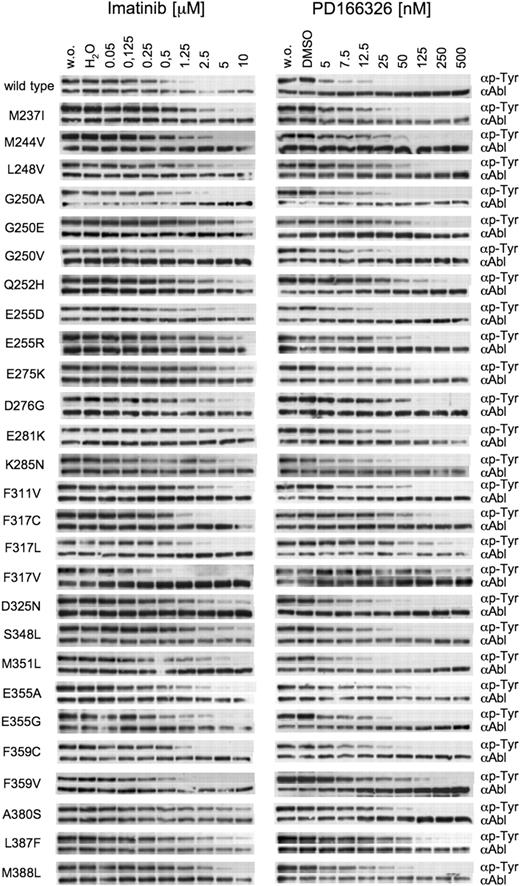
We next intended to examine the extent of cross-resistance of particular mutations to both inhibitors. We therefore cloned all Bcr-Abl mutations, transfected Ba/F3 cells, and established stably transformed cell lines for 27 different exchanges. For each cell line, we analyzed inhibition of Bcr-Abl autophosphorylation (Figure 3) and cellular proliferation (Table 1). In the presence of imatinib at 2.5, 5, or even 10 μM, Bcr-Abl autophosphorylation was preserved in the majority of mutations identified in colonies resistant to either imatinib or PD166326. Mutations where Bcr-Abl was effectively suppressed by imatinib included exchanges that displayed relatively low cellular IC50 values and did not appear in the presence of imatinib in our screen, such as G250A, G250V, and F317V. In contrast, mutations displaying Bcr-Abl autophosphorylation even at 10 μM imatinib and high cellular IC50 values, such as G250E, E255R, or E275K, were abundantly found in our screen in the presence of imatinib.
Inhibition of wild-type and mutant Bcr-Abl by imatinib and PD166326.Point mutations of Bcr-Abl that were identified in resistant colonies were cloned. Parental Ba/F3 cells were transfected with mutant constructs. Stable cell lines were incubated without (w.o.) and in the presence of imatinib (left column) and PD166326 (right column) at the indicated concentrations. Whole cell lysates were subjected to SDS-PAGE. Blots were probed for phosphotyrosine and Abl. Y253H, E255K, E255V, and T315I were previously reported.18
Inhibition of wild-type and mutant Bcr-Abl by imatinib and PD166326.Point mutations of Bcr-Abl that were identified in resistant colonies were cloned. Parental Ba/F3 cells were transfected with mutant constructs. Stable cell lines were incubated without (w.o.) and in the presence of imatinib (left column) and PD166326 (right column) at the indicated concentrations. Whole cell lysates were subjected to SDS-PAGE. Blots were probed for phosphotyrosine and Abl. Y253H, E255K, E255V, and T315I were previously reported.18
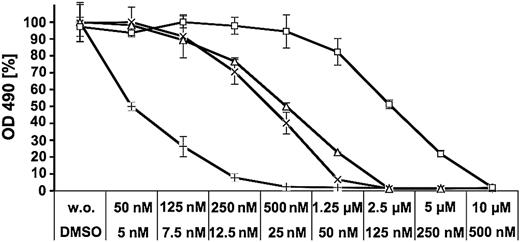
PD166326 at 125 to 500 nM was capable of suppressing resistance mutations originating from both the imatinib and PD166326 screen in most cases, including all P-loop mutations (Figure 3). A more complex picture emerged for mutations of sites that are known to contact imatinib. It has been shown previously that T315I causes a fully resistant phenotype to both inhibitors.6,16,18 Another contact site is F317. F317C and F317L displayed a resistant phenotype toward high PD166326 concentrations but showed an only moderate resistance to imatinib (Figure 3; Table 1). F317V was even more resistant to PD166326 but behaved like Bcr-Abl-wt in the presence of imatinib (Figure 4). Thus, mutations at F317 confer resistance to a pyrido-pyrimidine compound, but at the same time do not considerably interfere with inhibition by imatinib.
F317V causes resistance to PD166326 while preserving sensitivity to imatinib. Ba/F3 cells transfected with Bcr-Abl wild-type (wt) or Bcr-Abl F317V were incubated for 24 and 48 hours without and in the presence of imatinib (wt, ×; F317V, □) or PD166326 (wt, +; F317V, □) at the indicated concentrations. Proliferation was measured in an MTS-based assay. There were 2 independent experiments performed. Values are expressed as mean of triplicates. Bars: ± SE. Representative results of one experiment after 48 hours of incubation are shown. OD indicates optical density.
F317V causes resistance to PD166326 while preserving sensitivity to imatinib. Ba/F3 cells transfected with Bcr-Abl wild-type (wt) or Bcr-Abl F317V were incubated for 24 and 48 hours without and in the presence of imatinib (wt, ×; F317V, □) or PD166326 (wt, +; F317V, □) at the indicated concentrations. Proliferation was measured in an MTS-based assay. There were 2 independent experiments performed. Values are expressed as mean of triplicates. Bars: ± SE. Representative results of one experiment after 48 hours of incubation are shown. OD indicates optical density.
When C-helix mutations (D276G, E281K, and K285N) were engineered into Bcr-Abl and expressed in Ba/F3 cells, all 3 mutants displayed moderate-to-strong resistance to imatinib, but only low to moderate resistance to PD166326 (Figure 3; Table 1). Likewise, the SH2 contact mutations D325N, S348L, and M351L conferred resistance to imatinib, but behaved very similarly to wild-type Bcr-Abl in the presence of PD166326. Activation loop mutations (A380S, L387F, and M388L) gave rise to a moderate-to-high degree of resistance to imatinib, but as well caused a low-to-moderate resistance to PD166326.
Evidence for increased levels of active Bcr-Abl in resistant sublines with and without kinase domain mutations
Resistant sublines that were derived from the screen and did not carry point mutations displayed a higher level of phosphorylated Bcr-Abl and Stat5. This was accompanied by increased Bcr-Abl protein levels in some, but not all, cases (data not shown). Moreover, in contrast to the original sensitive Bcr-Abl-wt cells, phosphorylation of Bcr-Abl and Stat5 did not completely disappear when inhibitor was added in a concentration that corresponded to culture conditions that were applied during the screen. Similarly, many of the resistant sublines expressing mutant Bcr-Abl displayed only small increases of Bcr-Abl protein levels, but a tremendous increase of Bcr-Abl and Stat5 phosphorylation (Figure 5A). This was evident both in sublines harboring mutations that caused a low to moderate degree of resistance such as G250A and E355G, but as well in sublines with mutations mediating a high degree of resistance such as G250E or F317V (Figure 5A).
Increased amounts of phosphorylated Bcr-Abl and Stat5 protein in kinase inhibitor-resistant sublines growing in the presence of inhibitor. (A) Ba/F3-BA-wt cells (BA wt original), sublines derived from the PD166326 screen harboring mutations as indicated (screen), and Ba/F3 cells freshly transfected with Bcr-Abl E355G (E355G retransfected) were cultured without and in the presence of 25 nM PD166326. Whole cell lysates were analyzed for content of phosphorylated and total Bcr-Abl, phosphorylated Stat5, and actin. (B) Shift of dose-response of Ba/F3 cells transformed with engineered E355G in the presence of PD166326 compared with the subline derived from the screen expressing E355G. Ba/F3-BA-wt (+), Ba/F3 Bcr-Abl E355G screen (⋄), and Bcr-Abl E355G retransfected (▵) were processed as in Figure 4.
Increased amounts of phosphorylated Bcr-Abl and Stat5 protein in kinase inhibitor-resistant sublines growing in the presence of inhibitor. (A) Ba/F3-BA-wt cells (BA wt original), sublines derived from the PD166326 screen harboring mutations as indicated (screen), and Ba/F3 cells freshly transfected with Bcr-Abl E355G (E355G retransfected) were cultured without and in the presence of 25 nM PD166326. Whole cell lysates were analyzed for content of phosphorylated and total Bcr-Abl, phosphorylated Stat5, and actin. (B) Shift of dose-response of Ba/F3 cells transformed with engineered E355G in the presence of PD166326 compared with the subline derived from the screen expressing E355G. Ba/F3-BA-wt (+), Ba/F3 Bcr-Abl E355G screen (⋄), and Bcr-Abl E355G retransfected (▵) were processed as in Figure 4.
Bcr-Abl E355G, when retransfected into parental Ba/F3 cells, caused only a marginal resistance to an extent that may not allow survival in the presence of 25 nM PD166326 (Figure 3; Table 1). Consequently, proliferation (Figure 5B) and phosphorylation of Bcr-Abl and Stat5 (Figure 5A) were effectively suppressed at 25 nM PD166326. In contrast, the subline that was derived from the screen with 25 nM PD166326 expressing E355G displayed a far more resistant phenotype, as evidenced by survival and conservation of phosphorylated Bcr-Abl in the presence of 25 nM PD166326 (Figure 5A-B). Similar observations were made with G250A (Figures 3 and 5A, and data not shown). A PD166326-resistant subline expressing F317V also displayed an increased phosphorylation of Bcr-Abl and Stat5 (Figure 5A). However, in contrast to E355G and G250A, dose-response curves for the cell line expressing the engineered F317V mutation exactly matched the doseresponse curves of the subline derived from the screen (data not shown). This is in accordance with the observation that F317V was sufficient to cause a strong resistance to PD166326.
Discussion
The advance of targeted cancer therapy raised the issue of specific mechanisms of resistance to kinase inhibitors. In CML and Ph+ ALL, Bcr-Abl-dependent mechanisms of resistance to imatinib mesylate include overexpression of Bcr-Abl or amplification of the Bcr-Abl gene6,12 and, most importantly, point mutations within the Bcr-Abl kinase domain that interfere with drug binding.6-12,27,28 This phenomenon is not exclusive to Ph+ leukemia, since specific point mutations also emerged in FIP1L1-PDGFRalpha in patients treated with imatinib for hypereosinophilic syndrome29 and were detected in the cKit kinase domain in patients with gastrointestinal stromal tumors (GIST) and acquired resistance to imatinib.30-32 The same scenario may also apply to other neoplastic diseases such as resistance of lung cancer to therapeutic inhibition of the epidermal growth factor receptor (EGFR) and inhibition of FMS-like tyrosine kinase 3 (FLT3) in acute myeloid leukemia (AML). Indeed, mutations in the tyrosine kinase domain of FLT3 recently were found to be associated with resistance of FLT3-internal tandem duplication (ITD)-transformed cell lines to the FLT3 inhibitor SU-5614.33
We here describe a cell-based screening system for the prediction of specific kinase mutations that cause resistance to small molecule kinase inhibitors. This simple method enables the investigation of resistance mechanisms in targeted cancer treatment that has proved robust, can be performed with maintainable efforts in an academic setting, and most importantly, produces results that are clinically relevant and may thus direct future treatment strategies. Using imatinib, the pattern of exchanges within the Bcr-Abl kinase domain in this screen clearly reflected matters observed in imatinib-resistant patients. The results of the screen with PD166326 suggest that in contrast to imatinib, with the clinical application of pyrido-pyrimidines, mutations of P-loop, C-helix, SH2 contact, and A-loop would occur less frequently or would be manageable with moderate increases of dosage. Furthermore the screen with PD166326 identified mutations (F317) that caused strong resistance against PD166326 but mediated only a moderate (F317C and F317L) or no (F317V) resistance to imatinib. (The resistance profile for both compounds is summarized in Figure 6.) These results may be of clinical importance with alternative Abl inhibitors entering clinical trials. A switch to imatinib may successfully treat patients who become resistant to an alternative Abl inhibitor due to a mutation at position F317.
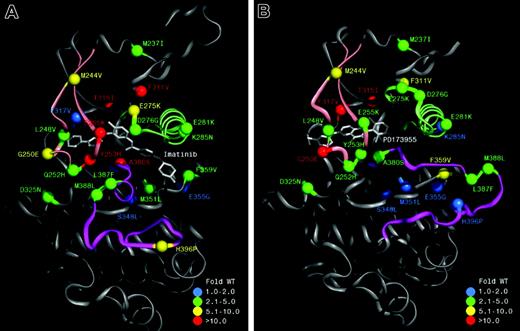
Extent of cellular resistance for mutations identified within the Bcr-Abl kinase domain to imatinib and PD166326. Ribbon representation of the c-Abl kinase domain in complex with imatinib (A; derived from Protein Data Bank 1IEP,34 with the A-loop in a closed conformation) and the pyrido-pyrimidine PD173955 (B; PDB 1M52,15 with the A-loop in an open conformation), with C-helix in light green, P-loop in dusky pink, and A-loop in magenta. Labels indicate the residue number of human c-Abl kinase type Ia. The colors of the spheres represent the degree of cellular resistance to imatinib (A) and PD166326 (B), expressed as fold cellular IC50 of wild-type Bcr-Abl in Ba/F3 cells.
Extent of cellular resistance for mutations identified within the Bcr-Abl kinase domain to imatinib and PD166326. Ribbon representation of the c-Abl kinase domain in complex with imatinib (A; derived from Protein Data Bank 1IEP,34 with the A-loop in a closed conformation) and the pyrido-pyrimidine PD173955 (B; PDB 1M52,15 with the A-loop in an open conformation), with C-helix in light green, P-loop in dusky pink, and A-loop in magenta. Labels indicate the residue number of human c-Abl kinase type Ia. The colors of the spheres represent the degree of cellular resistance to imatinib (A) and PD166326 (B), expressed as fold cellular IC50 of wild-type Bcr-Abl in Ba/F3 cells.
Our method can be applied to other Abl kinase inhibitors that are active in the low nanomolar range and display inhibitory properties in imatinib-resistant mutant forms of Bcr-Abl, such as AP2346435 or BMS-354825.36 Moreover, this cell-based method may be used for screening and investigation of resistance mechanisms of other oncogeneic tyrosine kinases (eg, cKit, FIP1L1-PDGFRalpha, FLT3, or EGFR) to investigational or clinically applicated kinase inhibitors.
Point mutations in imatinib-resistant colonies emerged at concentrations that correspond to and exceed imatinib plasma concentrations that were measured in patients (2.46 μM mean plasma through concentration at 400 mg per day).22 In our screen, at inhibitor concentrations that correspond to 10 times the cellular IC50 value of wild-type Bcr-Abl, mutant colonies still emerged with a frequency of 0.46 per million cells at 4 μM imatinib, opposed to 0.07 with 50 nM PD166326. Several groups have generated cell lines that display resistance to imatinib at concentrations up to 2.5 μM by exposing them to imatinib in gradually increasing concentrations. In these reports, resistance was accompanied by amplification of the Bcr-Abl gene, an increase in Bcr-Abl protein,37-39 and overexpression of P-glycoprotein.39 There is only one report of a point mutation (T315I) in a human myeloid blast crisis KBM5 cell line that was detected after prolonged drug exposure in vitro.40 In our work, specific resistance mutations became frequently apparent in a cell line that was transfected by Bcr-Abl and exposed to inhibitor beginning with the final concentration without gradual increase. With these conditions, specific resistance mutations to imatinib and PD166326 were a frequent event, demonstrating that the generation and selection of Bcr-Abl point mutations do not require in vivo conditions. It has been shown that Bcr-Abl point mutations in patients who later develop imatinib resistance can preexist prior to treatment with imatinib.10,23,41 In our screen, specific resistance mutations reproducibly appeared in different sublines of Ba/F3 cells that were transfected at different time points, including freshly obtained Ba/F3 cells that were examined immediately after transfection. There was no difference in time to growth of mutant colonies between different lines of Bcr-Abl-transformed Ba/F3 cells. Consequently, in Ba/F3 cells the generation of specific resistance mutations does not require long-lasting transformation by Bcr-Abl. Moreover, Ba/F3-BA-wt single cell-derived sublines again gave rise to the rapid development of a variety of different mutations excluding the possibility that we selected a fixed subset of preexisting mutations in each individual Ba/F3-BA-wt line. However, we cannot specify the exact time point at which the mutations occurred, since direct sequencing may have missed low-abundant mutant colonies that may have come up during the 7-day expansion period of the single cell-derived sublines before inhibitor was added.
It has been reported that pyrido-pyrimidine derivatives are more potent inhibitors of wild-type Bcr-Abl than imatinib,15 and are active against mutant forms of Bcr-Abl that cause resistance to imatinib, including P-loop mutations and the activation loop mutant H396P.16-18 For PD166326, 14 of 16 P-loop mutant colonies came up at 25 nM, a concentration that is below cellular IC50 values of P-loop mutations with PD166326,18 and 2 colonies at 50 nM, which is close to cellular IC50. Accordingly, no P-loop mutant colonies came up at 100 nM, which is above IC50 and close to IC95 values. This “cutoff” feature clearly distinguished PD1663326 from imatinib. In line with this, P-loop mutations were completely suppressed when the concentration of PD166326 was slightly increased. This is not the case with imatinib, where P-loop mutations shift dose-response curves above clinically relevant concentrations.8,16,17 The superiority of pyrido-pyrimidines with respect to inhibition of P-loop mutations may be of clinical relevance, since P-loop mutations account for almost half of all mutations that have been detected in patients.
T315I, a frequently occurring mutation in imatinib-resistant patients, was the position most frequently affected in PD166326-resistant colonies, and constituted the only mutation detected in 2 colonies growing at 100 nM PD166326. This was no surprise, since a change of this critical contact site to isoleucine was predicted to interfere with the binding of both imatinib and pyrido-pyrimidines15,42 and confers absolute resistance to both types of inhibitors.6,16,18 C-helix sites were affected in imatinib-resistant, but not PD166326-resistant, colonies. The C-helix mutation D276G has been reported in a single imatinib-resistant patient,24 whereas E281K and K285N have not been detected in patients so far. In line with the notion that pyrido-pyrimidines, in contrast to imatinib, do not intrude to the region of the binding pocket where C-helix is located,15 mutations of this part of Bcr-Abl strongly suppressed activity of imatinib. However, the activity of PD166326 was also slightly affected for D276G and E281K. Point mutations within the activation loop that were reported in patients (L387F,25 L387M,11 H396P,8 and H396R12 ) account for 6% of all mutations. In contrast to imatinib, pyrido-pyrimidine compounds do not require a specific conformation of the activation loop to bind,15 and may therefore not interfere with structural changes within this region. Accordingly, all activation loop mutations that were identified in our screen (A380S, L387F, and M388L) mediated resistance to imatinib. However, L387F was detected in a PD166326-resistant colony, and all 3 activation loop mutations, in contrast to H396P,18 mediated marginal but significant shift in dose-response toward PD166326, indicating that the activation loop may be critical for the binding of pyrido-pyrimidines as well.
M351T, located within E helix in a region making a contact to the SH2 domain of Abl,42,43 accounts for 15% of resistance mutations identified in patients. We observed an exchange to leucine at this position and exchanges at 2 more sites in this region (D325N and S348L) that did not occur in patients so far. As imatinib, in contrast to pyrido-pyrimidines, requires an inactive kinase conformation for binding,15 the mutation of SH2 contact sites may affect sensitivity to imatinib by changing the equilibrium between active and inactive conformations.44 Our observation that SH2 contact region mutations exclusively emerged in colonies resistant to imatinib, but not PD166326, supports the above-stated theoretical considerations.
Azam et al recently used a different in vitro approach to find exchanges in Bcr-Abl that confer imatinib resistance using an Escherichia coli strain that randomly introduces point mutations that subsequently were introduced in Ba/F3 cells, which then were selected for transformation and inhibitor resistance.44 Using this strategy, exchanges that were identified included mutations outside the kinase domain within the SH3 domain, SH3-SH2 connector, SH2 domain, and CD linker. We analyzed numerous resistant Ba/F3 sublines including 40 sublines displaying wild-type kinase domain sequence, but were not able to detect mutations outside the kinase domain. This may be due to properties inherent to such mutations that preclude formation or selection in Bcr-Abl-transformed cells or, alternatively, may be a matter of relative frequency. However, mutations outside the kinase domain, although not reported in patients so far, may be discovered when respective regions are routinely examined in resistant patients. In contrast to the method described by Azam et al, our method allows selection of resistant cells that use mechanisms of inhibitor resistance other than mutations of the Bcr-Abl gene. We observed an increase of phosphorylated Bcr-Abl in resistant sublines with and without mutations. This may complement kinase domain mutations that by themselves only marginally shift dose-response curves toward higher inhibitor concentrations, as observed for G250A and E355G. Moreover, increased Bcr-Abl activity may be of particular importance to allow single cells to survive in the presence of inhibitor until kinase domain mutations arise. Thus, this method allows the detection of distinct resistance mechanisms within one specific resistant subline (eg, a “weak” mutation) that is complemented by additional changes that together concert in a fully resistant phenotype.
We are currently applying this technique to different oncogenic kinases with compounds that are in preclinical or clinical investigation. We believe that using this or similar approaches will contribute important information that can be directly translated into sequential and combinatorial treatment strategies for targeted cancer treatment.
Prepublished online as Blood First Edition Paper, September 30, 2004; DOI 10.1182/blood-2004-06-2445.
Supported by a grant to J.D. and C.P. from the BMBF (Federal Ministry of Education and Research), German national genome project nos. 01-GS-0105 and 01-GS-0155.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Elisabeth Buchdunger (Novartis, Basel, Switzerland) and Harald Gschaidmeier (Novartis, Nürnberg, Germany) for providing imatinib.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal